Abstract
Traditionally, the pathology of human disease has been focused on microscopic examination of affected tissues, chemical and biochemical analysis of biopsy samples, other available samples of convenience, such as blood, and noninvasive or invasive imaging of varying complexity, in order to classify disease and illuminate its mechanistic basis. The molecular age has complemented this armamentarium with gene expression arrays and selective analysis of individual genes. However, we are entering a new era of epigenomic profiling, i.e., genome-scale analysis of cell-heritable nonsequence genetic change, such as DNA methylation. The epigenome offers access to stable measurements of cellular state and to biobanked material for large-scale epidemiological studies. Some of these genome-scale technologies are beginning to be applied to create the new field of epigenetic epidemiology.
Keywords: Epigenetics, Epidemiology, DNA methylation
Epigenetics
The term epigenetics was coined by the Cambridge University embryologist Conrad Waddington in a series of monographs, and he used it to describe his original view of developmental biology that the morphological and functional properties of an organism arise sequentially under a program defined by the genome under the influence of the organism’s environment [1]. The modern definition of epigenetics is modifications of the DNA or associated proteins, other than DNA sequence variation itself, that carry information content during cell division [2], although a few scientists take a more relaxed or stricter view, either including RNA modification or limiting to vertical (generational) transmission. Remarkably, the modern definition and Waddington’s have converged. That is because the epigenetic state of an organism progresses from gamete to zygote to somatic tissue, all of which have profoundly different epigenomes, while the DNA is the same. Furthermore, given that the developmental state of a cell can be completely reprogrammed by somatic cell nuclear transfer, or by specific genes in combination, the information specifying cell state is not the DNA alone but the epigenetic program layered on top of this genetic code and is heritable during cell division but ultimately reprogrammable.
The focus of this review is the specific epigenetic modification involving DNA methylation (DNAm), a covalent addition of a methyl (CH3) group to the nucleotide cytosine. DNAm is the only epigenetic modification whose mechanism for propagation is well understood biochemically. CpG dinucleotides show heritable methylation during cell division because the complementary strand shows the same sequence, and both cytosines are normally methylated. During DNA replication, the two daughter strands contain hemimethylated DNA, i.e., the parent strand is methylated and the daughter strand is not. The enzyme DNA methyltransferase I (DNMT1) has high affinity for this hemimethylated strand and adds a methyl group to the newly synthesized daughter cytosine at that site, likely within the same DNA replication complex. Dietary methionine and a cofactor synthesized from folic acid are necessary for the success of methylation maintenance, providing a strong link between the environment and the epigenome. Indeed in animals, the epigenome and gene expression can be modified by dietary manipulation of methylation precursors, and dietary deprivation of methionine leads to liver cancer in animals [3].
“CpG islands” are regions rich in CpG dinucleotides (formally defined as G + C content≥0.5 and CpGobs/CpGexp≥0.6)[4], and they are often described as uniformly unmethylated in normal cells, with the exception of the inactive X chromosome, and are near imprinted genes [5, 6]. However, the assumption that autosomal CpG islands (except for imprinted genes) are never methylated is clearly not the case [7-10]. It is also important to note that functionally important DNAm information is often not within conventionally defined CpG islands, e.g., the H19 and insulin-like growth factor II gene (IGF2) differentially methylated regions (DMRs) that regulate imprinting of IGF2 [11, 12].
Epigenetics of human disease
How can one identify disease-specific epigenetic differences? One would like to know that the epigenome varies normally in the population, is associated in particular ways with disease, and does not always simply reflect normal tissue-specific differences in gene expression. Individual gene data in support of this epigenetic variation were first reported in the 1980s [13]. Other genomic regions showing epigenetic variation in the population include X inactivation [14] and both familial and environmental determinants of IGF2/H19 imprinting, or parent of origin-specific gene silencing [14].
A common theme of disease epigenetics is the role of defects in phenotypic plasticity, the ability of cells to change their behavior in response to internal or external environmental cues; this was reviewed recently in detail [15]. For example, hereditary disorders of the epigenetic apparatus lead to developmental defects, a dramatic example being the Rett syndrome. This disorder involves loss of function of methyl-CpG-binding protein 2 (MeCP2), which recognizes DNAm. Children with Rett syndrome develop normally until 6–12 months and then gradually lose developmental milestones over years, due to a failure to maintain gene silencing in the brain. This process of delayed onset of disease is also a hallmark of bipolar disorder and schizophrenia.
The study of epigenetic changes in human cancers began with the discovery of widespread hypomethylation [16]. Cancer involves both hypomethylation and hypermethylation, attendant overexpression of oncogenes, silencing of tumor suppressor genes, and loss of imprinting. Here too, the mechanism by which epigenetic changes leads to cancer appears to involve disruption of normal phenotypic plasticity, in this case of the programming that leads a cell to differentiate normally within a given tissue compartment [2]. Moreover, epigenetic changes that arise constitutionally are associated with increased risk of common disease, such as loss of imprinting of the IGF2 gene in cancer, which has been shown in both human [17] and mouse [18, 19] studies. Prospective or nested case–control studies are needed to establish a cause and effect relationship in colorectal cancer.
Epigenetic alterations have long been linked to human disease, originally through disorders of genomic imprinting [20]. Defects in the epigenetic machinery also lead to developmental abnormalities, such as MeCP2 mutations in Rett syndrome [21] and DNMT3B mutations in immunodeficiency, centromeric region instability, and facial anomalies (ICF) syndrome [21].
Epigenetic alterations may also contribute to neuropsychiatric disease. Bipolar disorder shows several features consistent with an epigenetic contribution: lack of complete concordance in monozygotic twins; onset of illness in adolescence or adulthood rather than childhood, the often episodic nature of the illnesses, and the apparent relationship to environmental factors, such as stress [22, 23]. Stress has been shown to alter epigenetic marks including DNAm and histone modifications in the brain in animal models [24, 25]. Interestingly, three important bipolar disorder medications, the mood stabilizer valproate [24, 25], the antidepressant imipramine [25], and the antipsychotic haloperidol [26], have also been shown to induce epigenetic changes in the brain. More direct evidence in support of an epigenetic effect in bipolar disorder: is based on the identification of an excess of maternal transmission in some pedigrees [27]. The mounting evidence for epigenetic involvement in autism includes relationships with related phenotypes as well as direct evidence. For example, imprinted genes on the X chromosome are thought to be involved in social skills in girls because defects in these skills are found in Turner syndrome and in children lacking the paternal X chromosome but not the maternal X chromosome [28]. Both fragile X, a disorder with known phenotype overlap with autism, and ICF syndrome arise from malfunctions in the establishment of normal DNAm patterns [29, 30]. Rett syndrome, also associated with autistic features, is caused by mutations in the gene encoding DNA methyl-binding protein MeCP2, a protein important for interpreting DNAm and controlling the repression of gene transcription [31]. Patients with these three disorders exhibit mental retardation, demonstrating the importance for proper DNAm in the regulation of cognitive function. Furthermore, one mechanistic study has shown that abnormally hypomethylated CNS neurons were impaired functionally and were selected against in postnatal development [32]. Another suggests that neuronal activity can drive the transcription of genes important for controlling neurotransmitter release by regulating their DNAm status [33].
The potential for imprinting of autism-related genes could explain the lack of Mendelian inheritance in autism and the inconstant results across linkage and association studies that do not account for these features. Direct evidence for this idea comes from a study of the gene for contactin-associated protein-like 2 (CNTNAP2), identified by multiple studies as associated with autism spectrum disorders (ASD) [34-38]. In one of these studies, risk for ASD associated with the identified single nucleotide polymorphism (SNP) showed parent-of-origin specificity suggesting a role for imprinting [36].
Epigenetics of aging
Increasing evidence supports a role for epigenetics in the biology of aging. X-inactivated genes in the mouse show an increased frequency of reactivation with aging, consistent with age-related epigenetic change [39, 40]. The frequency of epigenetic changes in mice may be one to two orders of magnitude greater than the rate of somatic DNA mutation [41]. This fits with a role of epigenetics in late-onset disorders such as frailty, a syndrome of decreased resiliency and reserves, in which a mutually exacerbating cycle of declines across multiple systems results in negative energy balance, sarcopenia, and diminished strength and tolerance for exertion [42]. Accumulation of DNA sequence changes might not occur at enough high rate during the lifespan to induce common disease, but epigenetic changes may occur at a frequency that could contribute to this effect. Very few studies have demonstrated epigenetic changes in humans with age due to technical and biosample limitations. A recent study has shown differences in local and global methylation by age by examining the similarity in methylation patterns between MZ twins aged 3 years old and MZ twins aged 50. Although these analyses were not in the same individuals (the same twins were not followed longitudinally), the similarity in methylation patterns between young twins compared to the dissimilar patterns among older twins argues strongly for age-related changes in the epigenome [43]. Direct evidence comes from a recent study showing changes in DNA methylation in the same individual over time, described in more detail below.
Epigenomics
Epigenomics refers to genome-scale analysis of epigenetic marks. The term “methylome,” or genome-wide state of DNAm, was first introduced by the author in 2001 [44]. Despite the availability of an essentially complete genome sequence for several years, understanding the methylome has progressed more slowly, largely due to limitations in technology affecting sensitivity, specificity, throughput, quantitation, and cost among the previously used detection methods. All of the available methods involve trade-offs among these variables. Furthermore, all of these variables are themselves moving targets, particularly cost. The rule in genomic science generally is that increased demand substantially reduces cost because of three factors: fierce competition in the biotechnology sector; production efficiencies as methods are automated; and continued technological advances. It is also important to define clearly what is meant by genome scale. The term is commonly applied to any method not limited to specific predefined genes, but no epigenomic method in common use examines the entire epigenome. For the sake of this article, the discussion will be limited to DNAm analysis because of its particular suitability for pathological and epidemiological studies due to its stability in biobanked specimens.
The human genome contains ~3 × 109bp of DNA, of which there are ~3 × 107 CpG dinucleotides, and half of that is nonrepetitive single- or low-copy sequence [45]. CpG dinucleotides are the sites that can be methylated and the methylation in turn replicated faithfully during cell division by DNA methyltransferase 1. While non-CpG methylation exists, it is not currently considered epigenetic information since no mechanism is known for its propagation during DNA replication.
What are the methods in common practice for measuring genome-scale DNAm? While this review naturally is written from the perspective of our own approaches, there are several other excellent reviews of epigenomics [46, 47]. Most investigators are drawn to commercially available methods, particularly those that can be performed as a service, with only DNA needing to be prepared by the investigator. However, these methods are not necessarily the most comprehensive or most accurate. A method similar to array-based SNP analysis is the Illumina GoldenGate methylation assay [48], or its more recent cousin, the Illumina Infinium methylation platform. Both methods involve bisulfite conversion of unmethylated DNA to uracil, followed by polymerase chain reaction (PCR) which propagates a thymine residue at the converted base [49]. Methylated cytosine is unconverted and thus read as cytosine. Thus, the methylation state (C/mC) is transformed to a pseudopolymorphism (T/C, respectively). The readout is then as for any SNP and is semiquantitative, accurate within ~17% for the GoldenGate assay [50]. The major limitation of this approach is the relatively poor coverage of the genome by both methods, only ~1,500 CpG by GoldenGate and ~27,000 CpG by Infinium, thus representing 0.01% to 0.18% of the single-copy methylome. A second limitation is the choices involved in selecting CpG sites for analysis. The chips are designed based in part on the idea that functional CpG methylation lies within canonical gene promoters and CpG islands. CpG islands are defined algorithmically, i.e., based on a formula given above. The major rationale for this choice is literature showing hypermethylation of CpG islands in cancer. Yet, those studies are largely self-referential in design, and recent studies described below suggest that most variable DNAm occurs outside of these islands. Nevertheless, great advances have been made possible by these reagents and methods, and they show the promise of increasing efforts by many laboratories to improve the resolution of genome-scale technology.
Furthermore, there are comparatively few data supporting the choice specifically of promoters and CpG islands for studies of other diseases, or normal population variation. Indeed, a relatively small scale but very comprehensive study was performed by Stephan Beck at the Sanger Center on ~1.8 Mb of DNA including ~40,000 CpG sites across 12 tissues [10]. The study showed that most methylation variation was not at transcriptional start site-associated CpGs or at CpG islands [10]. One encouraging result from that study, for those who wish to use CpG chips as described above, was a high degree of correlation between CpG site methylation within a few hundred base pairs. However, the choice of one or two CpGs per candidate region seems precariously underrepresented.
A second approach in common practice is hybridization of antibody-purified methylated DNA to high-density genome arrays [51]. For example, NimbleGen offers of methylated DNA immunoprecipitation (MeDIP) to a ~2-Mb array tiled through gene promoters and CpG islands. The coverage of this array is much greater than the SNP-based arrays described above. However, choice of selection is still a significant issue given that complete tiling of the genome would currently require ten arrays, which is cost-prohibitive for large-scale epidemiological studies. Furthermore, MeDIP shows significant limitations in discriminating methylation differences in regions of medium- to low-density CpG content [52], and our recent study shows that that is exactly where many or most significant variation in DNAm occurs [53]. Another method focused on CpG islands is restriction landmark genome scanning [54]. There are emerging alternatives for methylation fractionation, including affinity purification of methylated DNA on methyl-CpG-binding protein [55], or affinity purification of unmethylated DNA [56].
Two promising methods for genome-scale analysis use methylated DNA fractionation based on restriction endonuclease digestion. One of these, developed by John Greally and colleagues at Albert Einstein College of Medicine in New York, is termed HELP for HpaII-tiny fragment Enrichment by Ligation-mediated PCR [57]. It takes advantage of the difference in sizes of Hpa-II fragments, which are generated from unmethylated DNA, and Msp-I fragments, which recognize the same cleavage site but are methylation-independent. While initial specificity was relatively limited, recent improvements involve additional methylcytosine sensitive endonucleases and allow representation of >98% of CpG islands and >90% of refSeq promoters, and it can also be combined with next generation sequencing for readout [58]. A second involves fractionation of the unmethylated component with McrBC, which recognizes methylated DNA if there are two methylcytosines preceded by purines and separated by ~40–100 b, an easy condition to meet for methylated DNA except at very low CpG density. This approach was first applied to specific chromosome analysis [59] but was subsequently extended to study of human cancer [60].
Rafael Irizarry and I with our colleagues developed an array-based readout method that is independent of methylation fractionation method and can be applied equally to McrBC, HELP, or antibody-based methods. This approach, termed CHARM for comprehensive high-throughput array-based relative methylation analysis, involves two essential components. First, the array is agnostic to presuppositions about the location of differential methylation and tiles through regions based only on the relative CpG content in decreasing abundance [52]. It, therefore, includes all CpG islands, but that represents only 38% of the CpG “real estate,” or available oligonucleotide probe positions for analysis on the array. One could use additional arrays or soon to be released higher density arrays to increase coverage, which is now about one fourth of the entire nonrepetitive methylome. The second component is genome-weighted smoothing, or correction for the hybridization properties of the target (i.e., sample) genome at each location, which is in turn calculated from empirical measurements of hybridization efficiency with regard to GC content, CpG density, and length of fragments [52]. A statistical suite of postprocessing algorithms, written in R, is termed CharmR and is continually revised. The arrays and CharmR are open access and open source (http://www.biostat.jhsph.edu/~maryee/charmR/). Thus, while not commercially available, this technology is readily transportable to core laboratories that have statistical and programming support.
Although my colleagues and I have developed one of the current approaches to epigenomic analysis, we gladly welcome the advent of second generation sequencing technology for DNAm analysis. There are multiple competing commercial platforms for massively parallel sequencing on slides, with throughput per machine >300 Gb per run at <1% of the cost of conventional automated sequencing [61, 62]. A particular advantage of sequencing-based methylation analysis is the ability to ascertain allele-specific methylation by virtue of DNA polymorphisms within the same sequencing read. This is particularly true as longer reads become cost-effective.
Nevertheless, whole genome bisulfite sequencing applied to humans is not presently cost-effective for epidemiological studies. Costs are in the many tens of thousands of dollars, compared to hundreds of dollars per sample for alternative less comprehensive methods. Therefore, sequencing-based methods all involve significant trade-offs. One method involves “reduced representation,” using restriction enzymes to limit the sequenced target to regions within CpG islands [63], which may miss significant normal variation in patient populations or across tissues [53]. Single molecule sequencing detection, such as in development by Pacific Biosystems, or other methods not widely discussed publicly, might reduce costs to the point of making whole genome shotgun sequencing inexpensive compared to other methods for epigenomic profiling. Until that day comes, however, a great deal can be learned about the methylome of human normal and disease populations using array or chip-based approaches.
The first genome-scale epigenetic analysis of human cancer: CpG island shores
We recently exploited CHARM methodology to perform the first genome-scale analysis of the human cancer methylome and to compare it to the normal tissue-varying methylome. A comparison was first made of DNA from five autopsy specimens, three matched tissues each representing the three embryonic lineages, brain, liver, and spleen. Surprisingly, most tissue-specific DNAm was not at CpG islands but at regions of intermediate CpG density located up to 2 kb from the islands, and which we termed “CpG island shores” [53]. Even though CpG islands accounted for 33% of the CpG real estate on the arrays, they only accounted for 6% of these tissue-varying differentially methylated regions, or T-DMRs. In contrast 76% of T-DMRs were in CpG island shores. Furthermore, the T-DMRs were located for the most part outside of promoters (96%), and more than half were >2 kb from the nearest annotated gene [53].
Next, comparing 13 colorectal cancers to matched normal mucosa from the same patients, 2,707 regions were identified showing cancer-specific differentially methylated regions, or C-DMRs, under a false discovery rate of 5%. These, too, were highly enriched at CpG island shores (67%), and islands were comparatively underrepresented (8% compared to 38% on the arrays). The data were highly reproducible, being validated by independent quantitative bisulfite pyrosequencing on a replicate set of 50 colon cancers and matched normal mucosa [53].
Remarkably, there was a comparable amount of hypermethylation as hypomethylation, even though the cancer literature is heavily biased toward the former. That may be because the CpG islands, even though underrepresented for C-DMRs overall, show hypermethylation when methylation is altered in cancer, while the shores away from the islands tend toward relative hypomethylation. In retrospect, this is not surprising, since CpG islands are protected against normal DNA methylation, and thus, the only direction they can commonly change in disease is toward relative hypermethylation. In any case, the common dictum that cancer shows repetitive DNA hypomethylation and gene-specific hypermethylation appears to be false. While the former is true, single genes are numerically comparably altered by hypomethylation and hypermethylation in cancer [53]. These results are illustrated in Fig. 1.
Fig. 1.
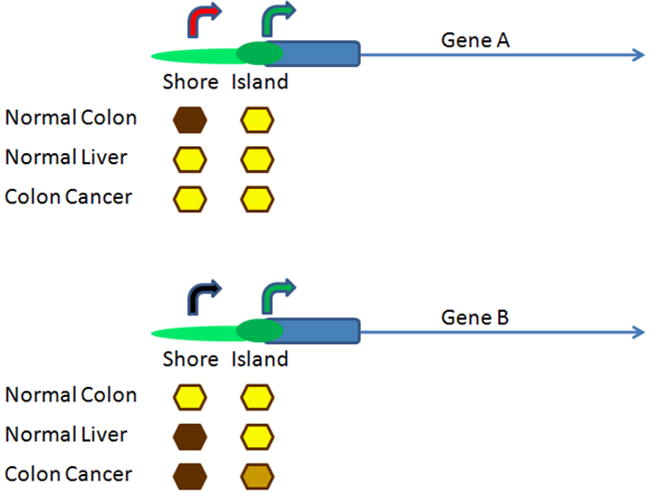
Altered DNA methylation of CpG island shores in human colon cancer. Shown are an example of hypomethylation (Gene A, top) and hypermethylation (Gene B, bottom) in cancer revealed by a genome-scale analysis of the cancer methylome. Gene A is normally methylated at the shore and not at the island, and it acquires a hypomethylated pattern at the shore in colon cancer, resembling that of the normal liver. Aberrant expression at an alternate promoter, or for an untranslated RNA, is activated at the shore. Gene B is normally unmethylated at both shore and island, and it acquires a hypermethylated island at the shore, resembling the normal liver, and potentially at the island as well. Aberrant silencing ensues at the shore and potentially at the canonical promoter
This comparative methylome analysis also showed that C-DMRs and T-DMRs largely overlap (65% using an F statistic). Indeed, if one performs supervised clustering to identify the C-DMRs that distinguish colorectal cancer from normal mucosa, those same DMRs in unsupervised clustering completely discriminate spleen from liver from brain [53]. Thus, the DMRs that regulate normal differentiation are involved in aberrant methylation in cancer, and this may occur generally across cancer, since they even distinguish tissues not of the type from which the cancer derives.
What do CpG island shores do? This is of great relevance to any investigator interested in the disease epigenome, since they were previously unapparent yet obviously at the heart of normal and abnormal variation. One clue comes from their localization, as they appear to be enriched in alternative transcriptional start sites for annotated genes, as well as unannotated RNAs. This localization was functionally supported by rapid amplification of complementary DNA ends experiments showing that hypomethylated CpG island shores in cancer show activation of alternative transcriptional start sites within them in the same tumor showing hypomethylation at these sites [53].
The new field of epigenetic epidemiology
Epidemiology is the study of disease in populations, and genetic epidemiology, or the relationship of genetic variation to disease, has exploded by taking advantage of the data generated by the HapMap project, a consortium effort to identify six million common polymorphisms across the genome and in the major human population groupings [64]. Genome-wide association studies have become the mainstay of human genetics research and have identified nearly 100 loci for over 40 diseases [65]. Concomitant copy number variant (CNV) analysis which can be performed on the same chip platform has also identified insertions or deletions that contribute to common disease [66]. However, epigenomics has not yet been integrated into the routine search for variation contributing to human disease susceptibility.
The new field of epigenetic epidemiology will measure and catalog such epigenetic variation within and across populations and to characterize the correlation properties of methylation, similar to the catalog of SNP/CNV variation and linkage disequilibrium. Epigenetic epidemiology can also provide a unique perspective on the environmental factors contributing to common disease. Several examples of environmentally mediated epigenetic effects have been documented, including the influence of methyl donors and folate from diet on methylation levels, smoking influence on methylation, and the effects of metallotoxins [67, 68]. DNA methylation occurs by homocysteine conversion to methionine, which is then converted to S-adenosylmethionine, the common methyl donor for DNAm. Animal work has shown that folate-deprived rats become hypomethylated locally, at particular genes [69, 70] and globally [71]. Human cell work has shown that specific genes may be hyper- or hypomethy-lated with reduced folate [72]. Also, clinical studies have shown a correlation between serum folate levels and hypomethylation [73], and epidemiologic interventions show older women put on folate-depleted diets result in increased plasma homocysteine and decreased methylation [74, 75]. A hypomethylation defect was associated with assisted reproductive technology in the conception of children with Beckwith–Wiedemann syndrome, a disorder of prenatal overgrowth, birth defects, and cancer [76], which has been borne out by several other groups [77, 78]. Thus, prenatal exposure can act through an epigenetic mechanism.
The prior focus of epigenomics on the simple interface between epigenetics and human disease phenotype variation has prepared us now to address the more complex task of including genetic variation in genome-scale analysis. Going forward, it is critical to develop genome-wide tools to determine the relationship between genetic variation, epigenetic variation, and disease simultaneously. This area of overlap, the hashed area in Fig. 2, is deliberately drawn as the larger fraction of the overlap between genetics and phenotype to emphasize that most genetic findings must be considered in an epigenetic context and to highlight that the full value of typical genetic epidemiology studies cannot be realized until the complementary epigenetic measures and statistical tools are developed and performed on these samples.
Fig. 2.
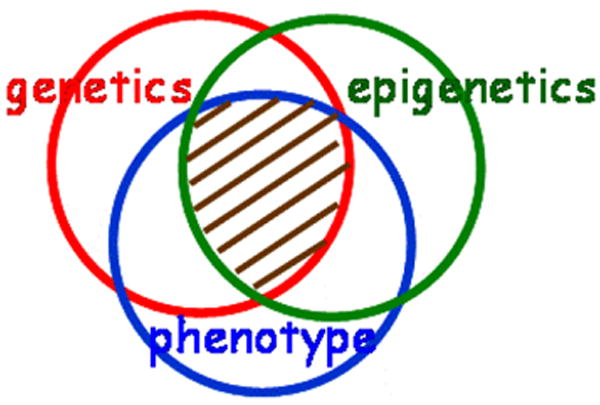
Epigenetic epidemiology. New genome-scale tools for epigenetic analysis will allow us to determine the relationship between genetic variation, epigenetic variation, and disease simultaneously. The area of overlap is deliberately drawn as the larger fraction of the overlap between genetics and phenotype to emphasize that most genetic findings must be considered in an epigenetic context and to highlight that the full value of typical genetic epidemiology studies cannot be realized until the complementary epigenetic measures and statistical tools are developed and performed on these samples
Fallin, Bjornsson, and I have proposed a common disease genetic and epigenetic (CDGE) model for human disease, which states that DNA sequence variation (traditional genetics), environment, and epigenetic mechanisms interact to cause or accelerate common disease, especially those of later onset [23, 79]. CDGE provides a model for understanding how one might integrate epigenetics into traditional studies genetics and the environment. For example, epigenetic marks such as DNAm may influence disease risk, either directly (such as aberrant DNAm turning a gene on/off inappropriately) or indirectly (through masking/unmasking DNA sequence variation that has disease consequences). This type of DNA variation, whose penetrance is dependent on epigenetic context, is denoted “gdep,” since it is only one risk factor in the context of certain epigenetic patterns. This type of sequence variation would be difficult to discover in traditional genetic association approaches, without knowledge of the epigenetic background. Parent-of-origin analyses in genetic epidemiology are aimed at accommodating this problem in the context of imprinting, but this is often low-powered and only addresses one kind of epigenetic model. Whether epigenotype (e.g., DNAm) acts directly or indirectly on disease risk, factors that control epigenotype are themselves critical risk factors. DNA variation that controls epigenotype such as DNAm may be located, for example, in genes that encode proteins in the one-carbon transfer pathway and may affect the cell’s ability to maintain DNAm. This type of risk-related DNA variation is denoted “gepg” to indicate that the multiple effects manifest through an epigenetic mechanism. With these thoughts in mind, at least three models can be considered for how DNA variation may contribute to risk: (1) independently of epigenetic mechanisms (gind), (2) as genetic mediators of epigenetic modifications of other genes (gepg), or (3) where the effect of the genetic variant depends on its epigenetic context (gdep). Only the first of these would be easily detected in current genetic association studies. Current genetic studies might have reduced power to detect gepg without epigenotype measures or knowledge of the factors that contribute to epigenotype. Current genetic studies would have virtually no power to detect gdep without epigenetic measurement [23, 79].
A critical clue to CDGE comes from a global genome-scale measurement of DNAm termed luminometric methylation assay (LUMA), a precise quantitative measure of Hpa II site methylation. Using LUMA, intraindividual change in DNA methylation was found over time with familial clustering. DNA from 111 participants in the AGES Reykjavik Study [80] was first analyzed. In this cohort, 8.1% of individuals showing changes greater than 20% in Hpa II methylation over time, and these were approximately equally divided between gains and losses of DNAm. Permutation analysis showed that the change observed was much greater than by chance (P<0.0001) [81]. Next examined was a second cohort of 126 individuals from a collection of Utah pedigrees that had been sampled twice over an average of 16 years. In this group, 11% of individuals show changes greater than 20% in Hpa II methylation over time. The Utah pedigrees showed high heritability, with a heritability estimate of 0.99 (P<0.0001) [81]. The familial clustering of methylation changes raises the possibility that methylation changes could be directly related to genetic variation, as suggested by CDGE. Another recent study shows associations of single nucleotide polymorphisms with nearby differences in DNA methylation [82], consistent with CDGE.
Future needs for epigenetic epidemiology will require advances in three areas. First, we must develop scalable, cost-effective approaches for population-level epigenetic profiling. This includes technical advances in measurement and quantification of DNAm. We must also develop even more comprehensive coverage of the epigenome. This can be done by increasing real estate on the arrays, in part perhaps by reducing coverage of highly comparable adjacent sequences on the tiled regions, or by increasing array density as will occur this year. A substantial advance will come from combining array-based advances with second generation sequencing technology. For example, one could capture the relevant epigenome target (identified by studies on arrays) using arrays or molecular inversion probe or other solution-based technologies and then perform bisulfite-based shotgun sequencing for single nucleotide resolution.
Second, we must further develop the statistical tools and concepts that are necessary to analyze, interpret, and compare population-level epigenetic data. A critical requirement for epidemiological analyses is the transformation of granular individual epigenotypes into the higher-level epidemiological data types without significant information loss. This will include developing new statistical tools for identifying the subset of variable DNAm regions relevant to human disease and developing methods to simplify granular methylation patterns into epigenetic “barcodes,” similar to the work by Irizarry on gene expression [83].
Third, we must integrate conventional genetic epidemiology with these epigenetic data to fully develop epigenetic epidemiology. We must answer fundamental questions about type, frequency, and properties of epigenetic variation within and across individuals, families, and populations. This can be done by relating genetic variation to epigenetic variation in normal populations, or by investigating epigenetic differences among monozygotic twins. A critical question is whether epigenetic marks are transmitted intact from parent to offspring and whether DNAm is allele-specific and covaries with allele-specific gene expression. For example, can we develop an epigenetic transmission test comparable to the transmission disequilibrium test used in genetic epidemiology? Finally, and most excitingly, we must begin to examine the epigenome comprehensively in large population-based epidemiological studies of disease. Such studies will greatly enhance cancer risk assessment and prevention and are already showing promise in better understanding common neuropsychiatric disease.
References
- 1.Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Poirier LA. The effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introduction. J Nutr. 2002;132:2336S–2339S. doi: 10.1093/jn/132.8.2336S. [DOI] [PubMed] [Google Scholar]
- 4.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 5.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 6.Riggs AD, Pfeifer GP. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 7.Strichman-Almashanu LZ, Lee RS, Onyango PO, et al. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song F, Smith JF, Kimura MT, et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiota K, Kogo Y, Ohgane J, et al. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- 10.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hark AT, Schoenherr CJ, Katz DJ, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 12.Cui H, Niemitz EL, Ravenel JD, et al. Loss of imprinting of insulin-like growth factor-II in Wilms’ tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001;61:4947–4950. [PubMed] [Google Scholar]
- 13.Silva AJ, White R. Inheritance of allelic blueprints for methylation patterns. Cell. 1988;54:145–152. doi: 10.1016/0092-8674(88)90546-6. [DOI] [PubMed] [Google Scholar]
- 14.Sandovici I, Naumova AK, Leppert M, et al. A longitudinal study of X-inactivation ratio in human females. Hum Genet. 2004;115:387–392. doi: 10.1007/s00439-004-1177-8. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 17.Cui H, Cruz-Correa M, Giardiello FM, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 18.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 19.Kaneda A, Wang CJ, Cheong R, et al. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proc Natl Acad Sci USA. 2007;104:20926–20931. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horsthemke B, Buiting K. Genomic imprinting and imprinting defects in humans. Adv Genet. 2008;61:225–246. doi: 10.1016/S0065-2660(07)00008-9. [DOI] [PubMed] [Google Scholar]
- 21.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 22.Petronis A, Gottesman II, Crow TJ, et al. Psychiatric epigenetics: a new focus for the new century. Mol Psychiatry. 2000;5:342–346. doi: 10.1038/sj.mp.4000750. [DOI] [PubMed] [Google Scholar]
- 23.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 25.Tsankova NM, Berton O, Renthal W, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 26.Shimabukuro M, Jinno Y, Fuke C, et al. Haloperidol treatment induces tissue- and sex-specific changes in DNA methylation: a control study using rats. Behav Brain Funct. 2006;2:37. doi: 10.1186/1744-9081-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon FJ, Stine OC, Meyers DA, et al. Patterns of maternal transmission in bipolar affective disorder. Am J Hum Genet. 1995;56:1277–1286. [PMC free article] [PubMed] [Google Scholar]
- 28.Skuse DH, James RS, Bishop DV, et al. Evidence from Turner’s syndrome of an imprinted X-linked locus affecting cognitive function. Nature. 1997;387:705–708. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- 29.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutcliffe JS, Nelson DL, Zhang F, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 31.Amir RE, Van den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 32.Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roohi J, Montagna C, Tegay DH, et al. Disruption of contactin 4 in 3 subjects with autism spectrum disorder. J Med Genet. 2008;46(3):176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakkaloglu B, O’Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arking DE, Cutler DJ, Brune CW, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alarcon M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 39.Wareham KA, Lyon MF, Glenister PH, et al. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- 40.Brown S, Rastan S. Age-related reactivation of an X-linked gene close to the inactivation centre in the mouse. Genet Res. 1988;52:151–154. doi: 10.1017/s0016672300027531. [DOI] [PubMed] [Google Scholar]
- 41.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandeen-Roche K, Xue QL, Ferrucci L, et al. Phenotype of frailty: characterization in the women’s health and aging studies. J Gerontol A Biol Sci Med Sci. 2006;61:262–266. doi: 10.1093/gerona/61.3.262. [DOI] [PubMed] [Google Scholar]
- 43.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feinberg AP. Methylation meets genomics. Nat Genet. 2001;27:9–10. doi: 10.1038/83825. [DOI] [PubMed] [Google Scholar]
- 45.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 46.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 47.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 49.Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 52.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costello JF, Smiraglia DJ, Plass C. Restriction landmark genome scanning. Methods. 2002;27:144–149. doi: 10.1016/s1046-2023(02)00067-1. [DOI] [PubMed] [Google Scholar]
- 55.Jorgensen HF, Adie K, Chaubert P, et al. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oda M, Glass JL, Thompson RF, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37(12):3829–3839. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada Y, Watanabe H, Miura F, et al. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–266. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ordway JM, Bedell JA, Citek RW, et al. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- 61.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shendure J, Porreca GJ, Reppas NB, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 63.Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39:S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 67.Sutherland JE, Costa M. Epigenetics and the environment. Ann N Y Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 68.Van den Veyver IB. Genetic effects of methylation diets. Annu Rev Nutr. 2002;22:255–282. doi: 10.1146/annurev.nutr.22.010402.102932. [DOI] [PubMed] [Google Scholar]
- 69.Pogribny IP, Basnakian AG, Miller BJ, et al. Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res. 1995;55:1894–1901. [PubMed] [Google Scholar]
- 70.Pogribny IP, Miller BJ, James SJ. Alterations in hepatic p53 gene methylation patterns during tumor progression with folate/methyl deficiency in the rat. Cancer Lett. 1997;115:31–38. doi: 10.1016/s0304-3835(97)04708-3. [DOI] [PubMed] [Google Scholar]
- 71.Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52:2071s–2077s. [PubMed] [Google Scholar]
- 72.Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001;60:1288–1295. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- 73.Fowler BM, Giuliano AR, Piyathilake C, et al. Hypomethylation in cervical tissue: is there a correlation with folate status? Cancer Epidemiol Biomarkers Prev. 1998;7:901–906. [PubMed] [Google Scholar]
- 74.Jacob RA, Gretz DM, Taylor PC, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 75.Rampersaud GC, Kauwell GP, Hutson AD, et al. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 76.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith–Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gicquel C, Gaston V, Mandelbaum J, et al. In vitro fertilization may increase the risk of Beckwith–Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74:599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bjornsson HT, Cui H, Gius D, et al. The new field of epigenomics: implications for cancer and other common disease research. Cold Spring Harb Symp Quant Biol. 2004;69:447–456. doi: 10.1101/sqb.2004.69.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, gene/environment susceptibility—Reykjavik study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intraindividual change in DNA methylation over time with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boks MP, Derks EM, Weisenberger DJ, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS ONE. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Methods. 2007;4:911–913. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]


