Abstract
Cytosine deaminase (CDA) from E. coli is a member of the amidohydrolase superfamily. The structure of the zinc-activated enzyme was determined in the presence of phosphonocytosine, a mimic of the tetrahedral reaction intermediate. This compound inhibits the deamination of cytosine with a Ki of 52 nM. The zinc and iron containing enzymes were characterized to determine the effect of the divalent cations on activation of the hydrolytic water. Fe-CDA loses activity at low pH with a kinetic pKa of 6.0 and Zn-CDA has a kinetic pKa of 7.3. Mutation of Gln-156 decreased the catalytic activity by more than 5 orders of magnitude, supporting its role in substrate binding. Mutation of Glu-217, Asp-313, and His-246 significantly decreased catalytic activity supporting the role of these three residues in activation of the hydrolytic water molecule and facilitation of proton transfer reactions. A library of potential substrates was used to probe the structural determinants responsible for catalytic activity. CDA was able to catalyze the deamination of isocytosine and the hydrolysis of 3-oxauracil. Large inverse solvent isotope effects were obtained on kcat and kcat/Km, consistent with the formation of a low-barrier hydrogen bond during the conversion of cytosine to uracil. A chemical mechanism for substrate deamination by CDA was proposed.
Cytosine deaminase (CDA1) catalyzes the hydrolytic deamination of cytosine (1), forming uracil and ammonia (Scheme 1). Amino acid sequence alignments and three dimensional crystal structures have identified this enzyme as a member of the amidohydrolase superfamily (AHS) with a single divalent cation in the active site (2). The amidohydrolase superfamily was first identified by Sander and Holm based upon the structural similarities of urease, phosphotriesterase, and adenosine deaminase (3). This superfamily has been partitioned into 24 clusters of orthologous groups (COG) by the NCBI (4). CDA is found within cog0402 along with guanine deaminase (5), adenosine deaminase (6), S-adenosyl homocysteine deaminase (7), N-formimino-l-glutamate deiminase (8) and two recently annotated enzymes: 8-oxoguanine deaminase (9) and isoxanthopterin deaminase (10).
Scheme 1.

All of the experimentally characterized members of cog0402 catalyze deamination reactions and share conserved active site residues and metal cofactor coordination schemes within a (β/α)8-barrel structural fold (10). For CDA, the amino acid residues that participate in binding the lone divalent cation are two histidine residues that follow the C-terminal end of β-strand 1 (His-61 and His-63), another histidine from the C-terminal end of β-strand 5 (His-214) and an aspartate from the C-terminal end of β-strand 8 (Asp-313). Two additional residues, His-246 from the end of β-strand 6 and Glu-217 in the loop that follows β-strand 5, participate in the activation of the hydrolytic water molecule and proton transfer reactions required for catalysis (1).
Porter established the metal activation requirements for cytosine deaminase from Escherichia coli and demonstrated that CDA purified from cells grown in a standard medium contains a mixture of Fe and Zn, and that a single equivalent of metal is required for full catalytic activity (11). The divalent metal can be removed with o-phenanthroline and the apo-enzyme reconstituted with Fe2+, Mn2+, Co2+ or Zn2+. However, oxidation of iron to the ferric state resulted in the inactivation of the enzyme (11). Fe-CDA has a reported kcat of ~185 s−1 and Michaelis constants for cytosine ranging from 0.20 to 0.32 mM (1, 11). In addition to the deamination of cytosine, CDA has been shown to utilize 5-fluorocytosine, 2-thiocytosine, 6-azacytosine 5-azacytosine, and creatinine as alternate substrates (12).
Multiple high-resolution structures have been determined for CDA from E. coli by Stoddard and colleagues (1, 13, 14). The first structure of bacterial CDA was determined with a single iron in the active site (PDB entry: 1K6W) while the second structure contained iron and 4-(S)-hydroxy-3,4-dihydropyrimidine2 (2) (PDB entry: 1K70) (1). Compound 2, a tight-binding inhibitor of CDA, is formed catalytically through the addition of water to pyrimidin-2-one (3). Other crystal structures of various mutants of CDA with enhanced catalytic properties for the deamination of 5-fluorocytosine have been determined (13, 14). CDA is a promising target for drug development since it can convert 5-fluorocytosine to 5-fluorouracil, which inhibits replication through the inactivation of thymidylate synthetase (15).
In spite of the large amount of kinetic and structural information available for CDA, there are a number of unresolved issues regarding the catalytic mechanism for the deamination of cytosine. The mechanism for the protonation of the leaving group ammonia and N-3 of the uracil product has not been adequately elucidated and the rate-limiting step for substrate turnover has not been identified. We report the crystal structure of E. coli CDA bound with zinc and phosphonocytosine, a potent mimic of the tetrahedral intermediate formed during the deamination of cytosine. This structure has been complemented with an interrogation of the CDA-catalyzed deamination of cytosine using a combination of metal ion substitutions, site directed mutations, substrate analogues, pH activity profiles, solvent isotope effects, and solvent viscosity effects.
Materials and Methods
Materials
All reagents were obtained from Sigma-Aldrich unless otherwise noted. 3-Oxauracil (4) was purchased from Research Organics Inc.. The E. coli CDA knockout strain was obtained from the Keio Collection of the National BioResource Project (NIG, Japan) (16). Kinetic assays were performed in a 96-well plate with a SPECTRAmax 384 Plus spectrophotometer from Molecular Devices. Protein concentrations were determined using the calculated extinction coefficient of 55,190 M−1 cm−1 at 280 nm (Protein Calculator v3.3 at http://www.scripps.edu/~cdputnam/protcalc.html).
Synthesis of Inhibitors
3-Methylcytosine was prepared according to the method of Brookes and Lawley (17). 4-Thiouracil was synthesized based on the procedure of Kaneko et al. (18). 4-Chlorouracil was constructed according to Kazimierczuk et al. (19). Phosphonocytosine (5) was prepared according to the method of Bartlett et al. (20) and phosphonouracil (6) was prepared by modification of the synthesis of 5. The structures of these and other compounds are presented in Scheme 2. The NMR and MS characterization of compound 6 is as follows: 1H NMR (300 MHz, D2O): 6.66 (1H, dd, J = 10.5, 41.1 Hz, PCH=), 5.18 ppm (1H, t, J = 11.1 Hz, NCH=). 31P NMR (121.4 MHz, D2O): 13.46 ppm. 13C NMR (75.4 MHz, D2O): 155.04, 137.47 ppm (d, JP-C = 4.3 Hz, NCH=), 92.26 (d, JP-C = 161.4 Hz, PCH=) ppm. MS (ESI positive mode) found: 149.0096 (M+H)+; MS (ESI negative mode) found: 146.9660 (M-H)− and C3H5N2O3P requires: 148.0038.
Scheme 2.
Preparation of Cytosine Deaminase
The gene from E. coli K12 encoding CDA was cloned into a pET-20a(+) expression vector. Mutants of CDA were prepared in accordance with the QuikChange mutagenesis kit instructions. The plasmid was transformed via electroporation into E. coli K-12 CDA knockout cells which were made electro competent and lysogenized with the λDE3 STAR lysogenization kit from Novagen. The cells were grown overnight on LB/agarose plates enriched with 100 μg/mL ampicillin. A single colony was selected for inoculation in 1 liter of LB enriched with 100 μg/mL ampicillin and 1.0 mM ZnCl2. After incubation for 18 hours with shaking at 20 °C, the cells were harvested by centrifugation. The cells were re-suspended and disrupted by sonication in 50 mM HEPES buffer, pH 7.5, containing 1.0 mM ZnCl2 and 100 μg/mL of the protease inhibitor phenylmethanesulfonyl fluoride. The solution was centrifuged to remove insoluble cell debris. A solution of protamine sulfate was added drop-wise to a final concentration of 2% w/v, for the precipitation of nucleic acids. The solution was centrifuged and then solid ammonium sulfate was added to a final concentration of 50% of saturation. The precipitated protein was removed by centrifugation and the pellet re-suspended with a minimal amount of 50 mM HEPES, pH 8.0. The solution was passed through a 0.45 μm syringe filter and further purified by gel filtration with a HiLoad 26/60 Superdex 200 preparatory grade column. The CDA containing fractions were pooled and further purified using a ResourceQ anion exchange column.
Protein Structure Determination
Crystals of E. coli cytosine deaminase complexed with Zn2+, and phosphonocytosine (5) were grown by hanging drop vapor diffusion by mixing equal volumes of protein and precipitant, and equilibrating over 1.0 mL of precipitant at room temperature. The protein solution contained wild-type CDA that was isolated from cells grown in the presence of 1.0 mM ZnCl2 (18 mg/mL) in 50 mM Tris buffer, pH 7.5, 1.0 mM ZnCl2 and 10 mM phosphonocytosine (5). The precipitant contained 35% pentaerythritol propoxylate, 0.05 M HEPES, pH 7.5, 0.2 M potassium chloride, and 1.0 mM ZnCl2. The crystals appeared in 8 - 9 days and exhibited diffraction consistent with the space group R32, with one molecule of protein per asymmetric unit. Prior to data collection, the crystals were transferred to cryoprotectant solutions composed of their mother liquor supplemented with 20% glycerol. After incubation for ~10 s, the crystals were flash-cooled in a nitrogen stream. Diffraction data were collected at the NSLS X4A beamline (Brookhaven National Laboratory) on an ADSC CCD detector. Diffraction intensities were integrated and scaled with the programs DENZO and SCALEPACK (21). The data collection statistics are given in Table 1.
Table 1.
Data collection and refinement statistics for CDA
| CDA·Zn2+ ·P-cytosine | |
|---|---|
| Data collection | |
| Space group | R32 |
| No. of mol. in asym. unit | 1 |
| Cell dimensions | |
| a, c (Å) | 145.212, 200.609 |
| Resolution (Å)a | 39.0-1.7 (1.76-1.70) |
| No. of unique reflectionsa | 86951 (8512) |
| Rmerge a | 0.096 (0.282) |
| I / σIa | 11.7 (3.1) |
| Completeness (%)a | 98.9 (96.5) |
| Refinement | |
| Resolution (Å)a | 39.0-1.7 |
| Rcryst | 0.173 |
| R free | 0.192 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.02 |
| No. atoms | |
| Protein | 3336 |
| Waters | 221 |
| Inhibitor | 9 |
| Zn2+ ions | 1 |
| PDB entry | 3O7U |
Numbers in parentheses indicate values for the highest resolution shell
The structure of the CDA complex was determined by molecular replacement with BALBES (22), using the E. coli CDA structure (PDB ID: 1K6W) as the search model. Subsequent iterative cycles of refinement were performed for the CDA complex including manual model rebuilding with COOT (23), refinement with PHENIX (24), and automatic model rebuilding with ARP (25). The model of the CDA·Zn2+·phosphonocytosine complex was refined at 1.7 Å with an Rcryst of 0.173 and an Rfree of 0.192 (complete refinement statistics are provided in Table 1). The final structure contains protein residues 4-425, one well-defined Zn2+, and one well-defined inhibitor molecule bound in the active site of CDA.
Measurement of Enzymatic Activity
Cytosine deaminase activity was determined at 30 °C using either a coupled or direct assay. For activity screening of substrate analogues with wild type and mutant enzymes, the coupled assay was used to detect the release of ammonia through the oxidation of NADH. This assay typically contained 7.4 mM α-ketoglutarate, 0.4 mM NADH, and 6.0 units of glutamate dehydrogenase in 100 mM HEPES, pH 8.5 (4). For activity assays at pH 7.5, a direct assay at 286 nm (Δε286 = −680 M−1 cm−1) was used as previously reported (26). For the development and implementation of a direct assay for use over a wide range of pH values, extinction coefficients for the deamination of cytosine to uracil were determined over the pH range 2.5-11.3. The compounds tested for catalytic turnover included the following: cytosine, isocytosine, creatinine, 3-oxauracil, 2,6-diamino-primidin-4-ol, 5-methylcytosine, 4,6-diamino-pyrimidine, 4-amino-2,6-dihydroxy-pyrimidine, 3-methylcytosine, 2-amino-4,6-dihydroxypyrimidine, 2,4,6-triamino-pyrimidine, 6-aminouracil, 2, 4-diamino-pyrimidine, 4-amino-pyrimidine, 2-amino-pyrimidine, 4,6-diamino-2-hydroxypyridine, 2,6-diamino-pyridine, 4-amino-pyridine, 2-amino-pyridine, 4-amino-imidazole, ammeline, 2-chloro-4,6-diamino atrazine, melamine, 4-thiouracil, 4-chlorocystosine, cytidine, CMP, CDP, and CTP.
Metal Analysis
Metal determination was performed with an Elan DRC II ICP-MS (inductively coupled plasma - mass spectrometer) from Perkin Elmer. The analogue detection mode was used with three averaged replicates per reading. External calibration standards were prepared through the serial dilution of a single 10 ppm stock mixture of Zn, Cd, Co, Cu, Mn, Ni and Fe in 2% nitric acid, purchased from Inorganic Ventures Inc.. Freshly prepared standards typically contained 2, 20 and 200 ppb of these metal ions in 1% Trace Select nitric acid from Fluka. The detection of Fe was determined using the dynamic reaction cell (DRC) mode, where NH3 gas was used to reduce interference from argon-oxide species. Protein samples were prepared for analysis by heating in concentrated nitric acid for 30 minutes, followed by dilution with water to 1% nitric acid.
Metal Chelation and Reconstitution
Apo-cytosine deaminase was prepared by dialysis of CDA with 10 mM o-phenanthroline in 50 mM MES, pH 5.75. The o-phenanthroline was removed from the protein by passage through a PD-10 column. Prior to use, the PD-10 column was washed with one column volume of 10 mM dipicolinate and then equilibrated with Chelex treated HEPES buffer, pH 8.0. The apo-protein was reconstituted overnight at a protein concentration of 1.0 mg/mL at 4 °C with varying amounts of ZnCl2 and FeSO4. The reconstitution with Fe2+ was performed anaerobically under argon.
pH Dependence of Enzyme Activity
The pH dependence of kcat and kcat/Km was determined at pH values ranging from 5.3 - 9.9 using 75 mM MES (pH 5.3 - 6.5), PIPES (pH 6.7 - 6.9), HEPES (7.1 - 8.1), TAPS (8.3 - 8.9) and CHES (pH 9.1 - 9.9) to control the pH. The pH values of the final solutions were measured after the completion of the assays. Direct kinetic assays were utilized in which the change in the extinction coefficient between cytosine and uracil was determined at each pH value.
Solvent Kinetic Isotope Effects and Solvent Viscosity Effects
Steady state rates were determined using the coupled assay for the detection of ammonia in D2O or H2O using 100 mM HEPES, pH 8.5, or 100 mM CHES, pH 9.0, at 30 °C. The effects of viscosity on kcat and kcat/Km were determined as previously described in CHES pH 9.0 at 30 °C (15).
Transition-State Inhibitors
Inhibition of CDA was measured using the phosphorus-based analogues of cytosine (5) and uracil (6). The inhibition constants were determined by incubating variable amounts of inhibitor with 2.5 nM enzyme for 30 - 120 minutes in 100 mM HEPES buffer, pH 7.5, in a volume of 245 μL. Residual activities were measured by initiating the reaction with 5 μL of 10 mM cytosine.
Data Analysis
All data were analyzed through fits to the corresponding equations using SigmaPlot 10. For determination of the kinetic parameters kcat, Km and kcat/Km, the data were fit using equation 1 where A is the substrate concentration, v is the initial velocity, kcat is the turnover number and Km is the Michaelis constant. For analysis of pH-rate profiles, plots of log kcat and log kcat/Km vs. pH that indicated the deprotonation of a single group required for maximum activity were fit with equation 2, where c is the maximum activity and Ka is the ionization constant for a group that must be unprotonated for activity. The pH-rate profiles showing a loss of activity at low pH and a wave-shaped loss of activity at high pH that transitions to a lower level of activity, were fit to equation 3. In equation 3, Kb is the ionization constant for the transition to the lower level of activity and d is the new plateau at high pH. pH rate profiles which showed only a wave-shaped plateau at high pH were fit using equation 4.
For evaluation of inhibition constants, Ki, equation 5 was used for plots of residual activity as a function of the inhibitor concentration. In this equation Et is the total enzyme concentration, I is the inhibitor concentration, vo is the activity of the enzyme in the absence of inhibitor, and vi is the residual activity of the enzyme in the presence of inhibitor.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Results
Purification and Metal Dependence
Wild type cytosine deaminase from E. coli was expressed in BL-21 λDE3 cells and purified to homogeneity. The purified enzyme had a turnover number of ~60 s−1. ICP-MS analysis indicated that the enzyme contained 0.56 equivalents of Zn and 0.20 equivalents of Fe. CDA was subsequently expressed in LB enriched with 1.0 mM ZnCl2 and the cells were lysed via sonication in 50 mM TRIS, pH 7.5, supplemented with 1.0 mM ZnCl2. The purified protein contained 0.79 equivalents of Zn, and 0.09 equivalents of Fe. Removal of the metal with o-phenanthroline reduced the catalytic activity to less than 1 s−1 and ICP-MS analysis verified that there were less than 0.05 equivalents of Fe, Zn, Mn, Ni, Cu, Cd or Co per subunit. Apo-CDA was reconstituted with either Zn2+ or Fe2+. The titration with increasing amounts of ZnCl2 demonstrated that CDA reaches maximum catalytic activity when one equivalent has been added (Figure 1). The reconstitution with Fe2+ was performed in a septum sealed vial under argon with 50 μM apo-CDA and three equivalents of FeSO4 in 20 mM HEPES at pH 7.5. The kinetic constants for the iron and zinc bound CDA are presented in Table 2.
Figure 1.

Reconstitution of apo-CDA (8.5 μM) with varying equivalents of ZnCl2. The catalytic activity was determined after dilution of the enzyme to 50 nM using 2.0 mM cytosine in 50 mM TRIS, pH 7.5 and 30 °C.
Table 2.
Metal Content and Kinetic Parameters for Wild-Type and Mutant Variants of CDAa
| enzyme | metal content | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| WT | 0.79 Zn, 0.09 Fe | 132 ± 2 | 0.97 ± 0.03 | (1.4 ± 0.1) × 105 |
| WT | Feb | 209 ± 15 | 0.33 ± 0.08 | (6.3 ± 1.6) × 105 |
| Q156N | 1.40 Zn, 0.04 Fe | < 0.005 | - | - |
| Q156A | 0.92 Zn, 0.04 Fe | < 0.005 | - | - |
| E217Q | 0.87 Zn, 0.15 Fe | < 0.005 | - | - |
| E217A | 0.90 Zn, 0.08 Fe | < 0.005 | - | - |
| H246Q | 0.31 Zn, 0.33 Fe | 0.18 ± 0.01 | 0.25 ± 0.03 | 720 ± 95 |
| H246N | 0.53 Zn, 0.29 Fe | <0.005 | - | - |
| H246A | 0.59 Zn, 0.33 Fe | < 0.005 | - | - |
| D313N | 1.05 Zn, 0.05 Fe | (7.5 ± 0.2) × 10−3 | 1.9 ± 0.2 | 4.0 ± 0.4 |
| D313A | 0.57 Zn, 0.32 Fe | < 0.005 | - | - |
The kinetic parameters were determined at pH 8.5, 30 °C from fits of the data to equation 1.
Apo-enzyme containing less than 0.05 equivalents of metal was reconstituted with 3 equivalents of ferrous sulfate.
Active-Site Mutants
Mutants of CDA were constructed in an attempt to determine the roles of specific residues in substrate recognition and catalytic function. The residues targeted for mutagenesis included Glu-217, Gln-156, His-246 and Asp-313. The kinetic constants for the mutants expressed in a medium containing 1.0 mM ZnCl2 are presented in Table 2. Most of the mutant enzymes possessed less than 0.01% of the activity of the wild-type enzyme and contained a mixture of zinc and iron bound in the active site. The H246Q and D313N mutants had low but detectable turnover numbers.
Substrate Specificity
A library of cytosine analogues was tested for activity with CDA at concentrations of 1.0 mM using 1.0 μM Zn-CDA in 75 mM HEPES, pH 8.5. The only compounds determined to be substrates for CDA in addition to cytosine were creatinine, isocytosine, and 3-oxauracil. The E217A mutant was able to catalyze the release of ammonia from 3-oxauracil with a turnover number of 0.18 s−1 but this mutant was unable to catalyze the deamination of cytosine. With wild-type CDA, one equivalent of ammonia was formed from the hydrolysis of 3-oxauracil. Malonate semialdehyde was the other product, as determined by negative mode ESI-MS from a prominent ion at m/z of 87. The proposed reaction for the hydrolysis of 3-oxauracil to malonate semialdehyde is shown in Scheme 3. In this mechanism we assume that the decarboxylation step and the subsequent loss of ammonia occur nonenzymatically. CDA was unable to catalyze the de-thiolation of 4-thiouracil (< 6 × 10−5 s−1) or the dechlorination of 4-chlorouracil (< 2 × 10−4 s−1). A summary of the kinetic constants for the active compounds are provided in Table 3.
Scheme 3.
Table 3.
Kinetic Parameters for Substrates with Wild-Type and Mutant Forms of Zn-CDAa.
| Enzyme | Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| WT | cytosine | 132 ± 2 | 0.97 ± 0.03 | (1.4 ± 0.1) × 105 |
| WT | isocytosine | 5.1 ± 0.1 | 0.46 ± 0.04 | (1.1 ± 0.1) × 104 |
| WT | creatinine | 5.6 ± 0.7 | 25 ± 4 | (2.2 ± 0.5) × 102 |
| WT | 3-oxauracil | 2.3 ± 0.1 | 4.1 ± 0.3 | (5.6 ± 0.5) × 102 |
| E217A | 3-oxauracil | 0.18 ± 0.01 | 0.85 ± 0.04 | (2.1 ± 0.1) × 102 |
The kinetic parameters were determined at pH 8.5, 30 °C from fits of the data to equation 1.
pH-Rate Profiles
The effects of pH on the catalytic activity of CDA were determined for the iron and zinc enzymes using the coupled and direct enzymatic assays. The absorbance maximum for the substrate changes from 275 nm at low pH to 267 nm at high pH due to the protonation of cytosine with a pKa of 4.5. The absorbance maximum of the product changes from 258 nm at low pH to 284 nm at high pH due to the ionization of uracil with a pKa of pH 9.5 (27). The differences in the extinction coefficients between cytosine and uracil (Δε) were determined at pH values between 5.0 and 11. The catalytic activity at pH values between 5.0 - 8.5 was followed at 286 nm. For pH values between 8.3 - 11, the reactions were monitored at 298 nm.
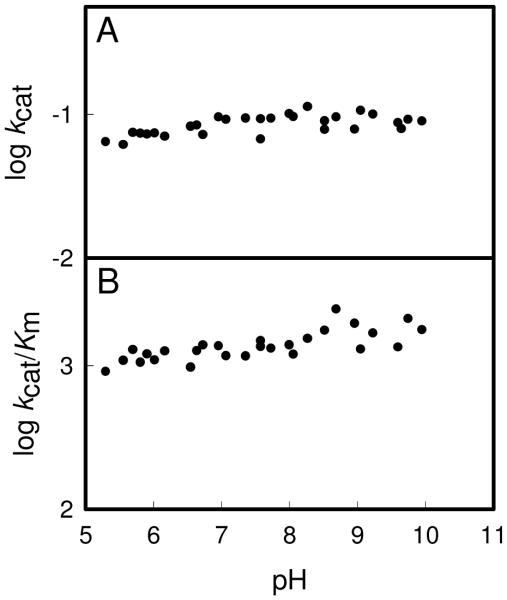
The pH-rate profiles for kcat and kcat/Km for the deamination of cytosine by Zn-CDA are presented in Figure 2. For kcat there is a loss of activity at low pH with an apparent kinetic pKa of 7.3 ± 0.1 and no loss of activity up to pH 10. The pH-rate profile for kcat/Km is more complex. There is a loss of activity at low pH and a wave-like transition to a lower plateau at high pH. A fit of the data to equation 4 gave a pKa for the loss of activity at low pH of 5.5 ± 0.2 and a pKb of 7.9 ± 0.4 at high pH. The pH rate profiles for Fe-CDA are presented in Figure 3. For kcat there is loss of activity at low pH with a pKa of 6.0 ± 0.1. For kcat/Km there is no loss of activity between pH 5 and 7.5 but there is a wave-like transition to a lower plateau between pH 7.5 and 10. The apparent pKb for the change in activity at high pH is 8.2 ± 0.2. The profiles for the H246Q mutant exhibit a low level of activity that is independent of pH between pH 5 and 10 for either kcat or log kcat/Km as shown in Figure 4. The results are summarized in Table 4.
Figure 2.
pH-rate profiles for the deamination of cytosine by Zn-CDA. (A) log kcat vs. pH profile for Zn-CDA. The solid line represents a fit of the data with equation 2. (B) log kcat/Km vs pH profile for Zn-CDA. The solid line represents a fit of the data with equation 3.
Figure 3.
pH-rate profiles for the deamination of cytosine by Fe-CDA. (A) log kcat vs pH profile for Fe-CDA. The solid line represents a fit of the data with equation 2. (B) log kcat/Km vs pH profile for Fe-CDA. The solid line represents a fit of the data with equation 4.
Figure 4.
pH-rate profiles for the deamination of cytosine by Zn-H246Q CDA. (A) log kcat vs pH profile for Zn-H246Q CDA. (B) log kcat/Km vs pH profile for Zn-H246Q CDA.
Table 4.
Kinetic pKa Values from pH-Rate Profiles of Zn and Fe Substituted CDA and Mutant.
| pKa | pKb | |||
|---|---|---|---|---|
| enzyme | metal | kcat vs pH | kcat/Km vs pH | kcat/Km vs pH |
| wild type | Zn2+ | 7.3 ± 0.1 | 5.5 ± 0.2 | 7.9 ± 0.4 |
| wild type | Fe2+ | 6.0 ± 0.1 | - | 8.2 ± 0.2 |
| H246Q | Zn2+ | - | - | - |
Solvent D2O Kinetic Isotope Effects
Solvent deuterium isotope effects were determined by comparing the rates of hydrolysis in D2O and H2O. Solvent isotope effects of 0.62 ± 0.01 and 0.40 ± 0.02 were obtained for Zn-CDA on kcat and kcat/Km, respectively, at pH(D) 9.0 (data not shown).
Effects of Solvent Viscosity
The effects of changing the solvent viscosity on the relative values of kcat and kcat/Km for the deamination of cytosine by Zn-CDA were measured with sucrose as the micro-viscogen at pH 9.0. Shown in Figure 5 are the relative values for the kinetic constants as a function of solvent viscosity. The slopes of these plots are 0.08 ± 0.01 and 0.09 ± 0.05 for the effects on kcat and kcat/Km, respectively.
Figure 5.

(A) Viscosity effects on the relative values of kcat using sucrose as the micro-viscogen at pH 9.0. (B) Viscosity effects on the relative values of kcat/Km for Zn-CDA using sucrose as the micro-viscogen at pH 9.0.
Transition-State Inhibitors
Two phosphorus-based analogues of the putative tetrahedral intermediate were synthesized as potential inhibitors of CDA. The effectiveness of these compounds was evaluated with Zn-CDA at pH 7.5. The inhibition constants were determined from fits of the data to equation 5 and the plots are presented in Figure 6 (28, 29). The racemic phosphonocytosine analogue (5) exhibits a Ki of 52 ± 5 nM while the phosphonouracil analogue (6) has a Ki of 84 ± 7 μM.
Figure 6.
Inhibition of Zn-CDA in 50 mM TRIS, pH 7.5, with (A) phosphonocytosine (5) and (B) phosphonouracil (6). Solid lines represent fits of the data with equation 5. Enzyme and inhibitor solutions were pre-incubated together for 30 minutes at 30 °C prior to initiating the reactions with 0.2 mM cytosine.
Structure Determination of Zn-CDA
The structure of Zn-CDA was determined to a resolution of 1.7 Å with a potent mimic of the putative tetrahedral intermediate bound in the active site. The overall protein structure is very similar to that previously determined by the Stodddard group for Fe-CDA bound with inhibitor 2 (1). The RMSD between the two structures is 0.5 Å. Shown in Figure 7A is the structure of Fe-CDA determined in the absence of a bound ligand (PBD code: 1K6W). The inhibitor shown in Figure 7B is a mimic of the tetrahedral intermediate formed upon the attack of hydroxide at C4 of the substrate cytosine (PDB code: 3O7U). The inhibitor was synthesized as a racemic mixture at the tetrahedral phosphorus center. In this structure it is assumed that the atom proximal to the metal ion is the phosphoryl oxygen and the distal atom is the amino group. This orientation is consistent with attack of hydroxide on the re-face of cytosine (1). Recognition of this analogue of the tetrahedral intermediate is governed by hydrogen bonds from Gln-156 with N1 and the C2 carbonyl oxygen. Glu-217 interacts with N3 and possibly with the exocyclic amino group of the inhibitor. Asp-313 interacts with the exocyclic amino group of the inhibitor and the phosphoryl oxygen. His-246 is 2.8 Å away from the phosphoryl oxygen of the inhibitor. An image of the observed residue-inhibitor interactions and the corresponding distances are presented in Figure 7B. The electron density for the inhibitor is presented in Figure 8.
Figure 7.
Structure of the active site of cytosine deaminase. (A) Fe-CDA in the absence of bound ligands. The coordinates were taken from PDB code: 1K6W. The three residues that facilitate the nucleophilic attack of water (Glu-217, His-246, and Asp-313) and the residue that facilitates the binding of cytosine (Gln-156) are shown. (B) The active site of Zn-CDA in the presence of the tight-binding transition state inhibitor phosphonocytosine (PDB code: 3O7U).
Figure 8.

The active site of Zn-CDA with bound phosphonocytosine. Omit electron density map (Fo-Fc) is contoured at 5.0 sigma. The ligand was omitted from the model and the remainder of the unit cell was subjected to a cycle of simulated annealing with PHENIX at 3000 degrees C.
Discussion
Mechanism of Action
The working model for the reaction mechanism for the deamination of cytosine is presented in Scheme 4. From the structure of CDA in the presence of the tight binding phosphonate inhibitor and the mutagenesis experiments, it is clear that this enzyme requires a single divalent cation in the active site for the coordination of the water molecule that will eventually attack cytosine. There are also three active site residues that are required for the activation of this nucleophilic water and for subsequent proton transfer reactions. These residues include Glu-217, His-246, and Asp-313. Mutation of any of these residues dramatically diminishes the catalytic efficiency of CDA. This same triad of active site residues is located within the active sites of adenosine deaminase (6, 30), guanine deaminase (2, 5) SAH deaminase (7), 8-oxoguanine deaminase (9) and isoxanthopterin deaminase (10)
Scheme 4.
In the mechanism proposed here, the metal bound water molecule is hydrogen bonded to Asp-313 and His-246. This is supported by the X-ray structure of CDA (PDB code: 1K6W) in the absence of an added substrate analogue (1). From the X-ray structures of CDA in the presence of inhibitor 2 (PDB code: 1K70) and inhibitor 5 (PDB code: 3O7U) it is clear that Glu-217 is positioned to deliver a proton to N3 of the product uracil. However, this residue appears to be too far from the metal bound water molecule for the direct abstraction of a proton from this molecule. The role of Glu-217 proposed here is also supported by the ability of the E217A mutant to catalyze the hydrolysis of compound 4 where protonation of the oxygen located in the comparable position of N3 is not necessary (Scheme 3). In the mechanism proposed here, His-246 functions to shuttle the proton from the metal bound water molecule to Glu-217. It is uncertain whether this proton transfer occurs prior to, or after, the binding of cytosine in the active site. Once the substrate is bound in the active site, the metal bound hydroxide attacks the re-face of cytosine. The stereochemistry of this reaction is supported by the structure of inhibitor 2 bound in the active site of Fe-CDA (1) and of inhibitor 5 bound in the active site of Zn-CDA.
The tetrahedral intermediate collapses after abstraction of a proton by Asp-313 and delivery of this proton to the nitrogen of the leaving group amine to generate uracil and ammonia. From the structure of inhibitor 5 bound in the active site of Zn-CDA, Asp-313 is the most likely residue positioned to facilitate the transfer of the proton from the attacking hydroxide to the leaving group amine. However, it cannot be certain whether this transformation is also assisted in some undefined manner by His-246. In many examples from the amidohydrolase superfamily the equivalent aspartate residues have been demonstrated to facilitate the protonation of the leaving group (2). The mutation of His-246, Asp313, or Glu-217 results in the dramatic loss in catalytic activity and thus each of these residues are significant for the proper functioning of this enzyme.
In the pH rate profiles with either the zinc or iron activated CDA there is a loss of catalytic activity as the pH is lowered toward 5. For kcat the kinetic pKa is lower for Fe-CDA than it is for Zn-CDA and for kcat/Km there is no loss of activity for Fe-CDA down to pH 5. It cannot be determined from the available data as to which of the three active site residues become protonated in this titration or whether this behavior reflects the protonation of the metal bound hydroxide. For kcat there is no loss of activity at high pH and for kcat/Km there is a small reduction in this kinetic constant to a slightly lower value with a new plateau.
The identity of the rate limiting step(s) was probed using solvent deuterium isotope effects and changes in solvent viscosity. No perturbations in the kinetic constants were observed when sucrose was used to change the solvent viscosity. This result indicates that a step in the reaction mechanism prior to product release limits the value of kcat and that a step after the binding of substrate limits the value of kcat/Km (31, 32). These step(s) could either be the actual bond making and breaking steps or conformational changes that precede or follow these steps.
Solvent deuterium isotope effects were used to probe the steps that involve proton transfer steps. These steps would include formation of the tetrahedral intermediate and collapse of this intermediate during the generation of the products, uracil and ammonia. For kcat and kcat/Km there is an inverse isotope effect of approximately 0.5 when D2O is used as the solvent. This inverse isotope effect is likely due to the formation of a low-barrier hydrogen bond between Glu-217 and N3 of cytosine and is indicative of a compressed hydrogen bond which forms due to the similarity in the pKa values of Glu-217 and N3, which are estimated to be about 4.5. Upon nucleophilic attack by hydroxide and formation of the tetrahedral intermediate, the pKa for N3 shifts to a much larger value as the proton is transferred from Glu-217 to N3 of uracil, which has a pKa of 9.5. Inverse isotope effects of comparable magnitude were previously reported for AMP deaminase (33) and adenosine deaminase (34, 35).
Footnotes
This work was supported in part by the NIH (GM 71790), the Robert A. Welch Foundation (A-840) and the Hackerman Advanced Research Program (010366-0034-2007). The X-ray coordinates and structure factors for cytosine deaminase have been deposited in the Protein Data Bank (PDB accession code: 3O7U).
Abbreviations: CDA, cytosine deaminase; AHS, amidohydrolase superfamily; COG, clusters of orthologous groups; ICP-MS, inductively coupled plasma mass spectrometry.
The stereochemistry of this compound was incorrectly designated as R in the original publication (1).
References
- 1.Ireton GC, McDermott G, Black ME, Stoddard BL. The structure of Escherichia coli cytosine deaminase. J. Mol. Biol. 2002;315:687–697. doi: 10.1006/jmbi.2001.5277. [DOI] [PubMed] [Google Scholar]
- 2.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 3.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 4.Xiang DF, Patskovsky Y, Xu C, Fedorov AA, Fedorov EV, Sisco AA, Sauder JM, Burley SK, Almo SC, Raushel FM. Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily. Biochemistry. 2010;49:6791–6803. doi: 10.1021/bi100897u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maynes JT, Yuan RG, Snyder FF. Identification, expression, and characterization of Escherichia coli guanine deaminase. J. Bacteriol. 2000;182:4658–4660. doi: 10.1128/jb.182.16.4658-4660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 7.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marti-Arbona R, Xu C, Steele S, Weeks A, Kuty GF, Seibert CM, Raushel FM. Annotating enzymes of unknown function: N-formimino-L-glutamate deiminase is a member of the amidohydrolase superfamily. Biochemistry. 2006;45:1997–2005. doi: 10.1021/bi0525425. [DOI] [PubMed] [Google Scholar]
- 9.Hall RS, Fedorov AA, Marti-Arbona R, Fedorov EV, Kolb P, Sauder JM, Burley SK, Shoichet BK, Almo SC, Raushel FM. The hunt for 8-oxoguanine deaminase. J. Am. Chem. Soc. 2010;132:1762–1763. doi: 10.1021/ja909817d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall RS, Agarwal R, Hitchcock D, Sauder JM, Burley SK, Swaminathan S, Raushel FM. Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase. Biochemistry. 2010;49:4374–4382. doi: 10.1021/bi100252s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter DJ, Austin EA. Cytosine deaminase. The roles of divalent metal ions in catalysis. J. Biol. Chem. 1993;268:24005–24011. [PubMed] [Google Scholar]
- 12.Porter DJT. Escherichia coli cytosine deaminase: the kinetics and thermodynamics for binding of cytosine to the apoenzyme and the Zn2+ holoenzyme are similar. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 2000;1476:239–252. doi: 10.1016/s0167-4838(99)00246-0. [DOI] [PubMed] [Google Scholar]
- 13.Fuchita M, Ardiani A, Zhao L, Serve K, Stoddard BL, Black ME. Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo. Cancer Res. 2009;69:4791–4799. doi: 10.1158/0008-5472.CAN-09-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahan SD, Ireton GC, Knoeber C, Stoddard BL, Black ME. Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy. Protein Eng. Des. Sel. 2004;17:625–633. doi: 10.1093/protein/gzh074. [DOI] [PubMed] [Google Scholar]
- 15.Huber BE, Austin EA, Richards CA, Davis ST, Good SS. Metabolism of 5-fluorocytosine to 5-fluorouracil in human colorectal tumor cells transduced with the cytosine deaminase gene: significant antitumor effects when only a small percentage of tumor cells express cytosine deaminase. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8302–8306. doi: 10.1073/pnas.91.17.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brookes P, Lawley PD. The methylation of cytosine and cytidine. J. Chem. Soc. 1962:1348–1351. [Google Scholar]
- 18.Kaneko K, Katayama H, Wakabayashi T, Kumonaka T. Pyrimidine-derivatives .1. The highly regioselective 4-thionation of pyrimidine-2,4(1h,3h)-dione derivatives with Lawesson reagent. Synthesis-Stuttgart. 1988:152–154. [Google Scholar]
- 19.Kazimier Z, Shugar D, Lipski M. Intermediates in the synthesis of purines and pteridines - Selective hydrolysis of chloropyrimidines. Acta Biochim. Pol. 1972;19:359. [PubMed] [Google Scholar]
- 20.Bartlett PA, Hunt JT, Adams JL, Gehret JCE. Phosphorus-containing purines and pyrimidines: a new class of transition-state analogs. Bioorg. Chem. 1978;7:421–436. [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 26.Porter DJ, Austin EA. Cytosine deaminase. The roles of divalent metal ions in catalysis. J. Biol. Chem. 1993;268:24005–24011. [PubMed] [Google Scholar]
- 27.Ganguly S, Kundu KK. Protonation/deprotonation energetics of uracil, thymine and cytosine in water from e.m.f/spectrophotometric measurements. Can. J. Chem. 1994;72:1120–1126. [Google Scholar]
- 28.Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 29.Sculley MJ, Morrison JF, Cleland WW. Slow-binding inhibition: the general case. Biochim. Biophys. Acta. 1996;1298:78–86. doi: 10.1016/s0167-4838(96)00118-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Quiocho FA. Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity. Biochemistry. 1998;37:8314–8324. doi: 10.1021/bi980324o. [DOI] [PubMed] [Google Scholar]
- 31.Brouwer AC, Kirsch JF. Investigation of diffusion-limited rates of chymotrypsin reactions by viscosity variation. Biochemistry. 1982;21:1302–1307. doi: 10.1021/bi00535a030. [DOI] [PubMed] [Google Scholar]
- 32.St Maurice M, Bearne SL. Kinetics and thermodynamics of mandelate racemase catalysis. Biochemistry. 2002;41:4048–4058. doi: 10.1021/bi016044h. [DOI] [PubMed] [Google Scholar]
- 33.Merkler DJ, Schramm VL. Catalytic mechanism of yeast adenosine 5′-monophosphate deaminase. Zinc content, substrate specificity, pH studies, and solvent isotope effects. Biochemistry. 1993;32:5792–5799. doi: 10.1021/bi00073a011. [DOI] [PubMed] [Google Scholar]
- 34.Weiss PM, Cook PF, Hermes JD, Cleland WW. Evidence from nitrogen-15 and solvent deuterium isotope effects on the chemical mechanism of adenosine deaminase. Biochemistry. 1987;26:7378–7384. doi: 10.1021/bi00397a027. [DOI] [PubMed] [Google Scholar]
- 35.Cleland WW. Low-barrier hydrogen bonds and low fractionation factor bases in enzymatic reactions. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]