Abstract
Autophagy and endocytosis are dynamic and tightly regulated processes that contribute to many fundamental aspects of biology including survival, longevity and development. However, the molecular links between autophagy and endocytosis are not well understood. Here, we report that BEC-1, the C. elegans ortholog of Atg6/Vps30/Beclin 1, a key regulator of the autophagic machinery, also contributes to endosome function. In particular we identified a defect in retrograde transport from endosomes to the Golgi in bec-1 mutants. MIG-14/Wntless is normally recycled from endosomes to the Golgi through the action of the retromer complex and its associated factor RME-8. Lack of retromer or RME-8 activity results in the aberrant transport of MIG-14/Wntless to the lysosome where it is degraded. Similarly, we found that lack of bec-1 also results in mislocalization and degradation of MIG-14::GFP, reduced levels of RME-8 on endosomal membranes, and the accumulation of morphologically abnormal endosomes. A similar phenotype was observed in animals treated with dsRNA against vps-34. We further identified a requirement for BEC-1 in the clearance of apoptotic corpses in the hermaphrodite gonad, suggesting a role for BEC-1 in phagosome maturation, a process that appears to depend upon retrograde transport. In addition, autophagy genes may also be required for cell corpse clearance, as we found that RNAi against atg-18 or unc-51 also results in a lack of cell corpse clearance.
Key words: C. elegans, autophagy, endocytosis, lysosomes
Introduction
Macroautophagy (hereafter autophagy) is a cellular bulk degradation process conserved in all eukaryotes.1,2 In autophagy, a portion of the cytoplasm is sequestered by a double-membrane organelle called the autophagosome, which then fuses with the lysosome to degrade the materials inside. Endocytosis is the vesicle-mediated process to internalize plasma membrane, and/or macromolecules from the outside environment. The endosomal system sorts the internalized molecules to their proper intracellular destination. In yeast, the vacuole is functionally equivalent to the lysosome in higher eukaryotes. In the yeast Saccharomyces cerevisiae, mutations in vacuolar protein sorting (VPS) genes result in secretion of proteins normally localized to the vacuole. Characterization of the VPS genes has provided insight into the mechanism of protein sorting and vesicle-mediated intracellular transport.
Genetic studies in the yeast S. cerevisiae have identified many genes that function in autophagy (ATG).3,4 Many of the ATG genes encode proteins that are required for the early formation of the autophagosome, and are thus localized to a perivacuolar structure called the preautophagosomal structure (PAS).5,6 Additional regulators of autophagy have been identified in screens for mutants in the cytoplasm-to-vacuole (Cvt) pathway.7,8 The Cvt pathway delivers vesicle-bound proteins to the vacuole from the cytoplasm. However, unlike autophagy which is degradative and mainly nonselective, the Cvt pathway is biosynthetic and selects specific cargo from the cytoplasm to be delivered to the vacuole.9
Yeast Atg6/Vps30 and its orthologs in multicellular organisms play a pivotal role in autophagy. The mammalian ortholog of Atg6/Vps30 is Beclin 1. In addition to regulating autophagy, Beclin 1 has been shown to be a tumor suppressor protein and the antiapoptotic protein Bcl-2 negatively regulates autophagy through binding with Beclin 1.10–14 C. elegans affords another genetic system for studying the role of Beclin 1. The C. elegans Beclin 1 ortholog bec-1 mediates autophagy and is essential for morphogenesis of the dauer larva and for life-span extension mediated by insulin signaling15 or dietary restriction.16–18 We have studied the role of C. elegans BEC-1 in autophagy and endocytosis.
In yeast, distinct roles of Atg6/Vps30 protein in autophagy and vacuolar protein sorting reflect two related but functionally different phosphatidylinositol-3-kinase (PI3K) complexes: the type I complex functions in autophagy and the type II complex functions in vacuolar protein sorting, respectively.19 The difference between the complexes is that complex I contains Atg14, whereas complex II contains Vps38, instead of Atg14. Vps38 is present on endosome and vacuolar membranes, and it is required for the targeting of Atg6/Vps30 and Vps34 to the endosome.20 In contrast, Atg14 localizes to the PAS and vacuolar membrane, and generates the autophagosome by recruiting Vps34 and Atg6/Vps30 to the PAS.20
Vps34, which corresponds to the mammalian class III PI3-kinase, is important for various membrane trafficking pathways such as the Golgi to lysosome pathway, internal vesicle formation in late endosomes and autophagy.21–26 Although it has been well established that the mammalian Beclin 1/Vps34 complex is required for autophagy, its involvement in other membrane trafficking pathways has been controversial in higher eukaryotes. Expression of C. elegans BEC-1 or human Beclin 1 in atg6-depleted yeast restores autophagy but not vacuolar protein sorting function in yeast.27 Although Beclin 1 and hVps34 were shown to co-localize in the trans-Golgi network (TGN) in HeLa cells, the role of Beclin 1 in vesicular trafficking in mammalian cells may be limited. RNA interference (RNAi) mediated suppression of beclin 1 impairs autophagy but does not affect fluid-phase endocytosis, endocytic sorting of the epidermal growth factor receptor (EGFR) or cathepsin D transport from TGN to lysosomes.28 In addition, the overexpression of a mutant version of human Beclin 1 that cannot bind to human Vps34 does not inhibit the ability to process cathepsin D, but significantly reduced autophagy activity and tumor suppressor functions in MCF7 cells.29 In contrast, expression of plant Atg6 in yeast atg6 mutants restores both autophagy and vacuolar protein sorting function.30 Furthermore, C. elegans BEC-1 has been shown to bind to VPS-34 and affect PI3P localization and general uptake of an endocytic marker.31 Biological functions of atg6/beclin 1 in higher eukaryotes suggest a role outside of autophagy. Arabidopsis atg6 mutants exhibit defects in autophagy and pollen germination, but the latter was not observed in other autophagy mutants, suggesting that Atg6 confers some autophagy-independent function.30 In mice, Beclin 1 knockout animals die at approximately embryonic day 7.5,13 whereas Atg5 and Atg7 knockout mice can survive until birth,32,33 thus Beclin 1 might have a more complex function in mice. Recently, possible orthologs of Atg14 and Vps38 have been identified in mammals,34,35 and shown to form distinct PI3-kinase complexes with different intracellular localization, suggesting that Vps34-Beclin 1 complexes may have different functions in metazoans similar to that in yeast cells.
Although C. elegans bec-1 had been previously implicated in endocytic trafficking,31 the nature of the contribution had not been described. Here, we showed that bec-1 mutant animals display defects in recycling of cargo molecules from the endosome to the late-Golgi. We observed that maternally rescued bec-1 mutants accumulate large vacuoles of mixed endosomal and lysosomal identity. Furthermore, we showed that bec-1 functions in endosome to Golgi retrograde transport. In particular, bec-1 mutant animals display defects in the recycling of MIG-14/Wntless cargo protein from the endosome to the trans-Golgi network. Recycling of the MIG-14/Wntless cargo is mediated by the retromer complex, a conserved cytoplasmic coat recycling complex that mediates the endosome-to-Golgi retrieval of vacuole/lysosome hydrolase receptors in yeast and mammals.36,37 We further showed that RME-8, a subunit of the retromer complex and BEC-1 colocalize in endosomes that are found adjacent to lysosomes in wild-type animals, and that RME-8 is redistributed to the cytoplasm in bec-1 mutants, indicating a role for bec-1 in retrograde transport. Finally, we report a lack of cell corpse clearance as observed by electron microscopy and an increased number of CED-1::GFP positive cells in bec-1 mutant gonads, similar to the phenotype in animals lacking the retromer subunit rme-8.38
Results
bec-1 has maternal and zygotic activity.
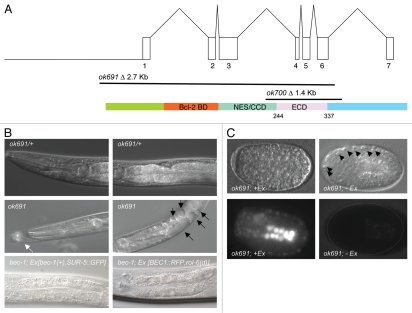
The C. elegans Gene Knockout Consortium generated two deletion mutations in bec-1, ok691 and ok700. The ok691 allele deletes the first six out of seven exons, including the ATG start codon (Fig. 1) and is thus a predicted molecular null allele.31 The ok700 allele deletes amino acids 168 to 314 of the “evolutionarily conserved domain” (ECD) in BEC-1 and also causes a frameshift that removes the remainder of the protein (Materials and Methods); this allele appears to be a strong hypomorph.
Figure 1.
bec-1 gene, protein, mutations and phenotypes. (A) At the top, the genomic structure of the bec-1 gene encompassing 7 exons, and encoding a 375 amino acid protein, shown below. In the middle, two deletions of bec-1 as provided by the C. elegans Gene Knockout Consortium. ok691 deletes most of the bec-1 open reading frame including the start ATG and exons 1–6, thus it is a molecular null. ok700 deletes exons 4–6 of bec-1 and renders the mRNA out of frame for exon 7. A cDNA was sequenced after RNA was extracted from an ok700 lysate, which shows that there is bec-1 mRNA transcribed in ok700 mutants that lacks the evolutionarily conserved domain of BEC-1 required for binding VPS-34. (B) Both deletions display an adult lethal phenotype with a striking accumulation of vacuoles (marked by black arrows in (B), middle, right panel). ok691 mutants (B, middle panel) segregating from a heterozygous parent (B, top panel). bec-1 homozygous mutant, either ok691 or ok700, appear superficially wild type during early larval stages but become increasingly uncoordinated during later larval stages and display molting problems (white arrow in (B), middle, left panel). Rescue of the bec-1 phenotypes including uncoordination, the accumulation of vacuoles and lethality was observed with a transgene containing the bec-1 wild-type genomic sequences and a ubiquitously expressed SUR-5::GFP marker (B, bottom, left panel) or a BEC-1::RFP construct (B, bottom, right panel). (C) Animals that carry the rescuing bec-1 genomic extrachromosomal array (Ex[bec-1(+), SUR-5::GFP]) (left bottom panel) are rescued for the bec-1 phenotype. Animals that have lost the array early in development are completely devoid of the GFP marker expression (C, right bottom panel) and lack the maternal and zygotic bec-1 expression. A hatched line delineates the periphery of the embryo (C, right bottom panel). These animals die during embryogenesis with an increase in the number of visible apoptotic cells (marked by black arrow heads, (C) top right panel).
Both ok691 and ok700 mutations were reported to display a highly penetrant embryonic lethal phenotype when segregating from a balanced heterozygote.31 In contrast, using a balancer marked with unc-5(e53) jcIs[ajm-1:gfp], which enables us to unambiguously identify bec-1 mutant animals during both embryonic and adult stages (see Materials and Methods), we found that ok691 and ok700 homozygous individuals derived from a bec-1(mutant)/unc-5(e53) jcIs1 heterozygous parent can reach adulthood (Table 1). However, these adults are sterile and die after the first day of adulthood. These results confirm that bec-1 activity is required for viability (Table 1). Further analysis suggests that bec-1 mutants segregating from heterozygous mothers reach adulthood because of maternal rescue.
Table 1.
Lack of zygotic bec-1 results in adult lethality
| Eggs | hatched | Reach adulthood | |
| bec-1(ok691)/unc-5; jcIs1 | 325 | 97.8% | 20.3% unc-5 homozygous |
| 53.1% heterozygous | |||
| 26.6% bec-1 homozygous | |||
| bec-1(ok700)/unc-5; jcIs1 | 270 | 94% | 24.6% unc-5 homozygous |
| 50% heterozygous | |||
| 25.4% bec-1 homozygous | |||
| N2 | 195 | 97% | 100% |
Both bec-1(ok691) and bec-1(ok700) homozygous mutants that segregate from the bec-1(mutant)/unc-5(e53) jcIs1 heterozygous parent appear superficially wild type during early larval stages but become increasingly uncoordinated during later larval stages and display a low penetrance molting defect. In addition, mutants accumulate vacuoles that become first visible during the L3 stage [Table 2: 70% of ok691 (n = 37), and 30% of ok700 (n = 40) contain at least one vacuole, Fig. 1B, middle panel]. These vacuoles are present in the intestine, hypodermis, pharynx and coelomocytes. Both the number and size of the vacuoles increases with age in larvae (Table 2). By L4 stage, 80% of the ok691 animals (n = 39), and 52% of ok700 (n = 40) contain at least one vacuole, and approximately 42% of ok691 mutants (n = 39), and 5% of ok700 mutants (n = 40) contain more than 10 vacuoles (Table 2). Lastly, both ok691 and ok700 mutant animals display growth retardation and reach the adult stage up to 1.5 days later than wild-type animals.
Table 2.
Lack of maternal and zygotic bec-1 leads to embryonic lethality
| Aberrant vacuole phenotype | ||
| % of Animals that contain at least one vacuole | ||
| L3 | L4 | |
| ok691 | 70% (n = 37) | 80% (n = 39) n.s. |
| ok700 | 30% (n = 40) | 52% (n = 40) n.s. |
| % of Animals that contain more than 10 vacuoles | ||
| L3 | L4 | |
| ok691 | 13% (n = 37) | 42% (n = 39)* |
| ok700 | 0 (n = 40) | 5% (n = 40) n.s. |
To confirm that the observed phenotypes are due to a mutation in bec-1, we injected a genomic sequence containing wild-type sequences of bec-1 into ok691 mutant animals (Ex [bec-1(+), SUR-5::GFP]) and obtained rescue of both the developmental defects and vacuolar phenotypes (Table 2 and Fig. 1B, bottom panel). Both the ok691 and ok700 bec-1 deletion alleles are likewise rescued by a functional BEC-1::RFP fusion construct (kindly provided by B. Bamber39) (Table 3). Interestingly, a significant number of transgenic embryos failed to hatch (Table 3). Only 15.1% of the embryos carrying the Ex[BEC-1::RFP, rol-6(d)] transgene and 38.6% of the embryos carrying the Ex [bec-1(+), SUR-5::GFP] transgene hatch. Since multicopy transgenic arrays are not efficiently expressed in the hermaphrodite germline,40,41 this observation suggested that bec-1 is maternally contributed.
Table 3.
bec-1 mutants accumulate aberrant vacuoles
| ok691; Ex array | +GFP Hatched | −GFP Hatched | Adults | n = eggs counted |
| Ex[BEC-1::RFP, pRF4] | 15.1% | 0 | 100% | n = 651 |
| Ex[bec-1(+), pTG96] | 38.6% | 0 | 92% | n = 185 |
To directly test this possibility, we analyzed the phenotype of bec-1 mutant embryos that originated from fertile homozygous ok691 mutant mothers carrying the rescuing transgene, Ex [bec-1(+), SUR-5::GFP]. The lack of the transgene in embryos can be visualized by the lack of GFP (Fig. 1C and right bottom panel). These embryos likely lack both maternal and zygotic expression of bec-1. We found that maternal and zygotic loss of bec-1 results in complete early embryonic lethality. As these embryos fail to hatch, we inferred that they were not rescued maternally. These embryos display classic morphogenesis defects with unattached cells floating outside the embryo (Fig. 1C and right top panel) and visible apoptotic cell corpses.31
bec-1 mutation disrupts the localization of PI3P.
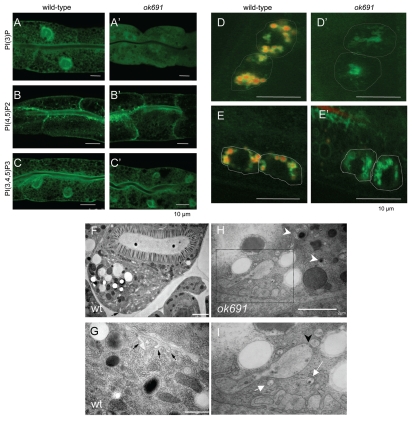
We analyzed the phenotype of bec-1(ok691), an unambiguous molecular null allele in detail. To understand the relationship of bec-1 and vps-34, and to test whether there is any dysregulation of phosphoinositides in bec-1 mutants, we monitored the expression and localization of PI3P-, PI(4,5)P2- and PI(3,4,5)P3-binding reporters42 in wild-type and bec-1(ok691) mutant animals. vps-34/let-512 encodes the class III phosphoinositide 3-kinase, a protein that regulates multiple steps in endocytosis.43 Since BEC-1 associates with VPS-34,31 we expected that the localization of PI3P might be affected in the bec-1(ok691) null mutant. PI3P was detected with a transgene encoding a tandem repeat of human Hrs FYVE domain, 2XFYVE::GFP. As shown previously,31 lack of bec-1 activity disrupts the localization of PI3P. In bec-1(ok691) mutants we observed a more diffuse localization of the 2XFYVE::GFP reporter (Fig. 2A') as compared to the same marker in control animals (Fig. 2A). The membrane of the enlarged vacuoles in mutant intestines was not labeled with the PI3P marker. Localization of PI(4,5)P2 was determined with a reporter that expresses the PH domain of phospholipase C delta. PI(4,5)P2 in the worm intestine is normally enriched apically with a less pronounced localization on basolateral membranes (Fig. 2B). The PI(3,4,5)P3 reporter consisted of the PH-domain of Akt fused to GFP, and was localized apically and basolaterally in the intestine (Fig. 2C). In ok691 mutants, the expression of the PI(4,5)P2 and PI(3,4,5)P3 reporters was largely unaffected when compared to that of wild-type animals (Fig. 2B' and C').
Figure 2.
bec-1 mutants display defects in endocytosis. The detection of the phosphoinositides PI3P, PIP2 and PIP3 in the intestines of wild-type (A–C), bec-1(ok691) mutants (A'–C'), animals is shown. Scale bars indicate 10 µm. In wild-type animals (A), PI3P is enriched in the apical membrane with weak basolateral labeling. In bec-1(ok691) (A') mutants very low and diffused expression of PI3P was observed. PIP2 expression is not affected in bec-1(ok691) mutants (B') when compared to wild-type animals (B). The PIP3 reporter labels apical membrane and strong basolateral expression in wild-type animals (C). The localization of PIP3 in bec-1(ok691) animals (C') was similar to that of wild-type animals (C). Micrographs of LMP-1::GFP positive lysosomes in coelomocytes 24 h after the injection of Texas Red BSA (a fluid phase marker for endocytosis) into the body cavity (pseudocoelom). A hatched line demarcates the periphery of the coeolomocytes. The red dye accumulates in lysosomes (LMP-1 positive) localized in the coelomocytes of the wild-type animals (D), but fails to accumulate in the lysosomes (LMP-1 positive) of bec-1(ok691) mutants (D'). Micrographs of LMP-1::GFP positive lysosomes in coelomocytes after 1 h of soaking in Texas Red BSA (a fluid phase marker for endocytosis). The red dye accumulates in lysosomes (green) localized in the coelomocytes of the wild-type animals (E), but fails to accumulate in the lysosomes (green) of bec-1 mutants (E'). Electron microscopy of an N2 wild-type (F and G) and bec-1(ok691) mutant young adults (H and I). (F) Low power electron micrograph of wild-type adult intestine in cross-section, fixed by high pressure freezing. Most of the basal membrane shows few signs of endocytosis, although there is some membrane infolding (black arrow in F) and a few small vesicles, particularly near the lateral membrane border between two intestinal cells on the extreme left edge of the tissue. Higher power view of adult wild-type intestine shows a few examples of membrane infolding (black arrows in G) representing a local zone where endocytosis is active. It is notable that the infolded membranes are rather large, and there are almost no vesicles nearby in the cytoplasm. In contrast, chains of vesicles (white arrows in I) and extensive infolded membranes are seen along the basal pole in the bec-1 mutant (H and I). Thus endocytotic events must be cleared relatively quickly in the wild-type intestine. In bec-1 mutant intestines, defective enlarged endosomes (marked by white arrow head in H) are seen that correspond to abnormal vacuoles of different sizes and contain either membranes or condensed material. (I) is a close up of the region marked in (H) and it shows enlarged vacuoles and a high number of early endosomes that appear to be fusing in some cases (black arrowhead in I). This phenotype is never seen in the intestines of wild-type animals (F or G).
bec-1 mutants have defects in endocytosis.
The endocytic nature of the vacuoles and defects in PI3P localization observed in bec-1 mutant animals prompted us to directly test whether bec-1 is required for endocytic trafficking in C. elegans. We concentrated our analysis on coelomocytes, which are large scavenger cells, situated in the pseudocoelomic space that continuously endocytose fluid from the pseudocoelom.44 To define more precisely the defect in bec-1 endocytosis, we performed fluid uptake assays by injection of Texas Red-coupled BSA (TR-BSA) into the body cavity of wild-type and bec-1 mutant individuals. We used LMP-1::GFP to visualize lysosomes and late endosomes. LMP-1 is similar to the vertebrate lysosome-associated membrane protein LAMP/CD68.45 LMP-1::GFP has been widely used as lysosomal marker.46,47 Injection of TR-BSA into the pseudocoelom (body cavity) can be monitored for its uptake, which is mediated by endocytosis at the basolateral cell membrane bordering the pseudocoelom.48 In wild-type animals, the fluorescent marker enters the coelomocytes by fluid phase mediated endocytosis and is transported through the different endocytic compartments to the lysosomes.46,49 After injection of wild-type animals, the fluid-phase marker is present in early endocytic compartments. After 15 min, it begins to accumulate in endosomes (LMP-1::GFP negative), and by 1 h it accumulates in lysosomes (LMP-1::GFP positive in Fig. 2d).46,50,51 In bec-1 mutants, the endocytosis of the fluid phase marker was found to be abnormal (Fig. 2D'). Texas Red BSA does not localize to LMP-1 positive endosomes even after 24 hours. After 1 h of soaking the animals with fluid markers (or allowing them to feed on it), the Texas Red BSA marker is clearly endocytosed into wild-type coelomocyte lysosomes (LMP-positive in Fig. 2D), but not into the bec-1(ok691) mutant coelomocytes (Fig. 2D'). These results indicate a role for BEC-1 in an early step of endocytosis.
The vacuoles in bec-1(ok691) mutants are of endocytic origin.
To characterize the abnormal vacuoles in bec-1 mutant animals and their origin, we used a combination of fluorescent and electron microscopy. In electron micrographs, the vacuoles are clearly surrounded by a membrane and, are either empty, filled with membrane whorls or electron dense deposits, features typical of late endosomes and lysosomes (Fig. 2H and I). We also often observed aggregation of endocytic structures (Fig. 2I, black arrow head).
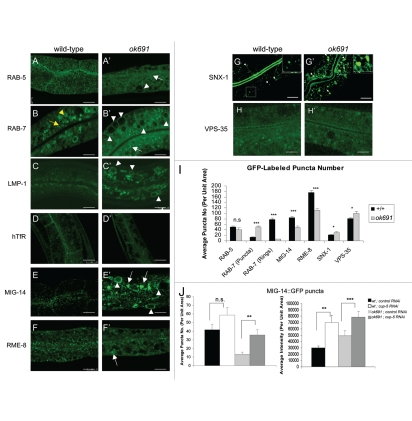
To better define the origin of the vacuoles, we crossed ok691 mutants with transgenic animals expressing markers for a variety of specific endocytic compartments. Wild-type intestines contain a large number of small GFP::RAB-5 positive punctate structures (Fig. 3A) near the basolateral and apical plasma membrane corresponding to early endosomes.52 Similarly a number of GFP::RAB-7 positive punctate structures, presumably corresponding to maturing endosomes, are observed near the plasma membrane53 in wild-type intestines (Fig. 3B). A number of larger ring-like GFP::RAB-7 positive structures localize deeper in the cytoplasm and are presumed to be mature late endosomes.47,53 We found that in ok691 mutants, the GFP::RAB-5 early endosomal marker labels the membrane of the abnormal vacuoles (Fig. 3A'). The GFP::RAB-5 is often specifically localized to selected regions of the abnormal vacuoles. However, the number and the intensity of GFP::RAB-5 punctate dots did not change in ok691 mutants when compared to wild-type (Fig. 3I). In contrast, the late endosomal markers, GFP::RAB-7 and LMP-1::GFP, label the membrane of the enlarged abnormal vacuoles in ok691, but only rarely the membrane of small abnormal vacuoles (Fig. 3B'). These results suggest that the vacuoles are aberrant endosomes and can be distinguished as two classes, perhaps representing different stages in the endosome maturation process. Unlike RAB-5-positive puncta, the small RAB-7-positive puncta overaccumulated in bec-1 mutant intestines, (Fig. 3B and B' and I). The accumulation of the small RAB-7-labeled structures appeared to be at the expense of the larger RAB-7-labeled rings that decreased in number (Fig. 3I). Markers for other endosome types such as apical recycling endosomes (GFP::RAB-11) and basolateral recycling endosomes (GFP::RAB-10) appeared normal (Fig. S1 and data not shown). Moreover, the abnormal vacuoles in bec-1 mutant animals were very faintly labeled or not at all labeled by GFP::RAB-10 (Fig. S1). Similarly, we noted that the expression of ALX-1::GFP in bec-1 mutants was not significantly different to that of wild-type animals (Fig. S1). ALX-1 is an Alix/Bro1p ortholog in C. elegans, a protein that together with RME-1 regulates recycling from endosomes to the plasma membrane.54 Taken together these results suggest a requirement for BEC-1 at a specific stage of endosome maturation.
Figure 3.
bec-1 mutants display a defect in retrograde transport. Confocal images in a wild-type background are shown for GFP::RAB-5 (A), GFP::RAB-7 (B), LMP-1::GFP (C), GFP::hTfR (D), MIG-14::GFP (E), GFP::RME-8 (F), GFP::SNX-1 (G), GFP::VPS-35 (H). Confocal images of bec-1(ok691) mutants are shown for GFP::RAB-5 (A'), GFP::RAB-7 (B'), LMP-1::GFP (C'), GFP::hTfR (D'), MIG-14::GFP (E'), RME-8::GFP (F'), GFP::SNX-1 (G'), GFP::VPS-35 (H'). (B) GFP::RAB-7 positive puncta (marked by yellow arrow) labels maturing endosomes and late endosomes (marked by yellow arrowhead). bec-1(ok691) mutants accumulate maturing endosomes marked with GFP::RAB-7, and display a decrease in late endosomes marked with GFP::RAB-7 (compare B and B'; See Fig. 2I). White arrowheads indicate enlarged intestinal endosomes (abnormal vacuoles) labeled by GFP::RAB-5 and GFP::RAB-7. In bec-1(ok691) animals, LMP-1::GFP also accumulates in the membrane of the enlarged endosomes (C'; marked by white arrowheads). In wild-type animals (D), the basolateral plasma membrane and basolateral endocytic compartments are labeled by hTfR::GFP (human transferrin receptor). The accumulation of hTfR::GFP is not affected in bec-1(ok691) mutants (D'), and the recycling endosome cargo hTfR::GFP does not accumulate in the enlarged endosomes (abnormal vacuoles) of bec-1(ok691) mutants. Confocal images of the worm intestine expressing the GFP-tagged endocytic transmembrane cargo marker MIG-14::GFP in wild-type (E), and bec-1(ok691) (E') animals. The retromer-dependent cargo protein Wntless MIG-14::GFP normally localizes basolaterally in the intestine and colocalizes with the early endosomal marker RAB-5.55 In bec-1 mutants, MIG-14::GFP transmembrane cargo protein accumulates in the enlarged endosomes (abnormal vacuoles; E'). We observed a decrease in the number of MIG-14::GFP positive puncta (I). The retromer subunit RME-8::GFP (F'), GFP::SNX-1 (G') and GFP::VPS-35 (H'), do not accumulate in the enlarged intestinal endosomes (abnormal vacuoles). A decrease in the number of GFP::RME-8 positive puncta is observed in bec-1(ok691) mutants (F'), and its intensity is markedly decreased. For all images, scale bars represent 10 µm. Quantification of endosome number as visualized by the positive labeling with endocytic markers is shown in (I). In the quantification of endosomal compartments, we saw no significant difference in RAB-5 early endosomes, we observed an increase in RAB-7 maturing endosomes, and a dramatic decrease in RAB-7-positive late endosomes. In addition, we observed a decrease in MIG-14 as well as RME-8 labeled compartments, and a slight increase in SNX-1 as well as in VPS-35 labeled compartments. Error bars represent standard deviation from the mean (n = 30 each, 10 animals of each genotype were sampled in three different regions of the intestine). In (J and K), we analyzed whether lysosomal degradation of GFP::MIG-14 occurs in bec-1 mutant animals. RNAi mediated depletion of lysosome biogenesis protein CUP-5/mucolipin1 increased the number and the intensity of MIG-14::GFP positive puncta (J and Fig. S5). After cup-5 RNAi, we saw a 4X increase in the number of MIG-14 positive puncta in bec-1 mutants, when compared to control RNAi, indicating that there is lysosomal processing of the MIG-14 cargo in bec-1 mutants (J, left panel). Similarly, we observed a significant increase in the intensity of MIG-14::GFP in bec-1 mutants after cup-5 RNAi (J, right panel). Asterisks indicate a significant difference in the one-tailed Student's t-test (*p < 0.05, **p < 0.005, ***p < 0.0005, n.s.: not significant).
As another control, we assayed the distribution of the human transferrin receptor (hTfR::GFP), transmembrane cargo that is recycled through the recycling endosome and not the Golgi.55–57 An equivalent version of this GFP-fusion protein was shown to be functional and traffic normally in mammalian cells or C. elegans.53 We found that the abnormal vacuoles in bec-1 mutants did not accumulate hTfR::GFP (Fig. 3D and D'), and its steady state localization was unaffected by a mutation of bec-1, indicating that hTfR transit through the recycling endosome is unaffected. From this and the experiment described using the RAB-10 and RAB-11 markers, we concluded that bec-1 is not required for the function of recycling endosomes.
Endosome to TGN retrograde transport is defective in bec-1 mutants.
In yeast, the identification of the yeast VPS34 gene encoding the PI3-kinase in a screen for vacuolar sorting mutants has demonstrated a direct role for PI3P in vesicular trafficking.52 Specifically, PI3P was shown to be required for the efficient sorting of proteins from the late-Golgi to the vacuole.58 Given the disruption in early endosomal trafficking that we saw in bec-1 mutants, we next asked whether the loss of bec-1 could affect endosome to Golgi retrograde trafficking.
In C. elegans, SNX-1 is the only ortholog of mammalian Sorting Nexin 1 and Sorting Nexin 2 proteins that function in endosome-to-Golgi retrograde transport along with Vps-26, Vps-29 and Vps-35, as part of the retromer complex.36,37 RME-8, a large protein with a DNA-J domain, binds to SNX-1.55 C. elegans GFP::SNX-1 and RME-8::GFP fusion proteins colocalize with early endosomal markers in living animals.55 In C. elegans, like all invertebrates, the Golgi network appears as dispersed ministacks throughout the cell rather than in one large juxtanuclear stack.59 To date, the only retromer dependent cargo protein known in C. elegans is MIG-14/Wntless.60,61 In the absence of retromer function, MIG-14/Wntless is depleted from the Golgi and missorted to the late endosome and lysosome.55,60–64 Endosome to Golgi transport and sorting of cargo protein MIG-14/Wntless depend on RME-8 and SNX-1.55 Thus, we reasoned that if BEC-1 functions in retrograde transport, bec-1 mutants should abnormally transport the MIG-14/Wntless cargo and have a similar phenotype to snx-1 or rme-8 mutants.
When compared to wild-type control animals, bec-1(ok691) mutants displayed an approximately four-fold reduction in the number of MIG-14::GFP-positive puncta. A similar reduction was observed in animals treated with RNAi against bec-1 and vps-34 (Fig. 3S). These results are consistent with the idea that BEC-1 functions with VPS-34 in retrograde transport (Fig. 3E and E'; Fig. 3S). However, we observed an increase in the total intensity of the MIG-14::GFP signal in contrast to what has been previously reported for MIG-14::GFP in rme-8 or snx-1 mutants.55,60,61 In bec-1 and vps-34 mutants, MIG-14::GFP is found in ring-like and intralumenal structures (Fig. 3S). The reasons for this difference are unclear, but one possibility is that MIG-14::GFP localizes to the membrane of the abnormal and enlarged vacuoles that accumulate in bec-1 mutants (Fig. 3E and E'; Fig. 3S), similar to the localization of GFP::RAB-7, and LMP-1::GFP. Another possibility is that the discrepancy between bec-1 and rme-8 phenotypes is due to the hypomorphic nature of the rme-8 mutation we used. These results suggest that there is a defect in the retrograde transport of MIG-14::GFP from the endosome to the Golgi in bec-1 mutants and that in bec-1 mutants, a substantial amount of MIG-14 is degraded by the lysosome, similarly to what has been described in animals with defective retromer subunit activity.
To determine if the decrease in MIG-14::GFP in bec-1 mutants is the result of missorting and lysosomal-mediated degradation, we monitored MIG-14::GFP levels in bec-1 mutants after RNAi depletion of cup-5/mucolipin1, a transmembrane protein required for normal lysosome biogenesis and normal levels of hydrolytic activity.46,55,65 Depletion of CUP-5 by RNAi increased the number of MIG-14::GFP positive puncta to nearly normal levels in the bec-1 mutant background (quantitation shown in Fig. 3J; Fig. 4S). In addition, a significant increase in the pixel intensity of MIG-14::GFP was observed in bec-1 mutants that were depleted for CUP-5 by cup-5 RNAi (Fig. 3J; Fig. 4S). These results confirm that lysosomal degradation of MIG-14::GFP indeed occurs in bec-1 mutants. A similar result was previously reported for retromer mutants, such as rme-8. In rme-8 mutants, CUP-5 depletion by RNAi restores the levels of MIG-14::GFP pixel intensity to nearly normal levels. Thus, loss of BEC-1 activity results in a similar phenotype to the retromer mutants, where the degradation of MIG-14 occurs through the missorting into the lysosomal pathway (Fig. 3J).
The increased degradation of MIG-14::GFP suggested that multivesicular body (MVB)-mediated transport of membrane proteins to the lysosome does not require BEC-1. To examine this point further, we assayed the degradation of CAV-1::GFP in early embryos.54,55,66 CAV-1 is a known transmembrane cargo protein that is degraded in the one cell embryo, right after fertilization, after the metaphase to anaphase transition.66–68 Degradation of CAV-1::GFP requires endocytosis and the endosomal sorting complex required for transport (ESCRT) machinery. CAV-1::GFP degradation was unaffected in animals treated with bec-1 RNAi (Fig. 2S), even after animals were grown as the second generation in RNAi plates, further suggesting that BEC-1, just like RME-8 and SNX-1,55 is not required for ESCRT-mediated degradation of integral membrane proteins. Similarly, we did not see a defect in CAV-1::GFP degradation after vps-34 RNAi (Fig. 2S).
Localization of RME-8, SNX-1 and VPS-35 in bec-1 mutant animals.
Since we observed a retrograde trafficking defect in bec-1 mutants, we considered that C. elegans BEC-1 may act similarly to Atg6/Vps34 in the yeast retrograde transport. In yeast, mutations in ATG6/VPS30 lead to a selective sorting and maturation phenotype of the soluble vacuolar protease CPY. Redistribution of Vps5p and Vps17p, two subunits of the retromer complex, was observed in strains deficient for Atg6/Vps30.69 To determine if the localization of retromer complex proteins is altered, we examined the localization of RME-8::GFP (Fig. 3F and F'), GFP::SNX-1 (Fig. 3G and G') and GFP::VPS-35 (Fig. 3H and H'). In bec-1 mutants, we found that the intensity and number of RME-8::GFP-positive puncta is decreased (Fig. 3 compare F to F'; Fig. 5S). Although RNAi against bec-1 did not have as dramatic an effect on RME-8::GFP as the bec-1 mutant, knockdown of vps-34 by RNAi also affected the number of RME-8::GFP positive puncta significantly (Fig. 5S).
RME-8 and SNX-1 have been shown to physically associate in vivo and this association is functionally relevant for the regulation of endosomal clathrin.55 We observed some increase in the intensity of clathrin-positive endosomes after RNAi treatment against vps-34 (visualized with a functional clathrin reporter, GFP::CHC-1;59 Fig. 6S), and no obvious changes upon knockdown of bec-1 activity. However, consistent with the decrease in number and intensity of RME-8::GFP positive punta observed in bec-1 mutants, we observed a significant increase in GFP::SNX-1 and GFP::VPS-35 positive puncta in bec-1 mutants when compared to wild-type animals (Fig. 3G' and H' quantitation in Fig. 3I). Moreover, the size of the GFP::SNX-1 positive puncta was significantly increased in bec-1 mutant animals (Fig. 7S). This increase in the size and distribution of SNX-1 positive endosomes in bec-1 mutants is similar to that observed in rme-8 mutants.55
Colocalization of BEC-1 and RME-8.
The retromer complex, including RME-8 and SNX-1, is found primarily on early endosomes, and many of the RME-8/SNX-1 positive endosomes are closely juxtaposed to the Golgi apparatus.37,55
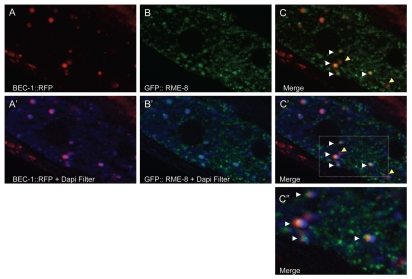
We found that in C. elegans BEC-1::RFP colocalizes well with a subset of RME-8::GFP labeled endosomes in the intestine, consistent with a direct role of BEC-1 in retrograde transport (Fig. 4C'). Interestingly, we note that BEC-1::RFP and RME-8::GFP double-positive endosomes are clearly adjacent to auto-fluorescent lipofuscin-positive lysosome-like organelles (in blue, Fig. 4C' and C”).
Figure 4.
BEC-1 colocalizes with RME-8. Representative images of intestinally expressed RFP-tagged BEC-1 (red, A, A') and GFP-tagged RME-8 (green, B, B') in wild-type intact living animals. Autofluorescent lysosome-like organelles are shown in blue (+ DAPI filter, D). White arrowheads indicate colocalization of the BEC-1::RFP and RME-8::GFP signals (C), observed as the yellow fluorescence. The blue autofluorescence is also shown with BEC-1::RFP (A') and with GFP::RME-8 (B') and in the merger of both in (C'). The colocalized BEC-1::RFP and GFP::RME-8 signals are found adjacent to the autofluorescent lysosomes which can be visualized with the DAPI filter (C'). A close-up of (C') is shown in (C”). Arrowheads in yellow show BEC-1::RFP that is not associated with RME-8::GFP. Magnification is 630x (A–C').
bec-1 has a role in germ cell corpse clearance.
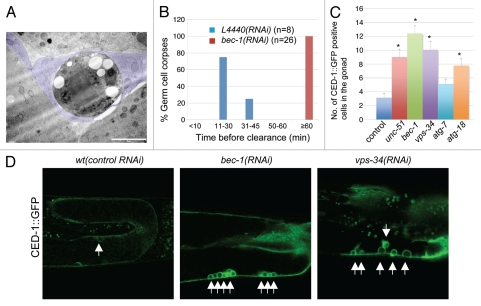
Loss of bec-1 leads to an increase in the number of visible apoptotic cell corpses.31 This phenotype could result from increased apoptosis or clearance defects in the execution of the cell death pathway or both. To distinguish between these possibilities, we analyzed germ cell corpses and their surroundings using transmission electron microscopy. Wild-type sheath cells are so efficient in clearing apoptotic germline cells that corpses are virtually never seen in a wild-type gonad by electron microscopy, although they have been observed in other endocytic mutants.70 In contrast, in bec-1(ok691) mutants, we found several examples where dying germ cells were engulfed by the sheath cell but not digested (Fig. 5A). We analyzed serial sections that span the entire diameter of each cell corpse to determine whether it is totally inside a sheath cell. In bec-1(ok691) mutants, all germ cell corpses were engulfed by gonadal sheath cells. These results suggest that the dying germ cell is still able to trigger the sheath cell to encircle it, but that bec-1 activity is required to promote the delivery of other components to the phagosome for the completion of cell degradation. To further corroborate these findings we followed individual apoptotic nuclei in animals with reduced bec-1 function and in wild-type animals and determined the time required for clearance. We found that all apoptotic nuclei in wild-type animals are cleared in less than 50 min, most within 30 min, whereas nuclei persist in animals with reduced bec-1 function for over 1 h (Fig. 5B). These observations indicate that the apparent increase in apoptotic nuclei reflects a defect in cell corpse clearance rather than an increase in the number of cells undergoing apoptosis.
Figure 5.
bec-1 mutants display lack of cell corpse clearance. (A) Micrograph of transmission electron microscopy (TEM) where poor degradation of apoptotic germ cell corpses is observed in bec-1 mutants. This TEM image was taken in a cross-section through the midbody. A rounded dying germ cell is shown with very dark cytoplasm and several large round vacuoles near the nucleus, which contains clumped chromatin. The dying cell is completely wrapped by the somatic sheath cell (tinted in purple) in an early phase of apoptosis. The sheath cell also wraps the normal germ-line (bottom two cells in the panel), separating this mesodermal tissue from the intestine (top cell in the panel). (B) Histogram indicating the distribution of the duration of germ cell corpses before they are completely degraded. The y-axis indicates the percentage of germ cell corpses that lasted for the period of time indicated in the x-axis before clearance. n is the number of cell corpses analyzed. (C) Representative images of CED-1::GFP positive labeled cell corpses in wild-type animals after treatment with control RNAi and RNAi against bec-1 and vps-34. (D) Quantification of germ cell corpses in wild-type animals after treatment with control RNAi, and RNAi against unc-51/Atg1, bec-1, vps-34, atg-7 and atg-18. Data derived from observing adults, 36 hours post larval L4 stage. Data were compared by unpaired t tests; 30 animals were analyzed for each experiment. Asterisks indicate a significant difference as a result from an analysis of variance (ANOVA). This analysis indicated that there are significant differences between all treatments and control, except for atg-7 RNAi (p < 0.001). Threshold for significance (alpha) in the t-tests was p < 0.01 using a “Bonferroni correction” for multiple corrections.
To investigate whether inactivation of other autophagy genes has an effect in cell corpse clearance, we counted apoptotic nuclei in the gonads of animals that had been treated with dsRNA against various autophagy genes. The phagocytic receptor cell death abnormal CED-1 is an engulfing cell-specific marker that recognizes cell corpses and clusters to the growing phagocyte.71 Using CED-1::GFP, we found an accumulation of germ cell apoptotic nuclei in the gonads of vps-34 RNAi animals (Fig. 5C), consistent with previous reports.71–73 In addition, we found that RNAi mediated knockdown of unc-51/ATG1, and atg-18 result in an increase of CED-1::GFP positive nuclei (Fig. 5D). Although we did observe an increase in CED-1::GFP positive nuclei in animals treated with RNAi against atg-7, after analysis of variance (ANOVA) for multiple comparisons, we found this result not to be significant.
Discussion
C. elegans bec-1 has been shown to have a role in organismal size, dauer morphogenesis, resistance to pathogens and the longevity of daf-2 (the insulin-like receptor) mutants, as well as the longevity of dietary restricted animals.15 daf-2 constitutive mutants at the restrictive temperature were shown to have an increase in autophagy, as marked by the GFP::LGG-1 autophagosome marker.27 GFP::LGG-1 localization in daf-2 insulin-like receptor mutants after bec-1 RNAi was observed to form large aggregates, rather than small punctate structures which represent “normal” autophagosomes in daf-2 mutants with wild-type bec-1 activity. This may be due to the lack of clearance of the GFP::LGG-1 protein due to a defect in the autophagolysosomal pathway15,16 but awaits further clarification. In addition to confirming a role for bec-1 in endocytic trafficking,31 we used a combination of endocytic markers to show that BEC-1 is involved in various steps of endocytosis. In particular, we have identified a role for BEC-1 in retrograde trafficking that resembles defects associated with loss of retromer activity, including defective recycling of MIG-14, known cargo for the endosome to Golgi retromer transport and a high number of CED-1::GFP positive cell corpses in bec-1 mutant gonads, as also observed in gonads from animals lacking retromer subunits.38
bec-1 has a maternal effect lethal phenotype.
Animals that completely lack maternal and zygotic BEC-1 activity die during embryogenesis. These embryos display a high incidence of apoptotic nuclei.31 Animals that segregate from a heterozygous parent, and have the maternally derived wild-type BEC-1 function, live to early adulthood. These homozygous bec-1 mutant animals display a striking accumulation of vacuoles in different tissues including the intestine and hypodermis. They are also uncoordinated in their movement.
Studies in mice have shown that lack of beclin 1 activity is also lethal. beclin 1 mutants die at day 7.5 of development,12,13 whereas atg5 and atg7 mutant mice die after birth in the period of starvation before the animals begin to breastfeed.33,74 Therefore, it is possible that the difference between the phenotype of beclin 1 and other autophagy genes is due to beclin 1 having additional defects in endocytosis in mammals.
bec-1 mutants display endocytic defects.
The lack of accumulation of endocytic tracers in coelomocytes indicates that the internalization step of endocytosis is defective in bec-1 mutants. This is consistent with previous reports of a pronounced defect in bec-1 mutants when assaying for the uptake of vitellogenin by oocytes,31 a process that requires endocytosis. Thus, bec-1 is required at an early step in endocytic trafficking.
We have analyzed the number and morphology of all major endocytic compartments in bec-1 mutant intestinal cells in vivo using a set of GFP-tagged markers that label each class of compartment. Our goal was to determine if there was a block in a particular transport step caused by the lack of bec-1 activity by measuring specific changes in endosome morphology, and intensity or number of GFP markers. We would expect that an endocytic compartment that normally receives cargo in a BEC-1 dependent fashion would be smaller in size or be lacking completely in intestinal cells lacking BEC-1 activity. We would also expect an accumulation or an enlargement of BEC-1 dependent endosomes or BEC-1 donor compartments in the bec-1 mutant intestines. Trafficking from the cell membrane to the lysosome requires the activity of Rab5 and Rab7 GTPases.75 Rab GTPases regulate intracellular trafficking by controlling the transport of vesicles between membrane compartments along endocytic transport. Rabs alternate between an “active” guanosine triphosphate (GTP)-bound state and an “inactive” guanosine diphosphate (GDP)-bound state. Active Rab5 localizes to early endosomes whereas Rab7 localizes to late endosomes. Rab5 and Rab7 regulate trafficking of cargo from the plasma membrane to the lysosomes.75
In bec-1 homozygous mutants, we found an accumulation of RAB-7 positive puncta structures and a decrease in RAB-7 ring-like structures. As RAB-7 positive puncta are presumed to be maturing endosomes, and the ring-like structures to be late endosomes, our results suggest a defect prior to full maturation of the late endosome. In addition, we found that the membranes of the abnormal vacuoles seen in bec-1 mutant animals were positive for RAB-5, RAB-7 and LMP-1, consistent with accumulation of incompletely matured endosomes. In yeast and mammalian cells, Ypt7p and Rab7 can mediate homotypic fusion of vacuoles and late endosomes/lysosomes.76,77 Thus, it is possible that the accumulation of RAB-7 and LMP-1 positive large vacuoles is the result of an enhancement of homotypic fusion consistent with an increase in RAB-7 activity. Distorted endosomal compartments were also observed by electron microscopy. These data demonstrate that the wild-type BEC-1 activity is required for the endolysosomal pathway. The fact that we see a lack of PI3P localization, the product of VPS-34, in the bec-1 null mutants indicates that the bec-1 phenotype may be due to the mislocalization of PI3P on endosomes.
In this study we show that the enlarged vacuoles in bec-1 null mutants result from a defect in maturing endocytic compartments. This phenotype has been previously documented for C. elegans intestines with defects in the recycling of endosomes as in rme-1 or rab-10 mutants, or when recycling is blocked by pharmacological agents in certain mammalian cell types.52,78 Similar vacuoles or enlargement of endocytic compartments have also been observed in phosphatidylinositol phosphate kinase 3 mutants, ppk-3, the C. elegans ortholog of the yeast PIKfyve/Fab1p.49 In these animals, enlargement of RAB-7-positive, and LMP-1 positive endocytic compartments occurs.49 Thus, there are multiple steps in endocytosis for which disruption may result in enlarged endocytic compartments. However the recycling marker RAB-10::GFP did not localize to the bec-1 mutant vacuoles, and the number of RAB-10::GFP puncta was not affected in bec-1 mutants, suggesting that the lack of bec-1 does not affect the recycling of endosomes.
BEC-1 functions in the retrograde transport.
Yeast Atg6/Vps30 has been implicated in early endosome to Golgi retrograde transport.69,79 We found that C. elegans bec-1 function is also required for retromer function. We show that BEC-1 and VPS-34 are required to rescue MIG-14 from degradation after its endocytosis. Our results show that aberrant sorting of MIG-14 in bec-1 and vps-34 mutants is similar, although not as dramatic as that in rme-8 or snx-1 mutants.55 It is not known whether MIG-14 may utilize any other alternative route of retrograte transport in C. elegans or in mammals.55 LMP-1 and MIG-14, two different transmembrane cargo proteins, labeled the abnormal vacuoles that accumulate in bec-1 mutant intestinal cells, whereas hTfR::GFP did not, indicating that LMP-1 and MIG-14 require BEC-1 for transport, while hTfR does not.
bec-1 mutants have a marked decrease in number of RME-8::GFP puncta expression. A similar phenotype was observed in animals with knockdown RNAi mediated against vps-34. Conversely, the number of GFP::SNX-1 and GFP::VPS-35 positive puncta increases in bec-1 mutants when compared to wild-type animals. For SNX-1, the size of the puncta is larger in bec-1 mutants. These findings are similar to those made in rme-8 mutants,55 although bec-1 and vps-34 mutants accumulate aberrant MIG-14::GFP positive vacuoles. We conclude that BEC-1 and VPS-34 function in retrograde transport, possibly in concert with RME-8. Since it is not known how RME-8 is recruited to the endosome, we hypothesize that BEC-1 and the product of VPS-34, PI3P, may facilitate the endosomal recruitment of RME-8. Further studies will be required to better understand the relationship between BEC-1, VPS-34 and RME-8, and the role of BEC-1 and VPS-34 in early endosome maturation and transport to the Golgi.
BEC-1 is required for cell corpse clearance.
We found that bec-1 loss of function mutant animals display a lack of cell corpse clearance in the hermaphrodite gonad rather than an increase in the incidence of apoptosis.31 Instead, we found that there is a significant lack of germ cell corpse degradation in bec-1 mutant animals when compared to wild-type. In addition, using transmission electron microscopy we do not see a major effect on engulfment, implying that loss of bec-1 activity does not appear to affect the signal from the dying cell. We cannot rule out a low-penetrance effect, but our in vivo results contrast with the previously reported requirement for autophagy genes such as beclin 1 and atg5 as an energy source to facilitate signaling from the dying cells to the phagocytic cell in embryoid bodies derived from ES cells.80
We also found that animals deficient in vps-34, which is required for RAB-5 recruitment to the nascent phagosome,71 show an increase in the number of germ cell nuclei. The degradation of cell corpses has been shown to require the sequential enrichment of early endosomes, late endosomes and lysosomes to the nascent phagosome (reviewed in ref. 81). RAB-7 and PI3P have been proposed to function as downstream effectors of the CED-1 pathway to mediate phagolysosome formation.82 Since we showed that RAB-7::GFP as well as PI3P localization is not normal in bec-1 mutants, the cell clearance defect of bec-1 mutants may also be due to the mislocalization of PI3P at the phagosome. Interestingly, we observed that knockdown of other autophagy genes by RNAi results in a similar accumulation of CED-1-positive engulfed germ cells in the hermaphrodite gonad. This result suggests that autophagosomes are also required in the process of phagosome maturation. Since a defect in cell clearance has also been reported for retromer subunit mutants, rme-8 and snx-1,38 at this point we do not know whether the lack of cell clearance defect in bec-1 mutants reflects a requirement for autophagy or for a bec-1-mediated retrograde transport function or both. This point should be addressed in future experiments.
Comparisons with mammals and yeast.
In yeast, two different Atg6/Vps30 complexes were found to function in autophagy and vacuolar protein sorting.19,79 The two complexes differed only in two proteins: Atg14 and Vps38. The complex consisting of Atg14, Atg6/Vps30, Vps15 and Vps34 has been shown to have autophagy function, and the complex consisting of Vps38, Atg6/Vps30, Vps15 and Vps34 was shown to be specific for vacuolar protein sorting, more specifically endosome to Golgi retrograde trafficking.69 Thus, all the subunits except for Atg14 and Vps38 are shared between the two complexes. The counterparts for these two complexes had not been found in other organisms, and orthologs for either Atg14 or Vps38 in higher eukaryotes were seemingly lacking, leading to the assumption that these molecular complexes only existed in yeast. Recently, using highly sensitive methods of purification, mammalian proteins were identified that interact with Beclin 1, the mammalian ortholog of Atg6/Vps30. In fact, three distinct Beclin 1 complexes have now been described and orthologs of yeast Atg14 and Vps38 have been found.34,83 The human ortholog of yeast Atg14 is Atg14L, and the human ortholog of Vps38 has been proposed to be Uvrag, since Uvrag primarily localizes to late endosomes, and shows weak homology with yeast Vps38.35 A complex consisting of Beclin 1, hVps34, hVps15 and Atg14L, functions in early autophagosome formation. Another complex consisting of Uvrag, Beclin 1, hVps34 and hVps15 appears to act in endosome to Golgi retrograde trafficking as well as in the fusion of lysosomes and autophagosomes. This is consistent with our findings. A third complex consists of Rubicon, Uvrag, Beclin 1, hVps34 and hVps15, where Rubicon negatively regulates the autophagosome maturation process, as well as endocytic trafficking.34,83 As Rubicon protein localizes to endosomes and lysosomes, it may be directly involved in the regulation of membrane fusion processes of endosomes/lysosome and autophagosomes. Uvrag has the capacity to bind to the Class C VPS complex, which is involved in the fusion process of autophagosomes and in endocytosis.84,85 In tissue culture experiments, the knockdown phenotypes of Atg14L and Rubicon are different, indicating that Beclin 1 has multiple roles in autophagy through the formation of different complexes. Orthologs to Uvrag and Rubicon have yet to be found in C. elegans by sequence homology, although an ortholog to Uvrag does exist in Drosophila.2 Our results support the idea that at least two BEC-1 complexes exist in C. elegans which function in autophagy and retrograde transport. Significant future work will be required to better understand the mechanisms by which BEC-1 activity functions in development, longevity and resistance to pathogens, and if these require BEC-1 autophagy function or its endocytic function or both.
Materials and Methods
C. elegans strains.
Standard procedures were used to culture C. elegans worms (Brenner, 1974). All strains were grown at 20°C, unless otherwise stated. The wild-type C. elegans strain N2 and the following mutant alleles were used: LGIV, bec-1(ok691) and bec-1(ok700) provided by the Knockout consortium, unc-5(e53) jcIs1 (Is[jam-1::GFP]) (gift from T. Schedl, University of St. Louis). The transgenes used: pwIs50[lmp-1::gfp unc-119(+)],46 pwIs72 [Pvha-6::gfp::rab-5, unc-119(+)], pwIs87[Pvha-6:: gfp::rme-1, unc-119(+)],47,53 pwIs170 [Pvha-6::gfp::rab-7, unc-119(+)]53 [Pvha-6::gfp::rab-10, unc-119(+)], pwIs69 [Pvha-6:: gfp::rab-11, unc-119(+)],53 pwIs90[Pvha-6::hTfR::gfp], izEx1[lgg-1promoter::gfp::LGG-1],27 izEx5[pAy39.1, bec-1promoter::BEC-1:: RFP],39 izEx6[pBEC-1, pTG96], [Pvha-6::RME-8::GFP, unc-119(+)], p[snx-1::GFP::SNX-1], izEx [Pvha-6::MIG-14::GFP, unc-119(+)],55 pwIs61 [GFP::cav-1, unc-119(−)]66 and bcIs39[P(lim-7)ced-1::GFP, lin-15(+)].71
Molecular analyses.
To characterize the nature of the bec-1 transcript present in bec-1(ok700) mutant animals, total RNA was isolated and subjected to RT-PCR using oligo dT primers. Determination of the DNA sequence for the RT-PCR product revealed a short transcript containing exons 1–4 resulting in a frameshift that removes the remainder of the open reading frame after exon 4.
Transgenes.
For rescue experiments of bec-1 mutants, a 14.0 Kb PCR fragment was amplified using wild-type genomic DNA as a template. This fragment contains the entire bec-1(+) gene plus 10 Kb of flanking 5′ DNA and 970 bp of downstream 3′ DNA. This PCR fragment was directly injected into bec-1(ok691)/nT1 mutant animals at 40 µg/ml together with the ubiquitously expressed cotransformation marker pTG96 (SUR-5::GFP) at 100 µg/ml. The rescuing BEC-1::RFP, pAR 39.1,39 was injected at 20 µg/ml along with the cotransformation marker pRF4 [rol-6(d)] at 100 µg/ml, and Rol lines were established.
Endocytosis assays.
To investigate endocytosis at the basolateral membrane of the intestine, young adult hermaphrodites were injected into the pseudocoelomic space with 0.1 mg/ml Texas Red-conjugated BSA (TR-BSA from Sigma), as previously described in reference 50. Briefly, TR-BSA is dissolved in egg salts (118 mM NaCl, 48 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, pH 7.4). We injected TR-BSA into the pseudocoelomic space in the pharyngeal region of adult worms that were immobilized on a dried agarose pad immersed in oil. Injected animals were rehydrated with M9 buffer and transferred to seeded NGM plates. At different time points, animals were mounted on agarose pads with a 1–2 µl drop of M9 containing 25 mM sodium azide to anesthetize them and view them on the confocal microscope. Animals were soaked in the same solutions to investigate endocytosis at the apical membrane.
RNA interference.
dsRNA-mediated gene interference experiments were performed by feeding bacteria expressing the dsRNA to larval L4 stage individuals, and scoring their progeny, unless described otherwise. L4 larvae were placed on plates containing NGM agar with 5 mM IPTG and HT115 (DE3) bacteria carrying double stranded RNA expression constructs and allowed to lay eggs for 24 h at 20°C, except in the case of GFP::CAV-1 expressing animals, which were incubated at 25°C. P0s were transferred every day for 3 days. F1 progeny were raised in the plates containing dsRNA bacteria and scored as 1-day-old adults. To determine if there is a lack of GFP::CAV-1 degradation, the second generation of RNAi animals were also assayed. In all experiments with bec-1 and vps-34 RNAi, the accumulation of vacuoles was monitored as the most obvious phenotype to determine that RNAi had been successful. RNAi clones were obtained from the Ahringer and Vidal genomic RNAi libraries86 (a gift from Dr. Malene Hansen).
Epifluorescence microscopic analyses.
We used a Leica TCS-SP5 laser-scanning confocal microscope to analyze the subcellular localization of fluorescent markers of endocytic and lysosomal compartments as well as endocytosis markers in mutant and wild-type animals. Images on the confocal were collected by a PMT (photomultiplier tube) detector, converted to Tiff format and cropped using Adobe Photoshop CS3. Quantifications were performed by counting punctate-positive structures in the wild-type and mutant intestines using Image J Software (National Institutes of Health). To quantify intensity of images, Metamorph software ver 6.3r2 (Universal Imaging) was used.
Live worms were mounted on 2% agarose pads with 10 mM sodium azide.87 For GFP::RAB-5, and RME-8::GFP puncta quantification, only structures smaller than 0.3 microns were counted in either wild-type or mutant animals. For MIG-14::GFP-positive puncta quantification, only structures smaller than 0.2 microns were counted in either wild-type or mutant animals. For quantification of GFP::SNX-1 and GFP::RAB-7, GFP-positive puncta are counted manually. Images taken with the DAPI filter were used to identify broad-spectrum intestinal autofluorescence caused by lipofuscin-positive lysosome-like organelles.47,88
Electron microscopy.
Animals were prepared for electron microscopic analysis as described previously in reference 15 and 27. Briefly, L4 animals were packed into the metal planchette using an excess of bacteria to avoid any empty space surrounding the animals. The animals were then frozen using a Bal-Tec HPM 010 high pressure freeze apparatus and freeze substituted in 1% osmium tetroxide in acetone beginning at −90°C. The samples were then infiltrated with epoxy resin and heat cured. These plastic blocks were thin sectioned, post-stained with uranyl acetate and lead citrate, and then examined by electron microscopy.
Quantitation of cell corpses.
The increase in the number of cell corpses was measured using the CED-1::GFP marker. Adult hermaphrodites at 36 h post-larval L4 stage animals were scored. Quantification of germ cell corpses in wild-type animals after treatment with control RNAi, unc-51/ATG1, bec-1, vps-34 and atg-18 RNAi animals. Data are derived from three experiments observing adults, 36 h post-larval L4 stage. Data were compared by analysis of variance (ANOVA) and five two-sample t-tests between control strain and the five treatments; 30 animals were analyzed for each treatment and control. Analysis of variance (ANOVA) indicated that there are significant differences between treatments and the t-tests found significant differences between the control treatment and all other treatments except atg-7 RNAi (p < 0.001). Threshold for significance (alpha) in the t-tests was p < 0.01 using a “Bonferroni correction” for multiple corrections.
Apoptotic cell clearance assay.
Germ cell corpses in the adult hermaphrodite gonads were scored under the Nomarski DIC microscope by their highly refractile appearance.89 A Zeiss ApoTome microscope equipped with Time Lapse software was employed to capture apoptotic nuclei. In addition, serial Z-section images of regions of the hermaphrodite gonad were recorded at 0.5 µm intervals every 5 min. Movement of cells and focus were closely monitored to ensure that the region of the gonad being recorded did not change. Animals were closely monitored for viability during the recording. Adult hermaphrodites 36–48 h post-larval L4 stage animals were scored.
Acknowledgements
We thank Drs. Johnny Fares, Hannes Bülow, Malene Hansen, Tim Schedl, as well as the C. elegans Knockout Consortium and CGC, for strains and reagents. We are grateful to Dr. Areti Tsiola and Dr. Nathalia Holtzman for assistance with the imaging and Image J program, and to the Core Facility for Imaging, Cellular & Molecular Biology at Queens College. We are thankful to Drs. Cathy Savage-Dunn, Hannes Bülow and Iva Greenwald for critical reading of this manuscript and for helpful discussions. This work was supported in part by grants from the NIH/NIA (AG024882-04S1) to Dr. Monica Driscoll (Rutgers University) and A.M., and from the NSF, Research Initiation Grant 0818802 to A.M. A.M. is an Ellison Medical Foundation New Scholar in Aging (AG-NS-0521-08). D.H.H. is supported by NIH RR 12596. B.D.G. is supported by NIH Grant GM067237.
Supplementary Material
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Cregg JM, Dunn W, Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 8.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teter SA, Klionsky DJ. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting and role of the vacuole in degradation. Semin Cell Dev Biol. 2000;11:173–179. doi: 10.1006/scdb.2000.0163. [DOI] [PubMed] [Google Scholar]
- 10.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Melendez A, Hall DH, Hansen M. Monitoring the role of autophagy in C. elegans aging. Methods Enzymol. 2008;451:493–520. doi: 10.1016/S0076-6879(08)03229-1. [DOI] [PubMed] [Google Scholar]
- 16.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A Role for Autophagy in the Extension of Lifespan by Dietary Restriction in C. elegans. PLoS genetics. 2008;4:24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 18.Meléndez A, Levine B The C. elegans Research Community, editor. WormBook. WormBook; 2009. Aug 24, Autophagy in C. elegans. doi/10.1895/wormbook.1.147.1, http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- 19.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 20.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol-3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown WJ, DeWald DB, Emr SD, Plutner H, Balch WE. Role for phosphatidylinositol-3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J Cell Biol. 1995;130:781–796. doi: 10.1083/jcb.130.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol-3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol-3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 24.Futter CE, Collinson LM, Backer JM, Hopkins CR. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson HW. Wortmannin causes mistargeting of procathepsin D. evidence for the involvement of a phosphatidylinositol-3-kinase in vesicular transport to lysosomes. J Cell Biol. 1995;130:797–805. doi: 10.1083/jcb.130.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson EE, Overmeyer JH, Gunning WT, Maltese WA. Gene silencing reveals a specific function of hVps34 phosphatidylinositol-3-kinase in late versus early endosomes. J Cell Sci. 2006;119:1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- 27.Meléndez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 28.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 29.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The Evolutionarily Conserved Domain of Beclin 1 is Required for Vps34 Binding, Autophagy and Tumor Suppressor Function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 30.Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M, Wang QJ, Holstein GR, Friedrich V, Jr, Iwata J, Kominami E, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 34.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. NatCell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 36.Collins BM. The structure and function of the retromer protein complex. Traffic (Copenhagen, Denmark) 2008;9:1811–1822. doi: 10.1111/j.1600-0854.2008.00777.x. [DOI] [PubMed] [Google Scholar]
- 37.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen D, Xiao H, Zhang K, Wang B, Gao Z, Jian Y, et al. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- 39.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26:1711–1720. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae YK, Kim E, L'Hernault SW, Barr MM. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr Biol. 2009;19:1599–1607. doi: 10.1016/j.cub.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roggo L, Bernard V, Kovacs AL, Rose AM, Savoy F, Zetka M, et al. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 2002;21:1673–1683. doi: 10.1093/emboj/21.7.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kostich M, Fire A, Fambrough DM. Identification and molecular-genetic characterization of a LAMP/CD68-like protein from Caenorhabditis elegans. J Cell Sci. 2000;113:2595–2606. doi: 10.1242/jcs.113.14.2595. [DOI] [PubMed] [Google Scholar]
- 46.Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, et al. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 49.Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J. The Phosphoinositide Kinase PIKfyve/Fab1p Regulates Terminal Lysosome Maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Grant B, Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsson L, Conradt B, Ruaud AF, Chen CC, Hatzold J, Bessereau JL, et al. Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics. 2008;179:375–387. doi: 10.1534/genetics.108.087247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apodaca G, Katz LA, Mostov KE. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J Cell Biol. 1994;125:67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi A, Pant S, Balklava Z, Chen CC, Figueroa V, Grant BD. A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burack MA, Silverman MA, Banker G. The role of selective transport in neuronal protein sorting. Neuron. 2000;26:465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- 57.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 58.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol-3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 59.Sato K, Ernstrom GG, Watanabe S, Weimer RM, Chen CH, Sato M, et al. Differential requirements for clathrin in receptor-mediated endocytosis and maintenance of synaptic vesicle pools. Proc Natl Acad Sci USA. 2009;106:1139–1144. doi: 10.1073/pnas.0809541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DY, Betist MC, Korswagen HC. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell. 2008;14:140–147. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 61.Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev Cell. 2008;14:132–139. doi: 10.1016/j.devcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, et al. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, et al. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol. 2008;10:170–177. doi: 10.1038/ncb1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Port F, Kuster M, Herr P, Furger E, Banziger C, Hausmann G, Basler K. Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat Cell Biol. 2008;10:178–185. doi: 10.1038/ncb1687. [DOI] [PubMed] [Google Scholar]
- 65.Schaheen L, Dang H, Fares H. Basis of lethality in C. elegans lacking CUP-5, the Mucolipidosis Type IV orthologue. Dev Biol. 2006;293:382–391. doi: 10.1016/j.ydbio.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17:3085–3094. doi: 10.1091/mbc.E06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bembenek JN, Richie CT, Squirrell JM, Campbell JM, Eliceiri KW, Poteryaev D, et al. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 2007;134:3837–3848. doi: 10.1242/dev.011361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato M, Grant BD, Harada A, Sato K. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J Cell Sci. 2008;121:3177–3186. doi: 10.1242/jcs.034678. [DOI] [PubMed] [Google Scholar]
- 69.Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- 70.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 72.Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, Wang X. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 2009;5:1000679. doi: 10.1371/journal.pgen.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 76.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Papini E, Satin B, Bucci C, de Bernard M, Telford JL, Manetti R, et al. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J. 1997;16:15–24. doi: 10.1093/emboj/16.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Weert AW, Geuze HJ, Groothuis B, Stoorvogel W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur J Cell Biol. 2000;79:394–399. doi: 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- 79.Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30 and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 81.Zhou Z, Yu X. Phagosome maturation during the removal of apoptotic cells: receptors lead the way. Trends Cell Biol. 2008;18:474–485. doi: 10.1016/j.tcb.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 84.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. NatCell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liang C, Sir D, Lee S, Ou JH, Jung JU. Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy. 2008;4:817–820. doi: 10.4161/auto.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 87.The C. elegans Research Community, editor. WormBook: Methods in Cell Biology. In: Shaham S, editor. WormBook. WormBook; 2006. Jan 2, doi/10.1895/wormbook.1.49.1, http://www.wormbook.org. [Google Scholar]
- 88.Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- 89.Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.