Abstract
Efficacy of siRNAs as potential anticancer therapeutics can be increased by their targeted delivery into cancer cells via tumor-specific ligands. Phage display offers an unique approach to identify highly specific and selective ligands that can deliver nanocarriers to the site of disease. In this study, we proved a novel approach for intracellular delivery of siRNAs into breast cancer cells through their encapsulation into liposomes targeted to the tumor cells with preselected intact phage proteins. The targeted siRNA liposomes were obtained by a fusion of two parental liposomes containing spontaneously inserted siRNA and fusion phage proteins. The presence of pVIII coat protein fused to a MCF-7 cell-targeting peptide DMPGTVLP in the liposomes was confirmed by Western blotting. The novel phage-targeted siRNA-nanopharmaceuticals demonstrate significant down-regulation of PRDM14 gene expression and PRDM14 protein synthesis in the target MCF- 7 cells. This approach offers the potential for development of new anticancer siRNA-based targeted nanomedicines.
Keywords: siRNA, liposomes, major coat protein, phage display, targeted delivery, breast cancer
Introduction
Small (short) interfering fragments of RNA (siRNA) are known to inhibit specific protein synthesis by suppressing target gene expression at the mRNA level by a mechanism called RNA interference (RNAi). They are considered as prospective anticancer drugs because of their high specific gene silencing efficiency and low toxicity1. However, systemic delivery of siRNAs into tumor cells is a challenging task because siRNAs themselves are unstable in blood stream and cannot penetrate through cellular membranes. For these reasons, other means of siRNA delivery to target cells have been devised. They include the encapsulation of siRNA in liposomes or other nanoparticles, viral and bacterial delivery of siRNA precursors, or stabilization of siRNA molecules via their chemical modification2.
Among various nano-carrier systems, PEGylated —stealth liposomes may be considered as ideal vehicles for siRNA delivery, mainly due to their biological inertness, non-toxicity and protection of siRNAs from nucleases3. Moreover, therapeutic efficacy of liposomes can be further increased by their coupling with tumor-specific ligands that enhance their selective interaction with tumors, or control unloading of their cargo within tumors29. For example, siRNA-loaded immunoliposomes targeted with anti-transferrin antibody produced specific inhibition of Her-2 expression in breast cancer animal models and inhibited tumor growth in pancreatic cancer animal model4. Attachment of cell-penetrating peptides (CPP), a family of peptides able to translocate across the cell membrane, was also used to deliver siRNA into cancer cells. It was shown that liposomes bearing a synthetic arginine-rich CPP are stable and can efficiently transfect lung tumor cells in vitro5. Recent study has shown that systemic delivery of siRNA encapsulated in targeted liposomes can efficiently depress expression of oncogenes in metastatic murine melanoma cells6. Taken together, targeted siRNA-containing liposomes represent a promising cancer treatment option. However, despite its promise, targeted liposome technology is not without difficulties. Preparation of the targeting ligands, such as antibodies, and their conjugation to lipids to make usable quantities of addressed vesicles, has proven troublesome, differing idiosyncratically from one targeted particle to another. Therefore, a new challenge, within the frame of this concept is development of highly selective, stable, easy to produce and physiologically acceptable ligands and their integration into targeted nanoparticulate formulations. These considerations and others led us to think of phage proteins as easily available targeting components of the drug carriers18,19,24.
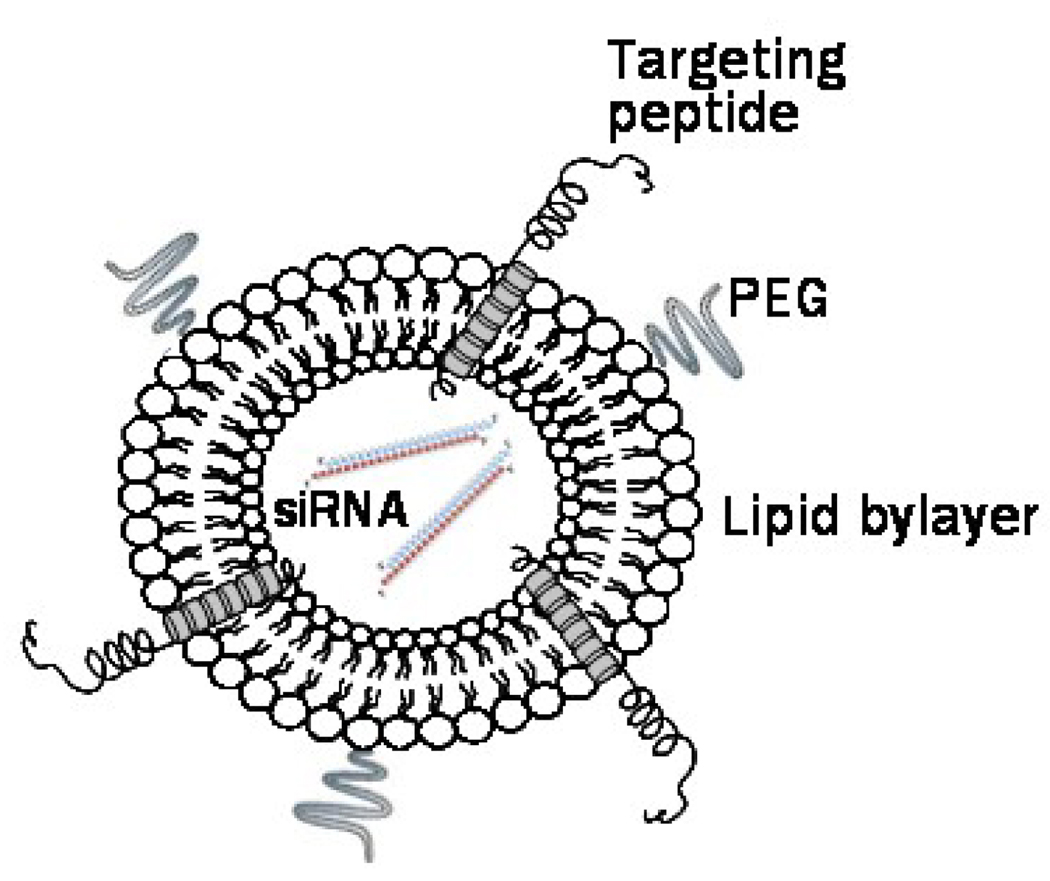
The integration of phage display technology with the nanocarrier-based drug delivery platforms emerged recently as a new drug targeting approach7–11. Evolved as a result of advances in combinatorial chemistry and phage display, phage technique provided a new way of dentification of tumor-specific peptide ligands in a high throughput fashion12–14. Initially, foreign proteins have been fused to the N-terminus of the minor coat protein pIII of filamentous phage yielding a chimeric —fusion phage, in which up to 5 copies of the foreign antigen were displayed on a tip of a virion15. The identified peptides can be then chemically synthesized and coupled to drug carriers16. A number of phage-borne peptides specific to various tumors have been identified by this way13 and some of these peptides have been successfully used to deliver nanocarriers to the diseased organs. In an alternative phage display format, a sequence encoding a foreign peptide was spliced in-frame into gene gpVIII encoding the major coat protein pVIII, leading to the landscape fashion of the phage display allowing thousands copies of guest peptide fused to the phage coat protein to cover whole virion surface17. The tumor-specific landscape phages can be affinity selected from multibillion clone libraries by their ability to interact very specifically with cancer cell surface receptors and/or penetrate into the cells11, 34. Fusion coat proteins pVIII, dominating in the landscape phage, were proposed themselves as easily available targeting ligands for pharmaceutical liposomes11,18, 19, 33, 35. This new approach is well justified by the ability of the phage coat proteins to spontaneously insert into the lipid bilayers20. The membranophilic property of major coat protein was exploited to insert target-specific peptides fused to the Nterminus of the phage coat protein into liposomes. The resulting liposomes demonstrated functional binding specificity towards model streptavidin-conjugated colloidal gold particles and target breast and prostate cancer cells11, 18, 19, 33, 35. The rationale behind this novel targeting concept is that a hybrid phage protein fused to a cancer cell-specific peptide serves a dual function in liposome targeting: it’s surface-exposed N-terminus navigates the liposomes to the target cellular receptors while hydrophobic C-terminus anchors the targeting peptides to the liposomal membrane, as illustrated in Figure 1 where the targeting peptide is DMPGTVLP.
Figure 1.
siRNA-loaded liposome targeted by phage protein fused with a MCF-7 cell-specific peptide DMPGTVLP. The hydrophobic helix of the protein is anchored in the lipid bilayer, whereas the N-terminal fusion peptide DMPGTVLP is displayed on the surface of the liposome. The siRNA molecules are pictured as strands inside the liposomes.
Here we adapted the phage-based targeting strategy for siRNA delivery to breast cancer cells. We applied for the first time the fusion of phage proteins with liposomes for construction of siRNA-loaded nano-vehicles specifically interacting with cancer cells. As a target for siRNA mediated silencing, we chose PRDM14 gene - a member of the family of genes that encode proline rich domain proteins (PRDM) and may play important role in breast cancer carcinogenesis21. Our studies showed that gene-specific siRNA duplexes, encapsulated in phage protein-targeted PEGylated liposomes, specifically inhibit the expression of the target gene in the breast cancer cells.
Materials and methods
Reagents
L-α-phosphatidylcholine (ePC); 1,2-dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt; DPPG); 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt; DOTAP); 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethyleneglycol)2000] (ammonium salt; PEG2000-PE); and cholesterol (CHOL) were from Avanti Polar Lipids Inc. (Alabaster, AL).
Sodium cholate, 2.5% CHAPS: 3-[(3-cholamidopropyl)dimethylammonio]-1-proanesulfonate, bovine serum albumin, phenylmethanesulfonyl fluoride (PMSF), and proteinase K were from Sigma (St. Louis, MO); 16% non-gradient tris-tricine polyacrylamide gel (Jule Inc.Milford, CT); Immobilon-P PVDF membrane from Millipore (Billerica, MA); NeutrAvidin™-HRP and BCA protein assay kits, and chemiluminescent substrate solution from Pierce (Rockford, IL); biotinylated-SP-conjugated Affinitipure goat antirabbit IgG from Jackson Immunoresearch (West Grove, PA).
Cells
The cell line MCF-7 (ATCC, HTB 22™) derived from plural effusion of human breast adenocarcinoma, human hepatocellular carcinoma HepG2 (ATCC, CRL-2235™) cells and human non-tumorigenic mammary epithelial cells MCF-10A (ATCC, CRL-10317™) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were grown as recommended by ATCC and incubated at 37°C, 5% CO2.
Phage display library and bacterial strain
Type 8 phage display landscape library f8/822, was used in selection procedures. In this phage library, foreign peptides are displayed as an extension of each major coat protein unit due to an in frame random oligonucleotide insertion in the gene gpVIII resulting in the display of ~4000 guest peptide units on the surface of a phage particle. The f8/8 library has random octapeptide inserts and a size of 1.4 × 109 clones. All general methods of handling phage, including propagation, purification, titering, production of pure phage clone, and isolation of phage DNA have been previously described 23. Escherichia coli (E. coli) strain K91BlueKan (Kanr) {Hfr C thi lacZΔ M15 lac Y∷mkh lacIQ} used for phage tittering and propagation was kindly provided by George Smith (University of Columbia-Missouri).
Selection of breast cancer cell-specific phage
The selection protocol previously described was used to identify phage clones homing to MCF-7 cells23. Binding specificity and selectivity of the phage DMPGTVLP (designated by the sequence of inserted foreign peptide) was determined in a phage capture assay23 adapted for 96-well culture plate format24. Selectivity of the phage towards target breast adenocarcinoma cells MCF-7 was tested in comparison with another breast ductal carcinoma ZR-75-1 cells, nonrelated hepatocellular carcinoma cells HepG2, normal human mammary cells MCF-10A, and serum. Unrelated phage clone with guest peptide VPEGAFSS (streptavidin binder25) was used for comparison as a negative control.
Mode of phage interaction with breast cancer cells
To determine a role of cell metabolism in association of the phage with alive cells, the incubation of phage with cells were carried out at room temperature without serum, at 37 °C without serum, and at 37 °C with serum. MCF-7 cells (2.0 × 105 cells/well) were grown in Leibovitz medium (L-15) in triplicate to confluence in three 96-well cell culture plates; serum-treated wells served as a negative control. In the cool experiment, the cells were serum-starved for 1h followed by incubation with phage clone (~106 cfu in blocking buffer (0.1% BSA in serum-free L-15 media)) for additional 1 h at room temperature. Subsequently, cells were washed with washing buffer (0.1% BSA and 0.1% Tween-20 in serum-free L-15 media) to remove unbound phage. Surface bound phages were recovered by treating wells with acid elution buffer (0.1 M glycine-Hcl, pH 2.2, 1 mg/ml BSA and 0.1 mg/ml phenol red), and the eluates were neutralized with neutralizing buffer (1 M Tris, pH 9.1). Wells were additionally washed twice with washing buffer and postelution washings were collected (fractions PEW-1 and PEW-2). To retrieve internalizing phages, wells were treated with lysis buffer (2.5% CHAPS) for 10 min on a rocker. The eluate, PEW-1, PEW-2 and lysate fractions were titered in E. coli K91 BlueKan cells and phage recovery was calculated as the ratio of output phage to input phage. The procedure was modified by carrying out the incubation of cells with phage at 37 °C instead of room temperature to study the effect of temperature on the binding and internalization of phages. In another experiment, the incubation was carried out at 37°C in the medium supplemented with serum.
Preparation and purification of the phage fusion coat protein
Phage fusion 55-mer coat protein ADMPGTVLPDPAKAAFDSLQASATEYIGYAWAMVVVIVGATIGIKLFKKFTSKAS (foreign 8-mer peptide shown with bold font) was prepared by stripping the DMPGTVLP phage in cholate buffer19. Briefly, the mixture of 350 µl phage in the TBS buffer (~1 mg/ml) and 700 µl of 120 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA, pH 8.0 and 27 µl chloroform was incubated at 37°C for 1h. The fusion protein was purified from the viral DNA and traces of bacterial proteins by size exclusion chromatography using a Sepharose 6B-CL (Amersham Biosciences) column (1 cm × 45 cm), which was eluted with 10 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0. The chromatographic profile was monitored by the Econo UV monitor (Bio-Rad, CA); 5 ml per fraction were collected and stored at 4°C.
Protein-liposome formulation
A lipid film composed of ePC, CHOL, DPPG, DOTAP and PEG2000-PE) (molar ratio 45:30:20:2:3) was prepared in a round bottom flask by removing chloroform. The film was further dried for 4 h under high vacuum26, 27 and then rehydrated in sterile PBS buffer pH 7.4 up to a final liposome concentration of 40 mg/ml. To obtain the —plain liposomes (liposome formulation without protein), the hydrated lipids were bath sonicated for 10–15 mins and finally extruded through 200 nm polycarbonate membrane. Phage protein was incorporated into the lipid formulation (10.3 mg/ml) by an overnight incubation at 37°C (1:200 wt phage-protein: liposomes) in 15 mM sodium cholate. The formulation was dialyzed overnight (dialysis membrane cutoff size 2000 Da) against PBS buffer pH 7.4 to remove the excess of sodium cholate.
siRNA-liposome formulation
A lipid film composed of ePC:CHOL:DOTAP:PEG2k-PE (60:30:10:2 molar ratio) was made in a round bottom flask removing the chloroform. The film was further dried for 4 h under high vacuum, and then rehydrated in sterile PBS buffer pH 7.4 (in nuclease-free water) up to a final liposome concentration of 10.3 mg/ml. The hydrated lipids were bath sonicated for 10–15 min and finally extruded through 200 nm polycarbonate membrane. Then, the plain liposomes (liposome formulation without siRNA) were incubated at room temperature for 3.5 h with a mixture of three siRNA fragments at a molar ratio DOTAP:siRNA/10:1.
Size distribution and zeta potential (ζ) analysis
All formulations were characterized by size, size distribution and zeta potential (ζ) using the dynamic light scattering (DLS) on a Zeta Plus instrument (Brookhaven Instrument Corporation) and Zeta Phase Analysis Light Scattering (PALS) with an ultrasensitive zeta potential analyzer instrument (Brookhaven Instruments, Holtsville, NY). A portion (5 µL) of each liposome suspension was diluted up to 1 ml in deionized water and then analyzed for the size distribution; for the zeta potential each sample was diluted in 1 mM KCl (5 µl/ 1.5 mL).
PicoGreen fluorescent assay
To check the amount of free siRNA in solution, fluorescent assay based on the interaction of the PicoGreen (Invitrogen, Carlsbad, CA) reagent and free (nonencapsulated in liposomes) siRNA was used. Fluorescence intensity in this assay is proportional to the amount of the siRNA. Only free siRNA is able to react with the probe and to emit fluorescence. The siRNA associated to the liposomes is shielded and not accessible to the probe. For analysis, siRNA-protein-liposomes (1 µl of preparation diluted in 10 µl nuclease-free water) was incubated with 990 µl of PicoGreen solution (1/200 dilution of the probe in TBE buffer) at 37 °C for 10 mins. The same amount of free siRNA in PicoGreen solution was used as a reference to determine the amount of siRNA not associated with lipids. As blanks, same dilution of protein-liposomes and plain PicoGreen solution were used and subtracted to the final sample fluorescence. PicoGreen-siRNA fluorescence intensity was detected at excitation wavelength of 480 nm and emission 520 nm by a Hitachi F- 2000 fluorescence spectrometer (Hitachi, Japan). The portion of the free siRNA was calculated according to the following formula: % siRNA in solution= (PicoGreen fluorescence liposomes/PicoGreen fluorescence free).
Knockdown of PRDM14 gene
siRNAs for human PRDM14:
5' CCAG UGAAGUGAAGACCUATT 3' (siPRDM14-F),
5' UAGGUCUUCACUUCACUGG TT 3' (siPRDM14-R),
5’ GGACAAGGGCGAUAGGAAATT-3’ (siPRDM14-2F),
5’ UUUCCUAUCGCCCUUGUCCTT-3’ (siPRDM14-2R),
5’ GGGAAAAUCUUCUCAGAUCTT-3’ (siPRDM14-3F), and
5’ GAUCUGAGAAGAUUUUCCCTT-3’ (siPRDM14-3R)
were purchased from Integrated DNA Technologies (IDT, Coralville, Iowa, USA). Negative control siRNA were purchased from Applied Biosystems (Foster City, CA). Human MCF-7cells were transfected with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. To study PRDM14 gene knockdown, 105 MCF-7 cells in 6-well culture plates were transfected with PRDM14-specific siRNA (40 nM) or scrambled siRNA (40 nM) mixed with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). For knockdown of PRDM14 gene by siRNA- DMPGTVLP phage fusion protein-liposomes, siRNA-DMPGTVLP-liposomes (50 µM), scrambled siRNA-DMPGTVLP-liposomes (50 µM), or siRNA-liposomes (150 µM) were mixed with 100,000 MCF-7 cells in the well of a 6-well culture plate and adjusted to 2 ml with L-15 media (10% FBS without antibiotics) resulting in 40 nM total concentration of siRNA. The plates were rocked gently at room temperature and incubated at 37°C for 72 h. The medium was changed every 24 h.
Analysis of PRDM14 gene expression by RT-PCR
Total RNA was extracted using the RNeasy Micro kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s protocol. Reverse transcription of total RNA and cDNA amplification by PCR was carried out using 25 ng of total RNA using one step Access RT-PCR kit according to the manufacturer’s protocol. For RT-PCR analysis of PRDM14 and GAPDH genes expression, the primers used were: PRDM14 sense
5’-GTGCGGTCCCGGGATGGCTCTAC, PRDM14 antisense
5’-GGGGCGGTGGAATTAAAGTGTCAG, GAPDH sense
5’-GGGGAGCCAAAAGGGTCATCATCT, and GAPDH antisense
5’-GACGCCTGCTTCACCACCTTCTTG (IDT, Coralville, Iowa, USA).
The primers for PRDM14 and GAPDH genes were used at final concentration 0.1 µM. One cycle of reverse transcription of isolated RNA at 48°C (45 min) and 94°C (2 min) was followed by 35 cycles of PCR at 62°C (30 sec), 68°C (1 min) and 68°C (7 min). Relative levels of gene expression were quantified using the KODAK imager.
Analysis of PRDM14 protein expression in MCF-7 cells using western blot technique
MCF-7 cells were treated with PRDM14 gene-specific siRNA and scrambled siRNA preparations (40 nM), encapsulated in phage protein-targeted liposomes or mixed with lipofectamine reagent. After 48 h, cells were lysed with RIPA buffer (Sigma, St. Louis, MO) containing protease inhibitor cocktail (Sigma, St. Louis, MO) and PMSF (2 mM final concentration). The protein concentration in whole cell lysate was measured by the BioRad DC protein assay (Hercules, CA). A portion (15 µg) of the whole cell extract were analysed by electrophoresis in 4–20% polyacrylamide gradient Tris-HCl gels (Hercules, CA) and transferred to PVDF membrane (Millipore, Billerica, Massachusetts). Proteins in the gel were transferred to an Immobilin-P PVDF membrane (Millipore, Billerica, Massachusetts) and resulting blots probed with polyclonal anti-PRDM14 antibody (Abcam, Cambridge, MA) (1:500 dilution) followed by incubation with peroxidase-conjugated Affinipure Goat Anti-rabbit IgG (1:5000) (Jackson ImmunoResearch, West Grove, PA) before being visualized using a chemiluminescent substrate solution (Pierce, Rockford, Illinois). Membranes were stripped using the western blot stripping buffer (Thermo Scientific, Rockford, IL) for 10 min and probed with monoclonal anti-GAPDH antibody (Abcam, Cambridge, MA )(1:2000) followed with incubation with peroxidaseconjugated Affinipure Goat Anti-mouse IgG (1:5000) (Jackson ImmunoResearch, West Grove, PA) and visualized using chemiluminescent substrate solution.
Statistical Analysis
Data from all experiments are expressed as mean ± standard error mean (SEM). Differences were determined using Student’s independent t-test (p < 0.05).
Results
Selection of breast cancer cell-specific phages
To obtain MCF-7 breast cancer cell-binding phages, the billion-clone library of phages harboring 8- mer peptides on all 4,000 copies of major coat proteins22 was used for in vitro selection.. A portion of the library containing 100 billion phage particles was depleted first for phages binding a cell culture flask and then was applied to MCF-7 cells. Unbound phages were removed, whereas bound phages were eluted with mild acid. The eluted phages were amplified, purified and used in subsequent selection rounds. The selection procedure was iterated until an essential enrichment of bound phages was reached at the fourth round of selection. Hundred clones randomly picked at the final selection round were amplified, and their DNA was sequenced and translated to reveal 44 phage clones displaying unique peptides.
Phage Characterization
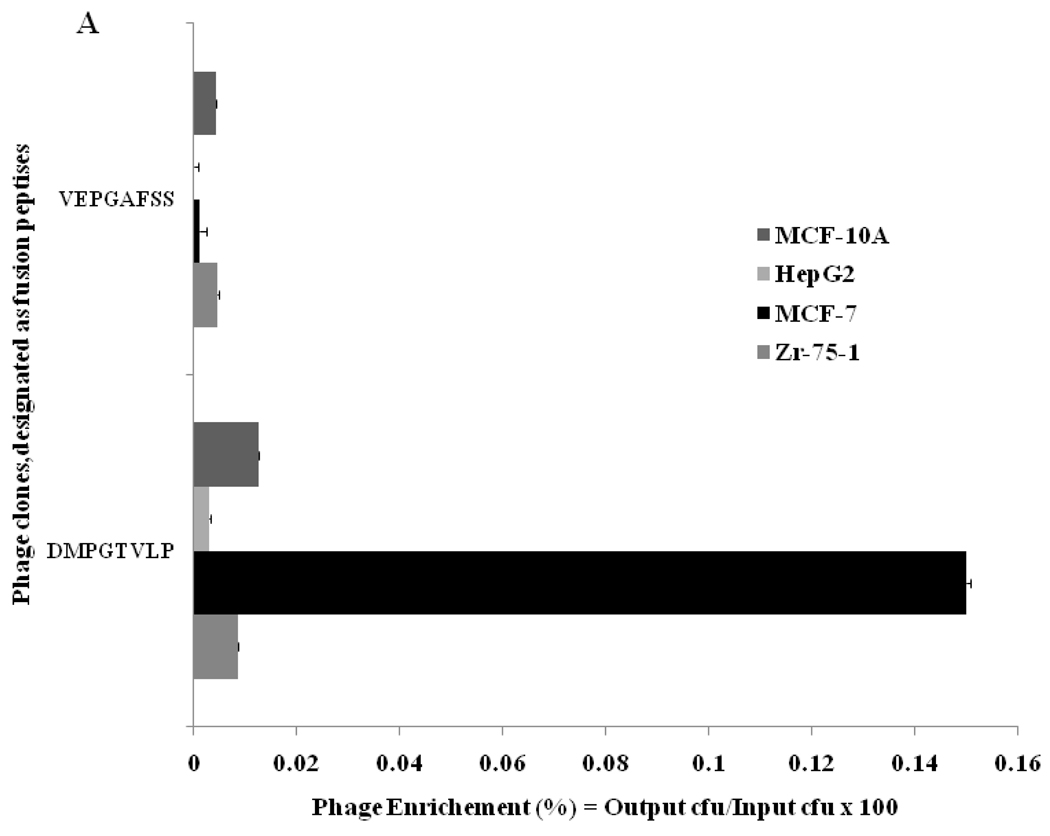
Selectivity of the phage was determined by measuring their binding to another breast ductal carcinoma ZR-75-1 cells, nonrelated hepatocellular carcinoma cells HepG2, normal human mammary cells MCF-10A, and serum. In this test, equal numbers of the phage particles were incubated with serum, the target and control cells in parallel on the same 96-well plate. After incubation, unbound phages were removed by multiple washing and cells were lysed with a mild detergent to recover cell-interacting phages. A relative level of phage binding to different cells and control serum was found by calculating the phage recovery—the ratio of output phage to input phage, determined by phage titering in host E. coli cells. At the same input phage concentration (~108 vir/ml) the phage recovery for MCF-7 cells was 17, 5,11 and > 200 times higher than for control cells ZR-75-1, HepG2, MCF-10A and serum respectively. Binding of the phage DMPGTVLP to MCF-7 was more than 2000 times higher than binding of a control nonrelated phage isolated from the same library. These data demonstrate high specificity of the selected phage towards the target breast cancer cells.
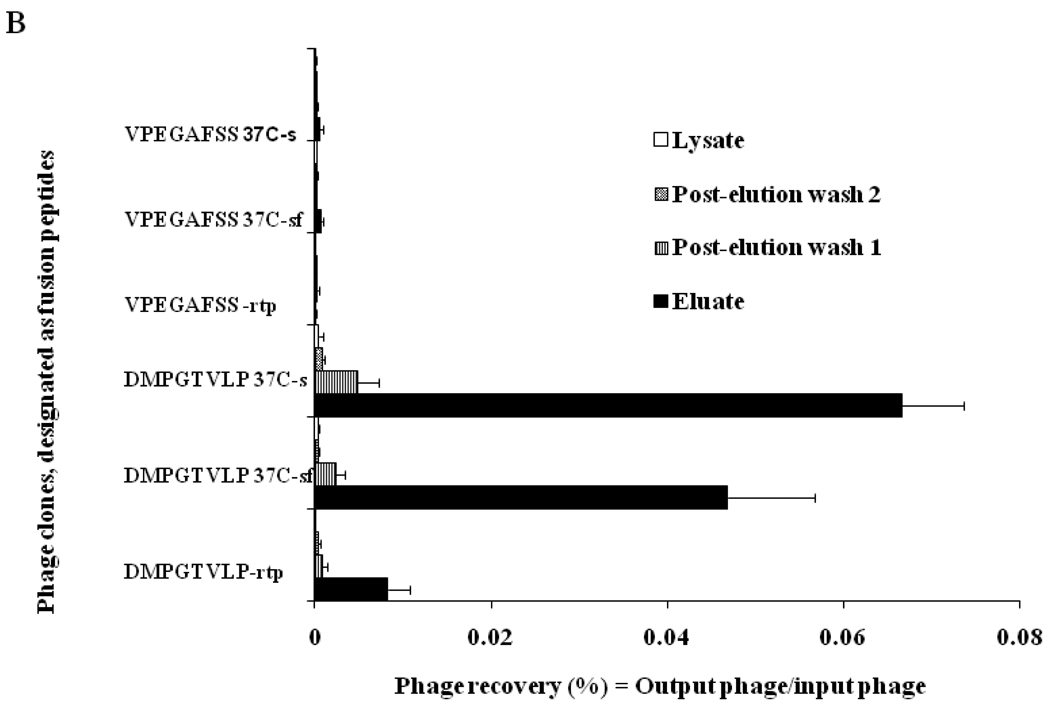
Selected phage was characterized also for its mode of association with the target cells. During association with target cancer cells, phage particles can remain attached to the surface cellular receptors, or they can penetrate into the cell through endocytosis or other mechanisms28. To determine a mode of phage interaction with MCF-7 cells, the binding of phage to cells was studied under three different conditions: a) in serum free medium at room temperature, b) serum free medium at 37°C, and c) serum-containing medium at 37°C. To determine a localization of the phage in the cells we used different ways of recovery of the cell-associated phage: with acid buffer for elution of surface-bound phage, followed by post-elution washing with neutral buffer (pH shock-released phages) and finally – with CHAPS buffer for recovery of cell-integrated and penetrated phage particles. The distribution of phage particles in various cell fractions (acid eluate, post-elution washes PEW1 and PEW2, and lysate) were determined by titering of the phage in host bacteria (Figure 2). The dominant portion of the phage was found in the acid fractions under all used conditions, showing that the selected phage remains bound to the cell surface and does not penetrate into the cell during its 1 h incubation with cells. The recovery of the phage incubated with cells in serum free medium increased more then six times when temperature was increased from 20 to 37°C, and then further increased by 40% when the incubation medium was supplemented with serum. The increased binding of the phage to the cell at elevated temperature in the presence of growth factors of the serum can be explained by an active role of cellular metabolism in the binding of the phage. The control phage recovery was negligible in all fractions under the treatment conditions thereby validating the specificity of interaction of selected phage DMPGTVLP with MCF-7 cells.
Figure 2.
A. Selectivity of phage DMPGTVLP towards breast adenocarcinoma cells MCF-7 in comparison with breast cancer ductal carcinoma cells ZR-75-1, normal breast cells MCF-10A and hepatocellular carcinoma cells HepG2. Phage selectivity was estimated as a percentage phage recovery: output (cell-associated) phage to input phage. The unrelated phage bearing the peptide VPEGAFSS was used as a control. B. Mode of interaction of DMPGTVLP phage with cells MCF-7 under three different conditions (description in the text). The mode of interaction was estimated as a percentage of phage recovery calculated as a ratio of output (cell-associated) phage to input phage. rtp depicts room temperature; sf - serum free medium; s – medium with serum.
siRNA- and phage fusion protein-containing liposomes
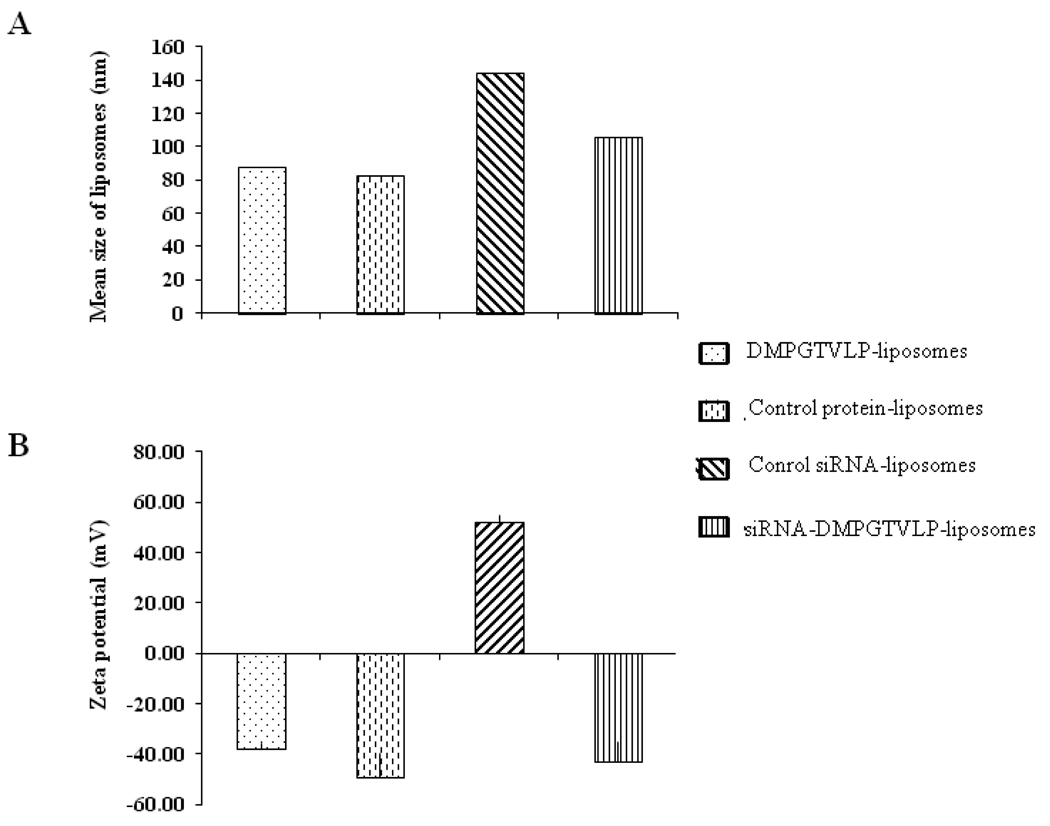
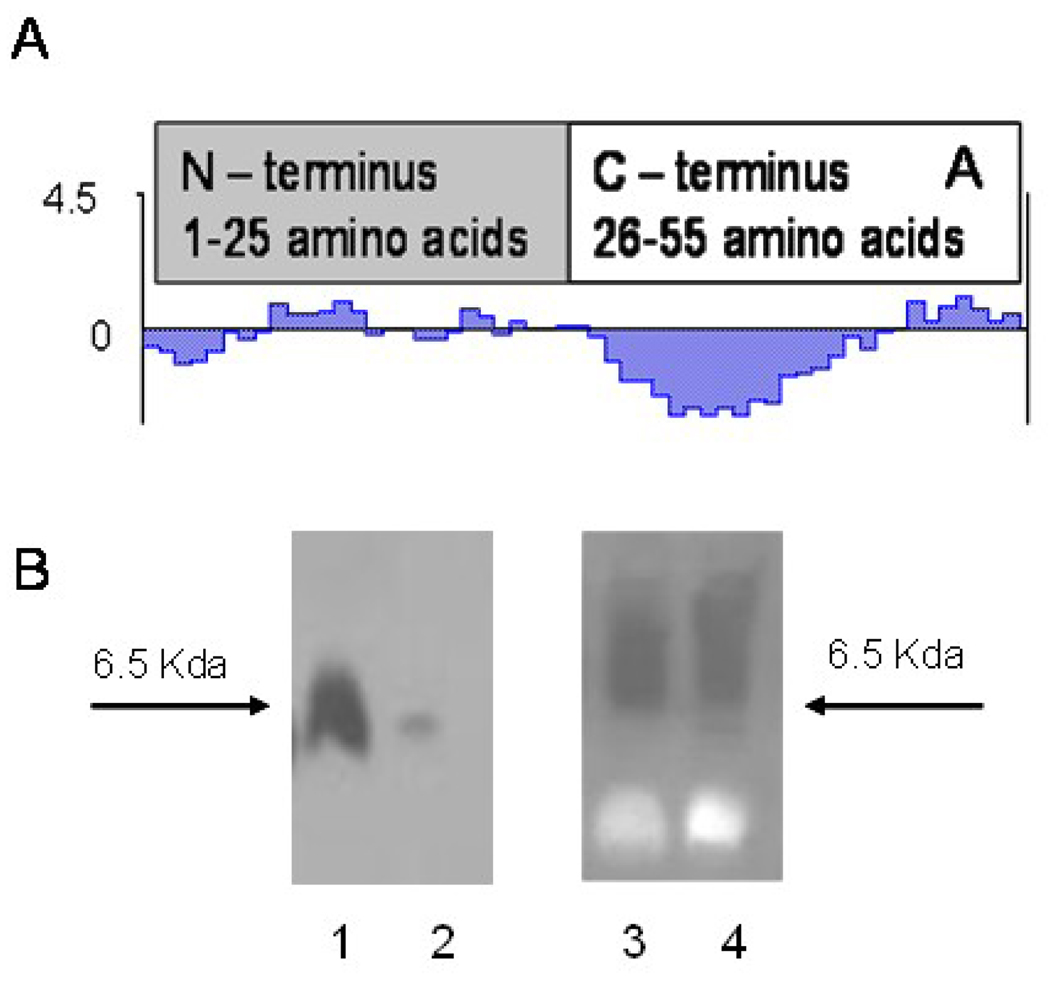
Previously, we developed a new approach to preparation of targeted liposomes that relies on the use of the phage fusion coat proteins as targeting ligands. In our approach, a cancer cell-specific phage protein is inserted into the liposome exploring its intrinsic —membranophilic properties11, 18, 19, 24. Fusion proteins carrying tumor-cell binding peptides inherit the major structural features of the wild-type major coat protein VIII (Figure 1). They have a positively charged Cterminus (amino acids 45–55), which navigates the protein through the liposome membrane, probably using the mechanisms intrinsic for cationic cell-penetrating peptides29. Highly hydrophobic —membranophilic segment (amino acids 27–40) allows the protein to accommodate readily in the membrane20, while amphiphilic N-terminus (amino acids 1–26), which is soluble in water, can interact with PEG residues on the surface of the —stealth liposomes and display the N-terminal cancer cell-binding octa- or nonamer on the liposome shell (Figure 1). Since spontaneous insertion of siRNA and fusion phage proteins into drug-loaded liposomes is driven by different mechanisms, we used three-step assembly of the targeted siRNA-nanomedicines. First, siRNAs was inserted into a liposome formed by a mixture of neutral and positively charged lipids (plus-liposome). This liposome has a positively charged interface that attracts the negatively charged siRNAs and drives their internalization30. Second, the fusion phage protein (DMPGTVLP) was inserted into a liposome formed by neutral and negatively charged lipids (minus-liposome). This liposome have a negatively charged interface that attracts C termini of the major coat protein, drives their translocation through the lipid bilayer and allows their anchoring at the internal liposomal interface, as was studied before20. Third, the plus liposomes loaded with siRNAs and minus-liposome loaded with phage protein were fused together to integrate into the protein-targeted particles containing siRNA. Liposome formulations were characterized by measuring size and size distribution and the surface charge (ζ). Protein-liposomes showed a mean size and surface charge comparable to their starting plain non-targeted formulation. The size and ζ of the plain formulation used to make siRNA-liposomes was also compared with the final siRNA-protein formulations. Although, protein-free siRNA-liposomes demonstrate a bigger size (144 nm) compared with protein-liposomes (87.7 nm, after the overnight incubation, fused formulations demonstrate size (105.9 nm) more close to that of initial protein-liposomes (Figure 3A). The surface charge of plain liposomes (−49.34 mV) is comparable to charge of protein-liposomes (−37.93 mV) suggesting that protein insertion did not affect the overall zeta potential of the formulation. The surface charge ζ of the fusion liposomes is comparable to the protein-liposomes (−42.8 mV and − 37.93 mV correspondingly) (Figure 3B) that may be due to the high level of shielding of siRNA in preparations. To check the amount of free siRNA in solution, fluorescent assay based on the interaction between the PicoGreen probe and siRNA was used. It was shown that the majority of the siRNA (90.5%) is encapsulated in the siRNA-protein–liposomes. The presence and topology of fusion coat protein pVIII in the final siRNA-protein liposomes was proved by western blot analysis. Proteinase K treatment of the liposomal preparation resulted in the dramatic decrease in signal intensity of N-terminus as revealed by anti-fd phage antibodies that bind specifically to N-terminus of major coat protein. On the other hand, proteinase K treatment did not change the intensity of C-terminus as probed by antibody specific for C-terminal region of major coat protein implying the orientation of Nout-Cin of the inserted peptide into the liposomes (Figure 4).
Figure 3.
Mean size (A) and Zeta potential (B) of liposome formulations. Liposomes modified with phage protein (DMPGTVLP-liposomes), liposomes without inserted phage protein (control protein-liposomes), liposomes without inserted siRNA (control siRNA-liposomes) and siRNA liposomes targeted with phage protein (siRNA-DMPGTVLP-liposome) are depicted in the picture.
Figure 4.
The presence and topology of phage fusion protein in the protein-modified liposomal preparations determined by Western blot analysis. A. Kyte and Dolittle hydrophillicity (hydropathicity) plot of fusion phage coat protein showing an amphiphilic N-terminus and an intensely hydrophobic segment of the C-terminus. B. Liposomal preparations were treated with proteinase K and then probed with antibodies specific for either N-terminus (Left) or C-terminus (Right) of the phage coat protein. Liposomal association does not provide protection from proteolytic degradation to the N-terminus region of the phage coat protein (Left) but the Cterminus is protected from degradation (Right). Lanes 1, 3 – liposomal preparations untreated with proteinase K, lane 2, 4 – liposomal preparation treated with proteinase K.
Gene PRDM14 silencing in breast cancer cells MCF-7 by protein-targeted siRNA-liposomes
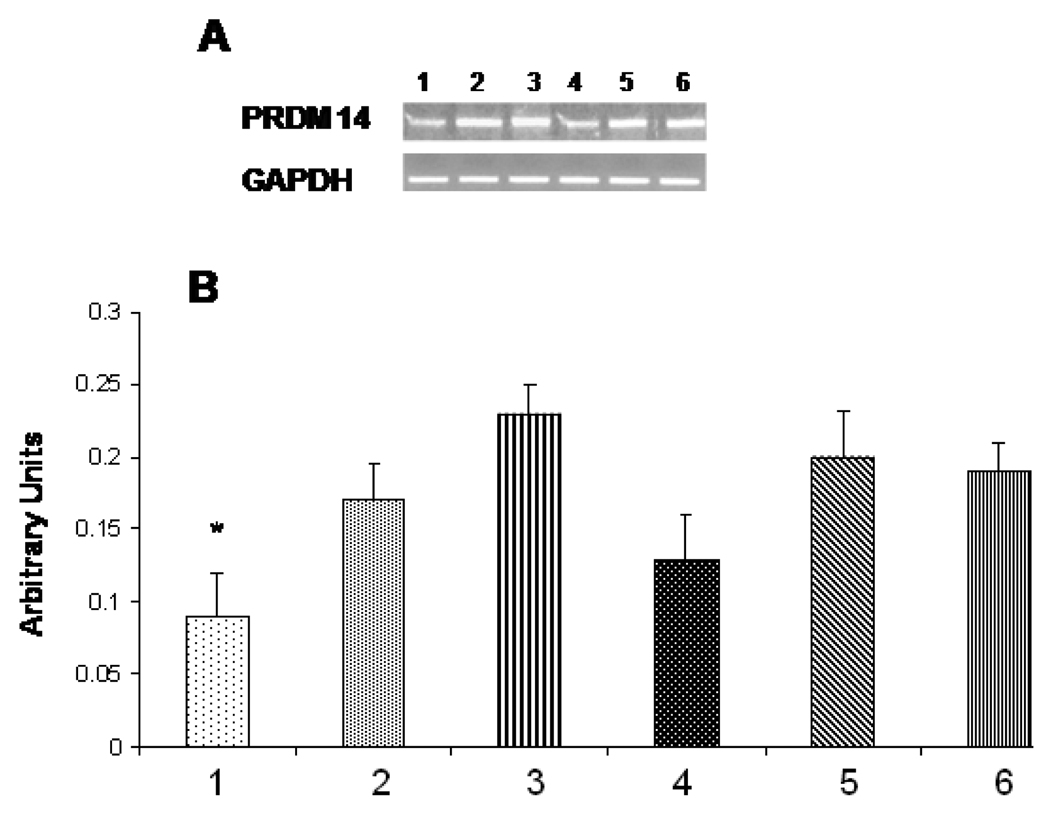
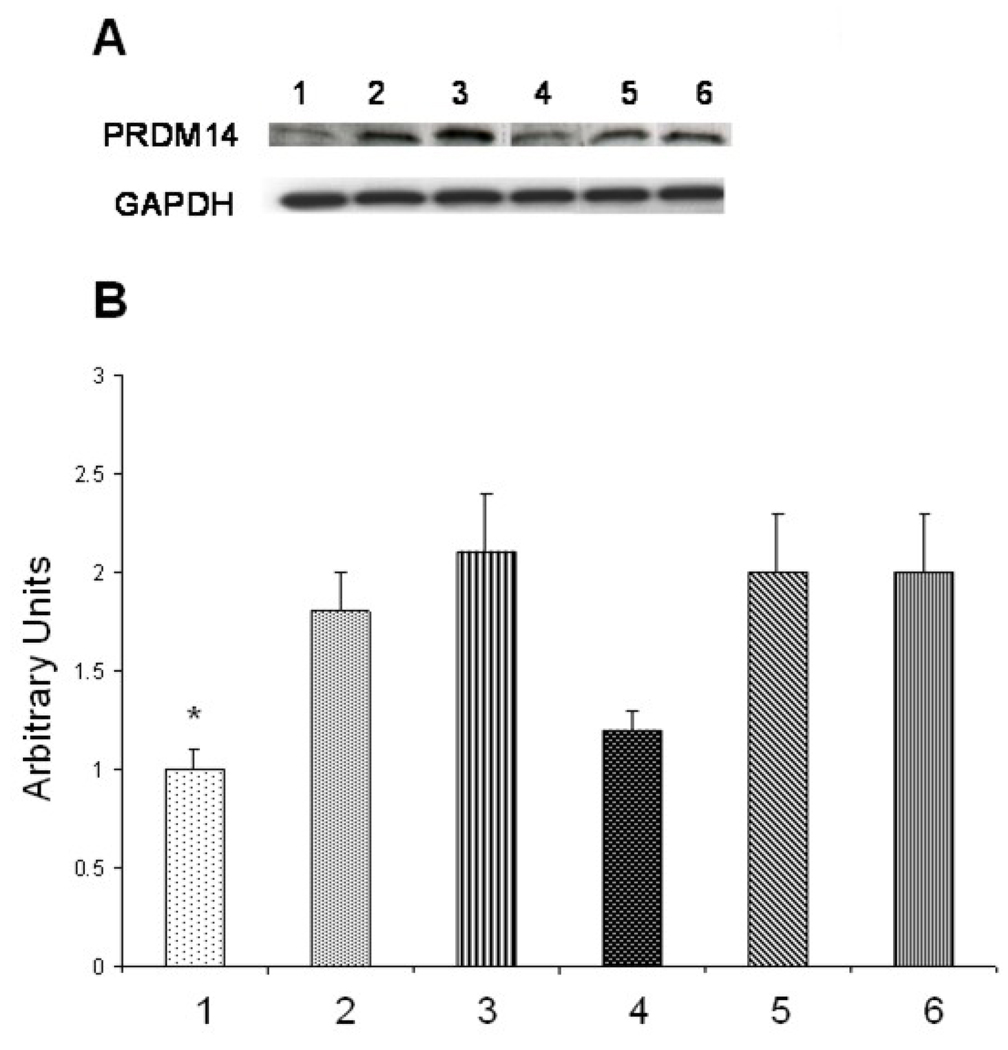
siRNA- protein-liposomes were tested for their ability to inhibit expression (silencing) of target PRDM14 gene and synthesis of its product—PRDM14 protein in breast cancer cells MCF-7. Cells were treated with siRNA–protein liposomes for 72 h, in parallel with control plane liposomes, siRNA-liposomes and phage protein-liposomes. Total RNA was isolated from treated cells and analyzed by RT-PCR with primers specific for PRDM14 gene. Relative expression of the gene was normalized against GAPDH gene. It was found that targeted siRNA- protein-liposomes down-regulated PRDM14 gene by 44% (p<0.01) close to the level of down-regulation of PRDM14 gene by siRNA-lipofectamine mix (46%, p<0.02) (Figure 5), while control liposomes had no effect on PRDM14 gene expression. Similarly, western blot analysis revealed that siRNA– DMPGTVLP phage protein liposomes down-regulate PRDM14 protein expression as compared to controls (Figure 6). These results prove the superiority of the selected phage protein and the targeting delivery of siRNA versus its non-targeting delivery.
Figure 5.
Analysis of PRDM14 gene transcription by RT-PCR. MCF-7 cells were treated with PRDM14 gene-specific protein-targeted siRNA–liposomes (40 nM) or protein-targeted scrambled siRNA-liposomes (40 nM) and incubated for 72 h. A. Relative transcription level of the target gene in cells treated with: 1. protein-targeted siRNA–liposomes, 2. protein-targeted scrambled siRNA-liposomes, 3. siRNA-liposomes, 4. siRNA-lipofectamine, 5. Scrambled siRNA-lipofectamine, 6. Control non-treated MCF-7 cells. B. The relative quantification was normalized against GAPDH using KODAK ID image analysis software. All data represent the mean ± S.D. * p<0.05, student-t-test.
Figure 6.
Analysis of PRDM14 protein expression by Western blot. MCF-7 cells were treated with PRDM14 gene-specific protein-targeted siRNA–liposomes (40 nM) or protein-targeted scrambled siRNA-liposomes (40 nM) and incubated for 72 h. A. Relative level of protein synthesis in cells treated with: 1. protein-targeted siRNA–liposomes, 2. protein-targeted scrambled siRNA-liposomes, 3. siRNA-liposomes, 4. siRNA-lipofectamine, 5. Scrambled siRNA-lipofectamine, 6. Control non-treated MCF-cells. B. Western blot band intensities quantified using Image J software (NIH). All data represent the mean ± S.D. * p<0.05, student-ttest.
Discussion
To enhance a potential anticancer efficiency of liposome-encapsulated siRNAs, we specifically targeted them via fusion with preselected phage protein specific for cancer cells MCF-7. To simplify the procedure and exclude any chemical conjugation reactions, we used spontaneous insertion of the phage proteins and siRNA into parental liposomes, followed by their fusion in mild conditions. The tumor-specific protein was isolated from the phage that was affinity selected from multibillion-clone landscape phage library f8/8 by their ability to bind very specifically breast cancer cells. The phage DMPGTVLP (designated by the structure of the borne foreign peptide) demonstrates high selectivity and specificity towards target cells versus control unrelated cells. The major coat protein of the selected phage was converted into the drug-loaded liposomal vesicles in which the phage spans the lipid bilayer displaying the tumor-binding peptides on the surface of the vesicles. This topology of the fusion protein in modified liposomes was confirmed by protease digestion experiments. Treatment of the siRNA-phage fusion protein liposomes with proteinase K resulted in complete loss of N-terminus signal while C-terminus was not destroyed by the enzyme, demonstrating that C-terminus translocated inside the liposomal membrane. The protein-liposomes efficiently shielded PRDM14 gene-targeted siRNA, as was shown by PicoGreen fluorescent assay. The size and size distribution of protein-targeted siRNA-liposomes did not change in 10% serum during a week demonstrating their stability. The novel siRNA-nanopharmaceuticals targeted to the breast cancer cells MCF-7 via their association with phage fusion proteins down-regulate PRDM14 gene expression and inhibite PRDM14 protein synthesis in MCF-7 cells as effectively as lipofectamine-siRNA complex considered as a —gold standard for delivery of siRNAs into cells in vitro. siRNAliposomes alone showed no effect on PRDM14 gene expression implying the specific celltargeting role of the fusion proteins through its anchoring to the cells and mediating delivery of siRNA-liposomes into the cells.
The new liposome targeting approach based on the use of landscape phage relies on powerful and precise mechanisms of selection, biosynthesis and self assembly of nanostructures. A culture of cells secreting filamentous phage is an efficient and convenient protein production system yielding up to 300 mg/liter of pure phage, with the major coat protein constituting 98% of the total protein mass. It can be easily isolated in a pure form using one-step chromatography. Phage itself and its coat proteins are not toxic and have been already tested for safety in preclinical trials31. Furthermore, the technique of polyvalent phage display in the major coat protein pVIII for construction of large (>109-clones) 8- and 9-mer landscape libraries has been developed22, 32. Hundreds of targeted phage probes against prostate, glial and breast tumor cells were successfully selected from these libraries using advanced biopanning protocols23. These and other current advances in siRNA tumor-targeted delivery can offer a new means for their intensive preclinical study as potential anticancer medicines.
Acknowledgements
This work was supported by the NIH grant# 1 R01 CA125063-01 and Calvert Research Institute LLC grants to Valery A. Petrenko. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of National Cancer Institute of NIH and Calvert Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There are no disclosures or any conflicts of interest with regard to this publication.
References
- 1.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 2.Kim SS, Garg H, Joshi A, Manjunath N. Strategies for targeted nonviral delivery of siRNAs in vivo. Trends in Molecular Medicine. 2009;15:491–500. doi: 10.1016/j.molmed.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng X, Vladau C, Zhang X, Suzuki M, Ichim TE, Zhang ZX, et al. A novel in vivo siRNA delivery system specifically targeting dendritic cells and silencing CD40 genes for immunomodulation. Blood. 2009;113:2646–2654. doi: 10.1182/blood-2008-04-151191. [DOI] [PubMed] [Google Scholar]
- 4.Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. doi: 10.1158/0008-5472.CAN-07-5810. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Tang N, Zhang CL, Liu XJ, Hu H, Zhang ZX, et al. Cell penetrating peptides enhance intracellular translocation and function of siRNA encapsulated in Pegylated liposomes. Yao Xue Xue Bao. 2006;41:142–148. [PubMed] [Google Scholar]
- 6.Li SD, Chono S, Huang L. Efficient Oncogene Silencing and Metastasis Inhibition via Systemic Delivery of siRNA. Mol Ther. 2008;16:942–946. doi: 10.1038/mt.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, et al. Tumor targeting with a selective gelatinase inhibitor. Nat Biotech. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 8.Lee TY, Wu HC, Tseng YL, Lin CT. A Novel Peptide Specifically Binding to Nasopharyngeal Carcinoma For Targeted Drug Delivery. Cancer Res. 2004;64:8002–8008. doi: 10.1158/0008-5472.CAN-04-1948. [DOI] [PubMed] [Google Scholar]
- 9.Medina OP, Soderlund T, Laakkonen LJ, Tuominen EKJ, Koivunen E, Kinnunen PKJ. Binding of Novel Peptide Inhibitors of Type IV Collagenases to Phospholipid Membranes and Use in Liposome Targeting to Tumor Cells in Vitro. Cancer Res. 2001;61:3978–3985. [PubMed] [Google Scholar]
- 10.Pastorino F, Brignole C, Di Paolo D, Nico B, Pezzolo A, Marimpietri D, et al. Targeting Liposomal Chemotherapy via Both Tumor Cell-Specific and Tumor Vasculature-Specific Ligands Potentiates Therapeutic Efficacy. Cancer Res. 2006;66:10073–10082. doi: 10.1158/0008-5472.CAN-06-2117. [DOI] [PubMed] [Google Scholar]
- 11.Petrenko V. Evolution of phage display: from bioactive peptides to bioselective nanomaterials. Expert Opin Drug Deliv. 2008;5:825–836. doi: 10.1517/17425247.5.8.825. [DOI] [PubMed] [Google Scholar]
- 12.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 13.Krumpe LR, Mori T. The Use of Phage-Displayed Peptide Libraries to Develop Tumor-Targeting Drugs. Int J Pept Res Ther. 2006;12:79–91. doi: 10.1007/s10989-005-9002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 16.Chang DK, Lin CT, Wu CH, Wu HC. A Novel Peptide Enhances Therapeutic Efficacy of Liposomal Anti-Cancer Drugs in Mice Models of Human Lung Cancer. PLoS ONE. 2009;4:e4171. doi: 10.1371/journal.pone.0004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilyichev AA, Minenkova OO, Kishchenko GP, Tat'kov SI, Karpishev NN, Eroshkin AM, et al. Inserting foreign peptides into the major coat protein of bacteriophage M13. FEBS Lett. 1992;301:322–324. doi: 10.1016/0014-5793(92)80267-k. [DOI] [PubMed] [Google Scholar]
- 18.Jayanna PK, Bedi D, Gillespie JW, Deinnocentes P, Wang T, Torchilin VP et al. Landscape phage fusion protein-mediated targeting of nanomedicines enhances their prostate tumor cell association and cytotoxic efficiency. Nanomedicine. 2010;6:538–546. doi: 10.1016/j.nano.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayanna PK, Torchilin VP, Petrenko VA. Liposomes targeted by fusion phage proteins. Nanomedicine. 2009;5:83–89. doi: 10.1016/j.nano.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soekarjo M, Eisenhawer M, Kuhn A, Vogel H. Thermodynamics of the membrane insertion process of the M13 procoat protein, a lipid bilayer traversing protein containing a leader sequence. Biochemistry. 1996;35:1232–1241. doi: 10.1021/bi951087h. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa N, Toyota M, Suzuki H, Honma T, Fujikane T, Ohmura T, et al. Gene Amplification and Overexpression of PRDM14 in Breast Cancers. Cancer Res. 2007;67:9649–9657. doi: 10.1158/0008-5472.CAN-06-4111. [DOI] [PubMed] [Google Scholar]
- 22.Petrenko VA, Smith GP, Gong X, Quinn T. A library of organic landscapes on filamentous phage. Protein Eng. 1996;9:797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]
- 23.Brigati JR, Samoylova TI, Jayanna PK, Petrenko VA. Phage display for generating peptide reagents. Curr Protoc Protein Sci. 2008;Chapter 18 doi: 10.1002/0471140864.ps1809s51. [DOI] [PubMed] [Google Scholar]
- 24.Wang T, D'Souza GG, Bedi D, Fagbohun OA, Potturi LP, Papahadjopoulos-Sternberg B, et al. Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein. Nanomedicine (Lond) 2010;5:563–574. doi: 10.2217/nnm.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrenko VA, Smith GP. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000;13:589–592. doi: 10.1093/protein/13.8.589. [DOI] [PubMed] [Google Scholar]
- 26.Pappalardo JS, Quattrocchi V, Langellotti C, Di GS, Gnazzo V, Olivera V, et al. Improved transfection of spleen-derived antigen-presenting cells in culture using TATp-liposomes. J Control Release. 2009;134:41–46. doi: 10.1016/j.jconrel.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Sawant RM, Hurley JP, Salmaso S, Kale A, Tolcheva E, Levchenko TS, et al. "SMART" drug delivery systems: double-targeted pH-responsive pharmaceutical nanocarriers. Bioconjug Chem. 2006;17:943–949. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poul MA, Marks JD. Targeted gene delivery to mammalian cells by filamentous bacteriophage. Journal of Molecular Biology. 1999;288:203–211. doi: 10.1006/jmbi.1999.2678. [DOI] [PubMed] [Google Scholar]
- 29.Tseng YL, Liu JJ, Hong RL. Translocation of liposomes into cancer cells by cellpenetrating peptides penetratin and tat: a kinetic and efficacy study. Mol Pharmacol. 2002;62:864–872. doi: 10.1124/mol.62.4.864. [DOI] [PubMed] [Google Scholar]
- 30.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, et al. Selection of Tumorbinding Ligands in Cancer Patients with Phage Display Libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 32.Kuzmicheva GA, Jayanna PK, Sorokulova IB, Petrenko VA. Diversity and censoring of landscape phage libraries. Protein Engineering, Design and Selection. 2009;22:9–18. doi: 10.1093/protein/gzn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T, Yang S, Petrenko VA, Torchilin VP. Cytoplasmic Delivery of Liposomes into MCF-7 Breast Cancer Cells Mediated by Cell-Specific Phage Fusion Coat Protein. Mol Pharm. 2010;7:1149–1158. doi: 10.1021/mp1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayanna PK, Bedi D, Deinnocentes P, Bird RC, Petrenko VA. Landscape phage ligands for PC3 prostate carcinoma cells. Protein Engineering, Design and Selection. 2010;23:423–430. doi: 10.1093/protein/gzq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, Petrenko VA, Torchilin VP. Paclitaxel-loaded polymeric micelles modified with MCF-7 cell-specific phage protein: enhanced binding to target cancer cells and increased cytotoxicity. Mol Pharm. 2010;7:1007–1014. doi: 10.1021/mp1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]