Abstract
M2 of the influenza virus is an intriguing transmembrane protein that forms a minuscule proton channel in the viral envelope. Its recognized function is to equilibrate pH across the viral membrane during cell entry and across the trans-Golgi membrane of infected cells during viral maturation. It is vital for viral replication and it is a target for the anti-influenza drugs, amantadine and rimantadine. Recently, high resolution structures of M2 channels of both flu A and B have been obtained, providing the desperately needed structural details for understanding the mechanism of proton conductance. In particular, the establishment of the functional solution NMR system of the proton channels enabled simultaneous high resolution structure characterization and measurement of channel dynamics coupled to channel activity. This review summarizes our current understanding of how protons are conducted through the M2 channel from a structural point of view, as well as the modes by which important channel gating elements function during proton conduction.
Keywords: M2, AM2, BM2, proton channel, influenza, flu
1. Introduction
Influenza is an enveloped, negative sense RNA virus belonging to the orthomyxoviridae family. There are three major serotypes: A, B, and C. The virion particle, of influena A, is round or oval in shape and approximately 90 nm in diameter [1]. The viral membrane is decorated with a large number of evenly spaced glycoproteins: hemagglutanin (HA) and neuraminidase (NA), at approximately four to one ratio, respectively [2]. The membrane also contains the matrix protein 2 (M2 or AM2 for influenza A M2), although in much lower abundance (1:10-100 M2:HA) [3] (Fig. 1A). Viral nucleocapside consists of eight separate segments of single-stranded negative-sense RNA, multiple copies of nucleoprotein (NP), and polymerase complex. Six of the eight segments of RNA (1-6) code for a single viral protein, and two strands (7 and 8) code for two proteins [1]. M2 protein is encoded on the seventh RNA segment together with the matrix protein 1 (M1). The ribonucleoprotein (RNP) and the lipid envelope are, presumably, connected through interaction with M1 (Fig. 1A). Similar to flu A, influenza B and C also contain the complex of three polymerase proteins and multiple copies of nucleoprotein (NP) [1]. However, unlike influenza A, flu B has four membrane proteins (HA, NA, BM2 (influenza B M2), and NB) and flu C contains only two (the CM2 (incfluenza C M2) protein and the hemagglutinin-exterase-fusion (HEF) protein that combines the functions of HA and NA) [1].
Figure 1.
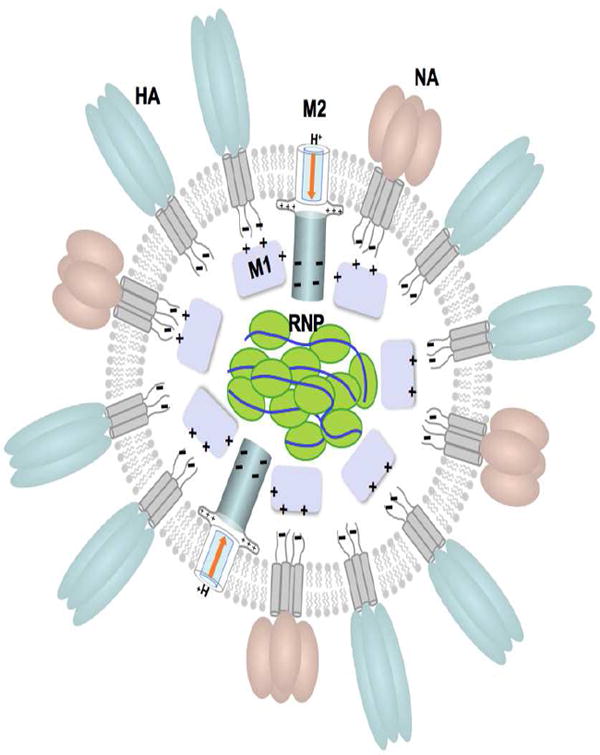
Viral particle of influenza with its membrane proteins: hemagglutinin – HA, neuraminidase – NA, matrix protein 2 M2; and associated proteins: matrix protein 1 – M1, ribonucleoprotein – RNP (A) A model of the viral particle. (B) Viral budding: HA of influenza A (PDB code: 5HMG), NA of influenza A (PDB code: 1ING), M2 of influenza B (PDB code: 2KIV and 2KJ1).
During infection the virus binds to an epithelial cell of the respiratory tract and enters the cell by receptor mediated endocytosis. In the endosome the virus experiences at least two pH changes, one from the extracellular pH to the early endosomal pH (~ 6) and later to the late endosomal pH (~ 5) [4]. The latter step is critical for the membrane fusion as it activates HA to catalyze the fusion of the viral envelope with the endosomal membrane [4]. Prior to membrane fusion, the low pH of the endosome activates the M2 channel to conduct protons across the viral envelope, which results in the acidification of the viral interior [5, 6]. It has been suggested that this acidification weakens electrostatic interaction between M1 and RNP complexes such that subsequent membrane fusion can release the uncoated RNPs into the cytosol [7]. Once in the cytosol, RNPs are trafficked to the host cell nucleus, where both mRNA and vRNA are synthesized [2, 7, 8]. The viral membrane proteins, HA, NA, and M2, are translated and inserted into the endoplasmic reticulum (ER) and then transported to the cell surface by trans-Golgi network (TGN). At this stage, M2 plays yet another role in the viral life cycle; it prevents the Golgi lumen pH from becoming too low so that nascent HAs do not undergo premature conformational rearrangement while they are transported to the plasma membrane of the infected cells [9, 10].
M2 of influenza A (AM2) is a 97-residue single-pass membrane protein with its N terminus directed toward the outside of the virus [6]. It consists of three segments: an extracellular N-terminal segment (residues 1-23), a transmembrane (TM) segment (residues 24-46), and an intracellular C-terminal segment (residues 47-97). Cross linking experiments showed that the channel is formed by four parallel monomers, in which the formation of inter-monomer disulfide bonds at Cys17 and Cys19 may stabilize, but they are not essential, the oligomeric assembly [10, 11]. Mutagenesis studies have identified two pore-lining residues important for channel function, the imidazole of His37 and the indole of Trp41. These residues determine proton selectivity and unidirectional conductance of the channel [12, 13].
BM2 of influenza B is a functional homolog of AM2. It is 109-residue long and, similar to AM2, forms a homotetramer in a membrane [14]. After being synthesized in infected host cells, BM2 is incorporated into the TGN membrane and transported to the cell surface for virus budding [15]. BM2 proton conductance has a pH profile similar to that of AM2 [16]. There are, however, significant differences between the two channels. Except for the HXXXW sequence motif in the TM domain that is essential for channel function, the two proteins share almost no sequence homology. Additionally, BM2 channel activity is higher than that of AM2 [16-18] and unlike AM2, the BM2 activity is completely insensitive to the anti-influenza drugs amantadine and rimantadine [16].
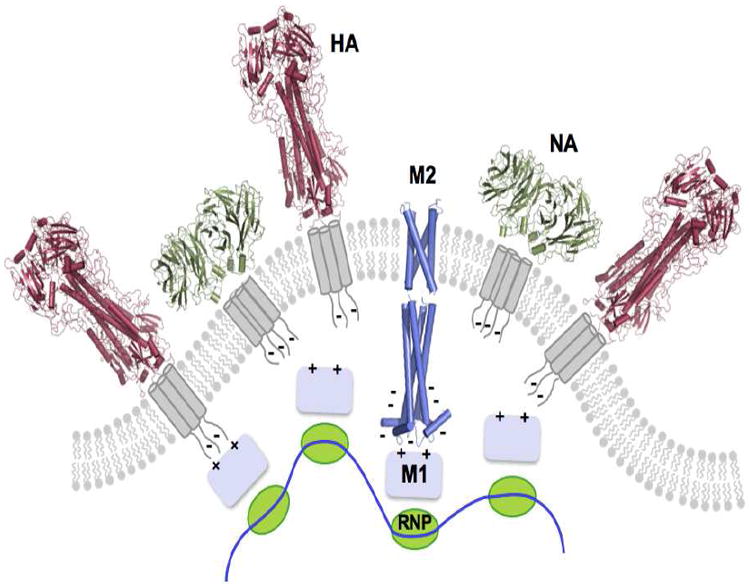
In addition to the membrane-embedded channel domain, AM2 and BM2 have large cytoplasmic regions. These regions have been suggested to play a role during viral assembly (Fig. 1B). Deletions and mutations of the AM2 cytoplasmic region cause incomplete incorporation of genomic RNA into the virion and defective virus budding. These defects may be attributed to the interrupted association between the cytoplasmic region of AM2 and the AM1 protein [19, 20]. Similar role in viral assembly has been suggested for the BM2 cytoplasmic domain. Deleting residues 51 – 80 or mutating residues 86-109 in the cytoplasmic region of BM2 decreases association of BM1 to the viral membrane and results in the failure of packaging of the RNP complex into the virion [21, 22]. Understanding the role of M2 cytoplasmic domains in virus assembly could offer new opportunities for developing anti-influenza therapeutics.
2. Structures of the AM2 channel
Solution structure of the closed state of AM2
Before the availability of high-resolution structures, a number of structural models of the channel region had been constructed based on biochemical experiments and computational modeling [23-26], site-directed infrared dichroism [27], and solid-state NMR (ssNMR) spectroscopy [25, 28-32]. Although the above models agree on the overall topology, they differ in the details essential for understanding the mechanism of channel opening, drug inhibition and drug resistance.
The closed state of AM2 was determined by solution NMR spectroscopy at pH 7.5 bound to the drug rimantadine. The structure includes residues 18 to 60, denoted here as AM2(18-60), which comprises the TM domain and 15 residues of the C-terminal extension [33]. Functional studies have shown that the C-terminal extension is crucial for a stable tetramer formation [33, 34], native-like conductance [35], and sensitivity to rimantadine [17]. In the closed conformation, AM2(18-60) contains an unstructured N terminus (residues 18-23), a channel-forming TM segment (residues 25-46), a flexible loop (residues 47-50) and a C-terminal amphipathic (AP) helix (residues 51-59) [33]. The TM helices form a four-helix bundle with a left-handed twist angle of ~23° and a well-defined pore [33]. The helices are tightly packed at the N terminus and they slightly splay out toward the C terminus (Fig. 2A). A ring of methyl groups from Val27 constricts the N-terminal side of the pore to ~3.1 Å, narrowing the entrance and restricting water molecules from penetrating the channel. Small motion or “channel breathing” may thus be required for water to enter the channel. It is widely accepted that water molecules are needed inside the channel pore for supporting proton conduction. Water nuclear Overhauser enhancement (NOE) experiment indicates the presence of water molecules near Ser31. The pore widens after Ser31 and becomes the widest at Gly34 position with an inner diameter of ~ 6 Å. The channel then narrows towards the C terminus as the sidechains of His37 and Trp41 constrict the channel to 1.7 and 1.4 Å (inner diameter), respectively. In particular, the four indoles from Trp41 pack within van der Waals (VDW) distance of each other. Moreover, Trp41 indole amine of one subunit is within hydrogen bonding distance of the Asp44 carboxyl carbon of the adjacent subunit. They likely form inter-subunit hydrogen bonds that lock the channel gate in the closed conformation. Arg45 is also in the vicinity for forming inter-subunit salt bridge with Asp44. The channel C-terminal side extends into a short loop (residues 47 – 50) that connects the TM domain to the C-terminal AP helix (residues 51 – 59). The AP helices also form a separate tetrameric domain with a head-to-tail assembly and a right-handed packing mode. Based on a pure structural analysis (Fig. 2A), the tetrameric assembly of the AP helices functions to further stabilizes the tetrameric state of TM helices. This is particularly important for the presumably less tightly packed open states of the channel.
Figure 2.
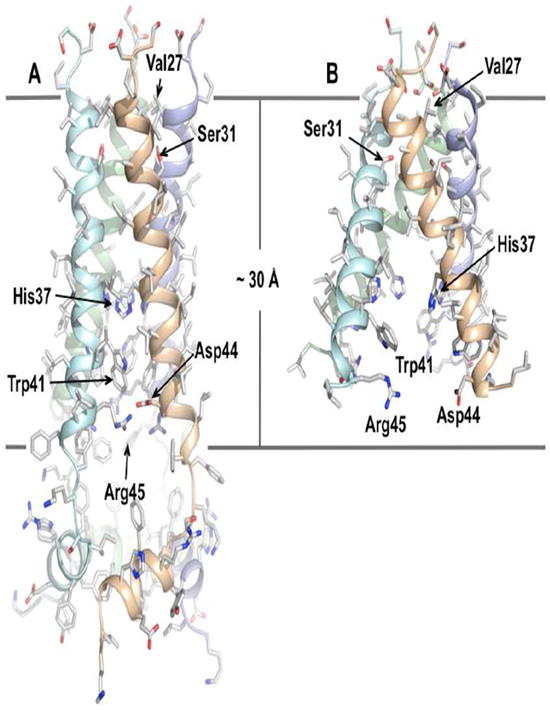
High resolution structures of the AM2 channel domain. (A) Solution structure of residues 18 – 60 in DHPC (1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine) micelles at pH 7.5 (PDB code: 2RLF) (Schnell and Chou 2008). (B) Crystal structure of residues 22 – 46 with the I33Se-Met mutation in OG (octyl-β-D-glucopyranoside) micelles at pH 7.3 (PDB code: 3BKD) (Stouffer, Acharya et al. 2008).
The drug rimantadine, which was important for stabilizing the closed state of the channel for NMR structure determination, binds at a lipid-facing pocket near the C-termininal end of the channel domain. This pocket is formed by Trp41, Ile42, and Arg45 from one TM helix and Leu40, Leu43, and Asp44 from the adjacent TM helix, and shows a unique amphipathic property. In this pocket, the amino group of rimantadine is in contact with the polar sidechain of Asp44 and possibly also Arg45. The polycyclic hydrocarbon cage of the drug forms hydrophobic interactions with Ile42 from one TM helix and Leu40 and Leu43 from the adjacent helix. However, the relevance of this binding site to channel inhibition is still under debate. In this review, we will focus on the mechanism of proton conductance and will not discuss drug binding and inhibition. An extensive review of the current different models of drug inhibition as well as their respective implication to developing new therapeutics is published elsewhere (Pielak and Chou, Protein & Cell, 2010).
Crystal structure of the TM segment of AM2
In a separate study, the crystal structure of a shorter construct AM2(22-46) in β-octylglucoside detergent was determined at 2.05 Å resolution [36]. As in the solution structure, the TM helices pack to form a left-handed four-helix bundle. However, while the helices are tightly assembled at the N terminus, they dramatically splay outward at an average angle of ~35° at the C terminus (Fig. 2B). As a result, the C terminus of the channel is wide open with no obvious structural features that support proton gating or selection (e.g., the Trp41 indoles from adjacent TM helices are on average ~9.5 Å apart). This wide opening of the channel is clearly inconsistent with the fact that the structure was determined at pH 7.3, which in principle supports the closed channel conformation. A plausible explanation for the large opening at the C terminus is that the TM peptide used for the crystallographic study does not contain the C-terminal region (residues 47 – 60), which is necessary for stable tetramer formation and native-like rates of proton flux [34, 35]. Alternatively, this unrealistic degree of splaying of TM helices could be due to crystal packing or combination of both. Therefore, in this review we will use the solution structure for discussing the mechanism of proton relay.
Properties of the open state of AM2
Although a well-defined structure of the open channel is not yet available, some features of the open state may be inferred from dynamics data obtained by solution NMR. Channel activation occurs upon protonation of the His37 imidazoles that are closely packed in the channel pore, which results in electrostatic repulsion that could substantially weaken helical packing in the TM domain. Presumably this widens the pore to admit water molecules. The NMR pH titration experiment showed that lowering the pH from 7.5 to 6.0 caused severe broadening of most of the TM NMR resonances due to exchange between multiple conformations [33]. The NMR chemical shift perturbation data suggest that the open state is not a unique structure like the closed state. AM2 lacks the extensive structural scaffolding observed in larger ion channels that undergo specific conformational changes between the open and closed conformation. Thus, AM2 appears to have evolved a two-state gating mechanism in which the closed state is structurally rigid, but the open state is dynamic with a loose quaternary structure.
In contrast to the TM domain, the NMR resonances of the AP helices are essentially unaffected by lowering the pH, indicating that their tetrameric assembly remains intact and may be needed to preserve the overall tetrameric state of the channel when the TM helices are destabilized in the open state. In addition to the AP helices, a pair of N-terminal cysteines, Cys17 and Cys19, have been shown to form intermolecular disulfides in vivo [10, 11]. Although these cysteines are not required for channel activity, they are conserved in nature and may play a role in keeping the tetramer together in the open state.
3. Solution structures of BM2
BM2 channel domain
Although the overall assembly of TM helices of BM2 is similar to that of AM2, e.g., both are left-handed four-helix bundle having a hydrophilic pore, the two channels differ substantially in details. Unlike AM2, the TM domain of BM2 shows strong coiled-coil characteristics with heptad repeats. It is the only known ion channel structure that adopts a coiled-coil assembly to conduct ions. The coiled-coil arrangement of BM2 allows the TM segment to form a stable tetramer by itself [18].
In the closed state, at pH 7.5, the BM2 TM domain, which includes residues 1-33, forms a coiled-coil tetramer with a packing angle of about −37° (Fig. 3A) [18]. The tetramer has a well-defined hydrophilic channel that is occluded by Phe5 and Trp23 at the N-, and C-terminal side of the pore, respectively [18]. The coiled-coil structure of the channel domain shows two heptad repeats: one from Leu8 at position g to Ile14 at position f, and the other from Leu15 at position g to Ile21 at position f (Fig. 3B). Positions a and d are occupied mostly by hydrophilic residues such as Ser9, Ser12, and Ser16, and constitute the core of the coiled-coil tetramer. His19 at position d and Trp23 at position a are also pore-lining, as expected of their roles in proton selectivity and gating (Fig. 3a). Positions g and e are occupied by leucines 8 and 15 and phenylalanines 13 and 20, respectively, to allow for peripheral hydrophobic interactions that stabilize the coiled-coil assembly [18] (Fig. 3a). The above amino acids in positions a, d, g, and e are completely conserved. The rest of positions, b, c, and f, of the heptad repeat are occupied by hydrophobic residues (with the exception of Ser11), which form the hydrophobic surface for membrane partition. This arrangement for coiled-coil assembly in membrane is an inverse of the water-soluble coiled-coil tetramers, in which positions a and d are typically hydrophobic residues and positions g and e are polar residues [37].
Figure 3.
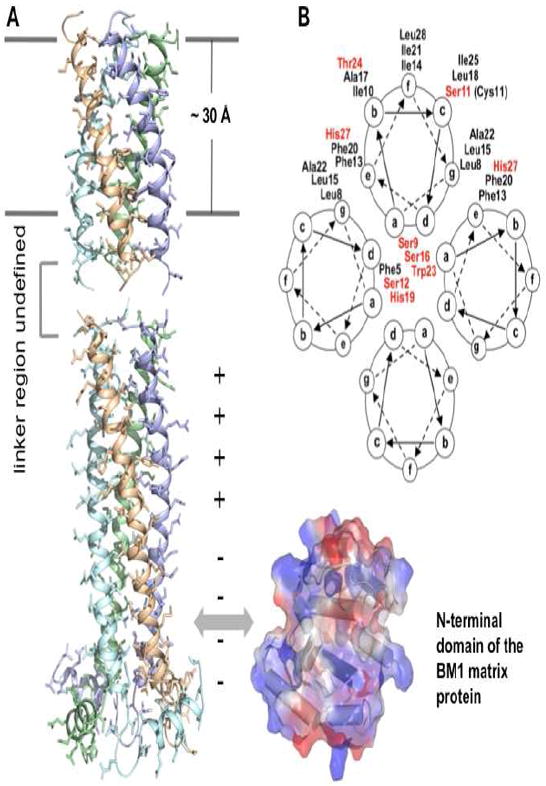
The BM2 structures. (A) Solution structures of BM2 channel (residues 1 – 33) and cytoplasmic (residues 44 – 103) domains. The channel domain (PDB code: 2KIX)(Wang, Pielak et al. 2009) is positioned relative to the hydrophobic region of the presumed lipid bilayer as defined by two lines. The structure of the linker region (residues 34 – 43) has not been defined. The cytoplasmic domain (PDB code: 2KJ1)(Wang, Pielak et al. 2009) interacts with the BM1 matrix protein, probably through the BM1 N-terminal domain shown on the right. (B) Helical wheel representation of the BM2(1-33) coiled coil tetramer. Polar resides are presented in red.
BM2 cytoplasmic domain
Residues 45 – 85 of the BM2 cytoplasmic domain form an uninterrupted helix that oligomerizes into a left-handed coiled-coil tetramer (Fig. 3A). A hairpin-like structure, consisting of residues 86 – 92, connects the coiled-coil structure to a short amphipathic helix that is roughly perpendicular to the coiled-coil helix. Although no inter-subunit NOEs were observed for residues 34 – 43, the region that connects the TM and cytoplasmic domain, residues 39-45 show NOEs to the glycerol protons of LMPG headgroup. The BM2 cytoplasmic domain shows a striking polarity in surface charge distribution, that the N-terminal half of the domain (residues 44-71) is almost entirely positive and the C-terminal half (residues 72 – 103) is almost entirely negative. This charge separation results in a large electrostatic dipole moment, 4215 Debyes, at neutral pH that is about 4 standard deviations above the mean of all proteins in the database [38].
The unusually strong dipole moment supports its interaction with the matrix protein BM1, which also has a strong dipole moment [39]. Perturbation of chemical environment of a defined region of the cytoplasmic domain by the M1 matrix protein indicates specific molecular recognition between the two proteins [18]. The perturbed region identified, from residues 84 to 108, is consistent with known deletions and mutations of BM2 that affect virus assembly. Viruses with BM2 deletion of 101-109 contained dramatically reduced RNP complex and affected membrane association of M1 [22]. Furthermore, alanine-scanning substitution of three consecutive residues showed that the 86-88A, 89-91A, 93-94A, and 95-97A mutants did not grow normally, and contained substantially reduced levels of M1 and nucleoprotein. The data from structural and reverse genetics studies indicate that the interaction between the cytoplasmic regions of the proton channels and matrix proteins play an important role in viral assembly.
During virus budding, the matrix proteins and RNPs must coat the plasma membrane such that budding would result in a properly assembled virus. Therefore, the membrane patch that is destined to bud out from the host cell must contain specific sites for recruiting the matrix protein and RNP complexes (Fig. 1B). The unusually strong electric dipole moment of BM2 cytoplasmic domain may serve to orient the M1 matrix protein, which also has a strong electric dipole moment, for specific association. Electric dipole facilitated molecular recognition is commonly observed in cellular signaling pathways, e.g., the interactions between the caspase recruitment domains [40]. The coating of M1 and RNPs to the virus membrane is likely achieved with cooperative interactions of M1 to the negatively charged membrane, the short cytoplasmic tails of HA and NA, and the cytoplasmic domain of M2 (Fig. 1B).
4. Mechanism of proton conductance
Protons are distinct from other ions in biological systems because they are in dynamic exchange with water, buffers, and titratable groups of lipids, proteins, and other cellular molecules. Therefore, moving protons across cell membranes normally require mechanism distinct from those adopted by other types of ion channels. Mobility of protons in aqueous solutions is about five times greater than that of monovalent cations similar in size and charge to hydronium ion (Reviewed in [41, 42]). It has been proposed that protons can move in water by hopping from one water molecule to another along water-wires, a chain of hydrogen-bonded water molecules, as opposed to random diffusion of hydronium ions. The water-wire mechanism, commonly referred to as the Grotthuss model, has been proposed to exist in the gramicidin A proton channel (Reviewed in [42, 43]). In other cases, such as bacteriorhodopsin, titratable amino-acid sidechains can establish similar hydrogen-bonded chain for proton conductance (Reviewed in [42]). Do the M2 channels use a hydrogen-bonded chain to conduct protons or other yet to be determined mechanism of proton relay?
Roles of His and Trp in channel activation
Probably the most important channel elements in M2 are histidine and tryptophan in the TM domain. The HXXXW sequence motif in the TM segment is not only absolutely conserved within the M2 variants but it is also the only sequence element shared between AM2 and BM2. Early whole cell recording experiments on AM2 showed that His37 is essential for proton selectivity and pH modulation. Currents of the H37G and H37E mutants were non-selective and did not exhibit pH dependence; they are also higher than those of WT [12]. Similar experiments showed that mutating Trp41 of AM2 to Ala, Cys, or Phe resulted in larger inward currents as well as outward currents that were not observed for the WT [13], indicating that Trp41 regulates unidirectional conductance. However, an alternative explanation of the data from Ref. 13 is presented in a recent review by Busath [44]. Based on these findings it was suggested that protonation of His37 may lead to the opening of the gate, thus allowing protons to pass to the C-terminal side of the channel [45]. Protonation of His37 was observed in ssNMR experiments, indicating three distinct pKa values 8.2, 6.3, and one below pH 5 [46]. It was then suggested that in the non-conductive state two pairs of histidines share protons (pKa 8.2) and the subsequent protonation (pKa 6.3) results in disruption of the histidine dimers that leads to conductive state [46]. Furthermore, it was suggested that protonation of histidines leads to cation-π interaction between the histidine imidazoles and the tryptophan indoles that may play a role in the channel opening [47, 48]. The proposed role of His/Trp in gating is consistent with the solution structure of the closed channel. The Trp41 indoles form a tight ring (1.4 Å in diameter) that prevents water molecules from accessing His37 from the C-terminal side of the pore (Fig. 4A&B). The 3-bond scalar coupling between backbone N and sidechain Cγ of Trp41 is 2.6 Hz, indicating that the χ1 rotamer is locked at the trans conformation. Moreover, Trp41 χ2 is also fixed at around -120° based on NOE and residual dipolar coupling (RDC) data. Thus the indoles of Trp41 are mostly locked in the closed channel. The solution NMR study also showed that these indole gates become unlocked as the channel is activated at lower pH. Relaxation-compensated Carr-Purcell-Meiboom-Gill (CPMG) measurement [49] of millisecond timescale dynamics of the Trp41 indole amine at different pH showed that as the pH was lowered from 7.5 to 6.0, the chemical exchange rate increased by more than four-fold, and that binding of rimantadine, an M2 inhibitor, decreased the exchange rate by about two fold [33]. Clearly, the dynamics of Trp41 indoles is closely coupled to channel activation, either through destabilization of TM helix-helix packing in the open state or through interaction with protonated His37 imidazoles, or both.
Figure 4.
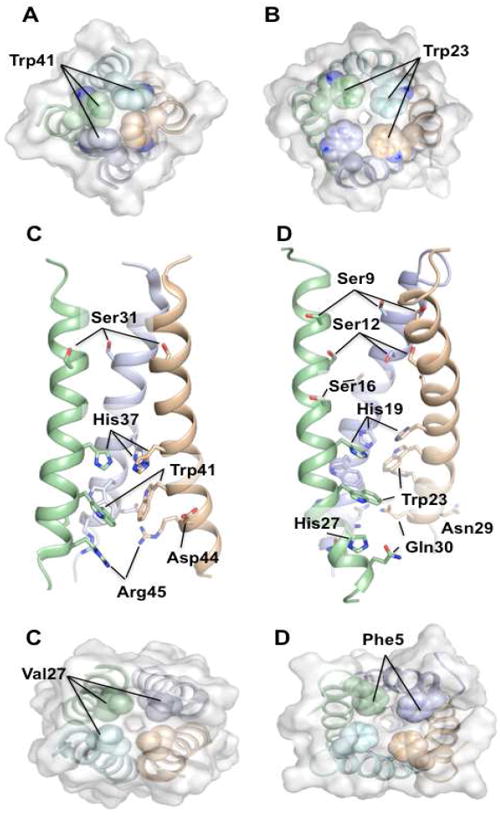
Structural elements of AM2 and BM2 important for channel function. (A) and (B) show the C-terminal tryptophan gates of the AM2 (PDB code: 2RLF)(Schnell and Chou 2008) and BM2 (PDB code: 2KIX)(Wang, Pielak et al. 2009) channels, respectively. (C) and (D) show the amino acid sidechains important for proton relay, selection, and gating in the TM domains of AM2 and BM2, respectively. (E) and (F) show the N-terminal constriction of the AM2 and BM2 channel, respectively.
Somewhat different results were obtained for the BM2 channel. Replacing His19 with either alanine or cysteine almost abolished channel conductance, whereas mutating Trp23 to either alanine, cysteine, or phenylalanine resulted in high non-selective currents [16, 50, 51]. It was also shown that protonation of His19 leads to cation-π interaction with Trp23 indoles, which is consistent with the AM2 data [50].
Roles of polar residues in proton conductance
Both AM2 and BM2 contain a number of polar residues lining the channel interior (Fig. 4C&D). These residues may be involved in channel hydration as a requirement for proton relay. Indeed, mutating these residues to more hydrophobic amino acids results in lower rates of proton flux [17, 18]. The BM2 channel is very hydrophilic, containing number of polar residues: Ser9, Ser12, Ser16, His19, His27, Asn29, and Gln30 (Fig. 4B). Replacing any of the serines on the N-terminal side of the His19 substantially impair activity (S9A: ~70%; S12A and S16A: ~55% of the WT rate of proton flux; rates were obtained at pH ~6.0) [18]. These residues may function collectively to mediate the entrance of protons and to relay them to His19, which together with Trp23 forms the gating element. Similarly, replacing polar residues on the C-terminal side of the gate also affects proton conduction (H27A and Q30A: ~75% of the WT rate of proton flux at pH ~6.0) [18], suggesting that they facilitate proton exit at the C-terminal side of the channel.
AM2 has only four residues within the TM region capable of relaying protons: Ser31, His37, Asp44, and Arg45 (Fig. 4a). Smaller number of polar residues may account for about two-fold slower rates of proton conduction as compared to BM2 [17, 18]. Mutating Ser31, which is on the N-terminal site of the gate, to alanine results in moderate decrease in rate of proton flux (90% of the WT at pH ~6.0); however, replacing Asp44, which may facilitate proton exit, with alanine dramatically lowers rates of proton flux (30% of the WT at pH ~6.0) [17]. The NMR structure of the closed channel shows that the region on the C-terminal side of the Trp41 gate is densely packed with hydrophobic phenylalanines, sealing the channel at the C-terminal side of the pore. Asp44 and Arg45 are the only polar residues in this otherwise hydrophobic portion of the channel. Therefore they most likely serve to assist proton exit by accepting protons or hydroniums and release them laterally to the hydrophilic head group region of the membrane. Asp44 is highly conserved (it is occasionally replaced by asparagine) whereas Arg45 is completely conserved. It is interesting to note that Asp44 is located at the center of the observed rimantadine binding pocket in the NMR structure. Drug binding at this position could inhibit channel activity by simply blocking the proton exit.
Mechanism of conductance
Although some of the functional results (e.g. conductance of histidine mutants above) differ significantly between AM2 and BM2, similar conclusions about the mechanism of proton relay can be drawn. Both AM2 and BM2 proton fluxes are driven by proton gradient and are unidirectional from N to C terminus. Both channels have their N termini physically constricted by hydrophobic sidechains – Val27 in AM2 (Fig. 4E) and Phe5 in BM2 (Fig. 4F), thus helical assembly dynamics or “channel breathing” may be required to facilitate entrance of water molecules or hydronium ions into the channel. Inside the channel, polar residues facilitate channel hydration, thus allowing protons to reach the histidines. Upon protonation, histidine imidazoles are positively charged and are in position to interact with tryptophan indoles through cation-π interaction. This interaction may play an important role in channel opening. While we have a rather clear understanding of a number of aspects of channel activation, in particular, those confirmed by the channel structure, a complete picture including both channel activation and proton replay is still missing. Based on the existing structural and functional data, two equally plausible models could be proposed. One is that M2 becomes a continuous hydrated pore at low pH, which allow protons to be conducted either by hopping along a water wire or by diffusion of hydronium ions. The alternative is a proton shuttling mechanism, where protons are transported from the N-terminal side to the C-terminal side of the His/Trp gating element by means of local conformational switch.
5. Conclusions and perspectives
We have summarized, based on the existing structural and functional data of AM2 and BM2, the possible mechanism of proton conduction through the M2 channels. The virus evolved two significantly different structural solutions for assembling a polar pore in the membrane with four TM helices for conducting protons. The AM2 structure shows a rather simple four helix bundle packing mode with the N-terminal half of the helices being tightly packed but the C-terminal half loosely interacting, and hence it requires the tetrameric packing of the AP helices on the cytoplasmic side for maintaining the structural integrity of the channel. On the other hand, BM2 TM adopts a distinct coiled-coil packing strategy, which is an inverse of a typical water-soluble coiled-coil structure (e.g., the GCN4 tetramer). This coiled-coil assembly provides strong inter-helical interaction, which results in a more stable TM tetramer. Despite the different structural solutions adopted by these two channels, the result is similar. Both tetramers form well-defined hydrophilic pores with residues constricting the N and C termini of the pore. The two channels also have similar arrangements of the pore-lining histidines and tryptophans for modulating flux rates in a pH-dependent manner and for regulating the directionality of the proton flux. In the closed state, the channel lumens do not appear to have enough structured water molecule to support the formation of a proton-conducting wire. It is possible, however, that a transient wire forms upon channel opening. Another possibility is that the protons are simply translocated by diffusion of hydronium ions, or by a shuttling mechanism, where protonation of histidine results in a conformational change that exposes it to proton acceptors (water) near the C-terminal channel exit. Regardless of the mechanism, the slow rates of proton flux suggest a high activation barrier at some point of the proton relay. Finally, the flu viruses do not use the M2 proteins only for proton conductance. Their relatively large cytoplasmic region are well structured, as was observed for BM2 and can specifically interact with the M1 matrix protein. Therefore, these “proton channels” have the dual functionality of conducting protons and recruiting other proteins during viral assembly.
Although the above structures illustrate how these simplistic four-helix bundle structures support proton conduction, the exact mechanisms by which protons are translocated through the channels remain to be understood and many questions await answers. Does histidine protonation merely activate the channel by electrostatic repulsion, or instead the imidazoles actively participate in proton transfer? Do protons reach the histidines by diffusion of hydronium ions or they travel along a water wire? Why did this enigmatic protein evolve to conduct protons in such a slow pace? Either a pure water wire or simple diffusion of hydronium would provide much faster proton flux rates. However, the virus seems to prefer this miniscule conductance. Finally, what is the mechanism of the high proton selectivity of M2 and why is it necessary for the virus? All of these questions are of high importance not only to influenza biologists but also to increase our general understanding of proton conducting channels, since so little is known about the mechanisms of proton translocation across cell membranes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. Williams & Wilkins; 2007. [Google Scholar]
- 2.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 7.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 8.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugrue RJ, Bahadur G, Zambon MC, Hall-Smith M, Douglas AR, Hay AJ. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 14.Paterson RG, Takeda M, Ohigashi Y, Pinto LH, Lamb RA. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S, Imai M, Ohara Y, Odagiri T. Influenza B virus BM2 protein is transported through the trans-Golgi network as an integral membrane protein. J Virol. 2003;77:10630–10637. doi: 10.1128/JVI.77.19.10630-10637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mould JA, Paterson RG, Takeda M, Ohigashi Y, Venkataraman P, Lamb RA, Pinto LH. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 17.Pielak RM, Schnell JR, Chou JJ. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci U S A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai M, Watanabe S, Ninomiya A, Obuchi M, Odagiri T. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J Virol. 2004;78:11007–11015. doi: 10.1128/JVI.78.20.11007-11015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai M, Kawasaki K, Odagiri T. Cytoplasmic domain of influenza B virus BM2 protein plays critical roles in production of infectious virus. J Virol. 2008;82:728–739. doi: 10.1128/JVI.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauer CM, Pinto LH, Cross TA, Lamb RA. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 25.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Expression and initial structural insights from solid-state NMR of the M2 proton channel from influenza A virus. Biochemistry. 2002;41:11294–11300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- 27.Kukol A, Adams PD, Rice LM, Brunger AT, Arkin TI. Experimentally based orientational refinement of membrane protein models: A structure for the Influenza A M2 H+ channel. J Mol Biol. 1999;286:951–962. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: an intrinsic property. J Mol Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Kim S, Kovacs F, Cross TA. Structure of the transmembrane region of the M2 protein H(+) channel. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovacs FA, Cross TA. Transmembrane four-helix bundle of influenza A M2 protein channel: structural implications from helix tilt and orientation. Biophys J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Z, Kovacs FA, Wang J, Denny JK, Shekar SC, Quine JR, Cross TA. Transmembrane domain of M2 protein from influenza A virus studied by solid-state (15)N polarization inversion spin exchange at magic angle NMR. Biophys J. 2000;79:767–775. doi: 10.1016/S0006-3495(00)76334-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobler K, Kelly ML, Pinto LH, Lamb RA. Effect of cytoplasmic tail truncations on the activity of the M(2) ion channel of influenza A virus. J Virol. 1999;73:9695–9701. doi: 10.1128/jvi.73.12.9695-9701.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 38.Felder CE, Prilusky J, Silman I, Sussman JL. A server and database for dipole moments of proteins. Nucleic Acids Res. 2007;35:W512–521. doi: 10.1093/nar/gkm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 40.Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 41.DeCoursey TE, Cherny VV. Common themes and problems of bioenergetics and voltage-gated proton channels. Biochim Biophys Acta. 2000;1458:104–119. doi: 10.1016/s0005-2728(00)00062-1. [DOI] [PubMed] [Google Scholar]
- 42.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 43.Roux B. Computational studies of the gramicidin channel. Acc Chem Res. 2002;35:366–375. doi: 10.1021/ar010028v. [DOI] [PubMed] [Google Scholar]
- 44.Busath DD. Influenza A M2: Channel or Transporter? In: Leitmannova Liu AI, editor. Advances in Planar Lipid Bilayers and Liposomes. Vol. 10. Academic Press; 2009. [Google Scholar]
- 45.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 46.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci U S A. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeuchi H, Okada A, Miura T. Roles of the histidine and tryptophan side chains in the M2 proton channel from influenza A virus. FEBS Lett. 2003;552:35–38. doi: 10.1016/s0014-5793(03)00781-6. [DOI] [PubMed] [Google Scholar]
- 48.Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza a virus. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 49.Loria JP, Rance M, Palmer AG., 3rd A relaxation-compensated carr-purcell-meiboom-gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 50.Otomo K, Toyama A, Miura T, Takeuchi H. Interactions between histidine and tryptophan residues in the BM2 proton channel from influenza B virus. J Biochem. 2009;145:543–554. doi: 10.1093/jb/mvp009. [DOI] [PubMed] [Google Scholar]
- 51.Betakova T, Hay AJ. Comparison of the activities of BM2 protein and its H19 and W23 mutants of influenza B virus with activities of M2 protein and its H37 and W41 mutants of influenza A virus. Arch Virol. 2009 doi: 10.1007/s00705-009-0483-9. [DOI] [PubMed] [Google Scholar]


