Abstract
Most linkage and population genetic studies that use microsatellites assume that the polymorphism observed at these loci is due simply to variation in the number of units of a single repeat. Variation is far more complex, however, for the numerous microsatellites that contain interruptions within the repeat or contain more than one type of repeat. We observed that for D18S58, a compound microsatellite containing (CG)m, as well as (CA)n repeats, the apparent length of certain alleles varied between genotyping experiments. Similar results were obtained with other (CG)m–(CA)n repeats. Sequencing demonstrated that the D18S58 alleles demonstrating variable mobility contained longer (CG)m stretches than those alleles whose length did not appear to vary between experiments. These results suggest that (CG)m repeats, which are frequently present in compound human microsatellites, are prone to form an unusually stable secondary structure. We discuss the relative frequency of different classes of compound microsatellites identified through database searches, as well as their patterns of sequence and variation. Further characterization of such variation is important for elucidating the origin, mutational processes, and structure of these widely used, but incompletely understood, sequences.
Microsatellites are tandemly repeated sequences of 1–6 bp (Tautz 1993). They have been used extensively for genetic mapping and forensic and population studies. However, much remains unknown about the possible functions microsatellites may have in the genome and about their patterns of sequence variation and mutation. A premise behind the use of microsatellites as genetic markers is that their alleles differ only in the number of units that they contain of a single repeat (Guyer and Collins 1993). Several studies, however, show that sequence variation at microsatellites is frequently complex (e.g., Eichler et al. 1994; Hirst et al. 1994; Kunst and Warren 1994; Snow et al. 1994; Urquhart et al. 1994; Blanquer-Maumont and Crouau-Roy 1995; Estoup et al. 1995; Garza and Freimer 1996; Grimaldi and Crouau-Roy 1997; Brinkmann et al. 1998a,b; Lin et al. 1998). Variation in the sequence of microsatellite alleles may affect the interpretation of genetic mapping and population studies in which microsatellites are used.
Complexity in interallelic variation takes three main forms. First, microsatellite alleles can vary due to small insertion–deletion or single base pair mutations in the sequence immediately flanking the repeat, in conjunction with differences in the number of repeat units (Grimaldi and Crouau-Roy 1997; Brinkmann et al. 1998a; Lin et al. 1998). Second, base substitutions or small insertions or deletions (i.e., imperfections) may occur within the repeat (Eichler et al. 1994; Hirst et al. 1994; Kunst and Warren 1994; Snow et al. 1994; Urquhart et al. 1994; Blanquer-Maumont and Crouau-Roy 1995; Estoup et al. 1995; Garza and Freimer 1996; Brinkmann et al. 1998a,b; Lin et al. 1998). As base substitutions do not change the length of alleles, this type of variation is usually hidden, and sequencing of many alleles may be required to appreciate it.
A third type of variation can occur at compound microsatellite loci (i.e., those containing stretches of two or more different repeats), which appear to comprise ∼10% of microsatellites (Weber 1990). Alleles at such loci can vary in the length of either or both repeats (Urquhart et al. 1994; Garza and Freimer 1996; Brinkmann et al. 1998b). Therefore, with compound microsatellites, as with imperfect ones, sequencing can reveal differences between alleles that are identical in length.
We present here a detailed examination of a compound dinucleotide repeat from the Genethon genetic map (Gyapay et al. 1994). This marker, D18S58, contains a long (CA)n repeat and a short (CG)m repeat. We report observations from genotyping and sequencing experiments that show that both repeats vary in length and that the length of the (CG)m repeat likely affects the structure of the microsatellite. Results obtained from additional (CG)m–(CA)n markers suggest that this phenomenon occurs with some other (CG)m–(CA)n repeats. These experimental results, and sequence analysis of additional compound repeats identified in sequence databases, extend prior suggestions of the importance of complex variation in understanding the biology of microsatellites.
RESULTS
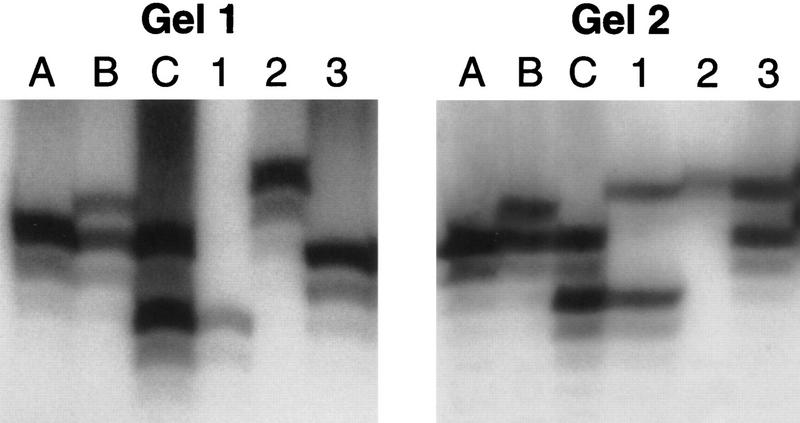
D18S58 is a compound microsatellite repeat containing a long (CA)n repeat and a short (CG)m repeat. The published sequence is G(CG)5(AC)18 (GenBank accession no. Z16735). While using this marker in a linkage study, we found that certain individuals carried alleles at D18S58 for which size could not be reliably established using standard genotyping procedures (termed “variable-mobility alleles”). In the linkage study these individuals had been typed with >400 microsatellite markers, and no unexpected allele sizes had been noted (McInnes et al. 1996). The apparent size of these variable-mobility alleles, determined relative to that of several nonvarying control alleles, differed between experiments, indicating that mobility on a denaturing acrylamide gel does not consistently reflect the actual size of these alleles (see Fig. 1; Table 1A). Occasionally, evidence suggested that the band representing the variable-mobility allele did not amplify by PCR to a detectable level (see Fig. 1, gel 1, lane 1).
Figure 1.
Sample D18S58 data. D18S58 data for six samples, run on two different standard gels, are shown. Samples A, B, and C are control individuals, who do not carry variable-mobility alleles. (Sample A is CEPH control 1347-2). Both of the gels were scored as follows (in bp): A (149/149), B (151/149), and C (149/145). Samples 1–3 are from individuals who carry one stable-mobility and one variable-mobility allele each. On gel 1, these samples were scored as follows (in bp): 1 (145/145), 2 (153/153), and 3 (149/149). On gel 2, they were scored (in bp) as 1 (152/145), 2 (153/152), and 3 (152/149). These three individuals were genotyped numerous times (see Table 1), and their D18S58 alleles were sequenced (see Table 2). Gel 1 shows an example of a case in which it is particularly likely that the variable-mobility allele simply did not amplify to a detectable level. Once on a standard gel (shown) and once on a formamide gel, sample 1 appeared homozygous for the 145 allele. The variable-mobility allele never appeared to be this small in samples 2 or 3, suggesting that, in this case, it simply did not amplify from sample 1. Whether the results for samples 2 and 3 on gel 1 are due to lack of detectable PCR of the variable-mobility allele or to comigration of the stable- and variable-mobility alleles cannot be determined.
Table 1.
Allele Mobility in Standard and 40% Formamide Gels
| Sample | A. Standard gels apparent size range (bp) | B. Formamide gels apparent size range (bp) | C. (CG)7-8 allele sequence length (bp) | D. (CG)5 allele sequence length (bp) | ||
|---|---|---|---|---|---|---|
| varying allele | stable allele | varying allele | stable allele | |||
| 1 | 145–153 | 145 | 145–155 | 145 | 155 (153, 151)a | 145 |
| 2 | 150–153 | 153 | 150–155 | 153 | 155 | 153 |
| 3 | 149–152 | 149 | 149–155 | 149 | 155 | 149 |
Apparent sizes of the variable- and stable-mobility alleles in the three individuals heterozygous for variable-mobility alleles are shown, as determined using standard (column A) and 40% formamide (column B) gels. Three control individuals without variable-mobility alleles were also typed as references. The relative mobility of their alleles did not change (see Fig. 1 for examples). The length of the (CG)7–8 and (CG)5 alleles, as determined by sequencing, are also shown. The variable-mobility allele present in sample 1 appeared to be 145 bp in size only twice and never electophoresed so rapidly in either sample 2 or 3; as this individual has a stable allele of 145 bp, this result may be due to lack of PCR of this allele in these two instances.
Due to variation in number of repeats in the sequenced clones of this allele, the allele length is uncertain but most likely is 155 bp.
Sequencing of Alleles
We identified seven individuals in whom a D18S58 allele varied in apparent size (these individuals each possessed one “variable” and one “stable” allele). We PCR-amplified D18S58 from genomic DNA of three of these individuals (1, 2, and 3) and cloned the products. Six to 10 clones from each individual were sequenced, and sequence was obtained for both alleles present in each person. In each of the three individuals whose alleles were sequenced, the length of the (CG)m repeat differed between their two alleles: The allele demonstrating stable migration patterns carried G(CG)5C, similar to the published sequence, whereas the variable-mobility allele contained G(CG)7–8C (see Table 2). No other differences in sequence were found between the stable and variable-mobility alleles. In each case, the length of the stable allele determined by sequencing agreed with that predicted by genotyping. In contrast, the variable-mobility alleles demonstrated a greater gel mobility than that predicted by their length, as determined by sequencing.
Table 2.
Sequence of D18S58 Alleles
| Sample | Sequence |
|---|---|
| Published sequence | (AT)3GCGCGCGCGCG-------(AC)18 |
| 1 stable allele | (AT)3GCGCGCGCGCGC------(AC)16 |
| 1 varying allele | (AT)3GCGCGCGCGCGCGCGCgc(AC)17–18a |
| 2 stable allele | (AT)3GCGCGCGCGCGC------(AC)20 |
| 2 varying allele | (AT)3GCGCGCGCGCGCGCGCGC(AC)18 |
| 3 stable allele | (AT)3GCGCGCGCGCGC------(AC)18 |
| 3 varying allele | (AT)3GCGCGCGCGCGCGCGCGC(AC)18b |
The sequence of the stable and variable-mobility alleles of the three individuals whose alleles were sequenced are shown and compared with the previously determined sequence of D18S58. Lowercase letters denote uncertainty in the sequence. One consistent difference was found between all six of the alleles sequenced and the published sequence: The published sequence has G(CG)5, whereas in all of the alleles sequenced here a C follows the last CG dinucleotide [i.e., G(CG)≥5C]. The last C may have been unreadable in the original sequence, due to compression. For five of the six alleles sequenced, a minimum of four independent clones were sequenced.
Due to variation in the number of repeats in the sequenced clones of this allele, the allele length is uncertain but most likely is 155 bp.
For one allele in sample 3, only one clone was obtained and sequenced.
Sizing of Alleles
We hypothesized that the longer (CG)m repeat contained in the variable-mobility alleles was responsible for their inconsistent electrophoretic mobility. Given the propensity of (CG)m repeats to form hairpins (Gacy et al. 1995), we further hypothesized that this inconsistency could be due to formation of an unusually stable secondary structure in the single-stranded denatured PCR product. Subtle differences between experiments could affect denaturation of the secondary structure, causing variations in allele mobility. Variability was observed, regardless of which primer was labeled, indicating that the structure formed on both strands of DNA.
We examined whether electrophoresing the PCR products on a gel with stronger denaturing properties might maintain the products in a more completely denatured state and yield the mobility expected, given their actual length. We amplified DNA from the seven individuals who carried a variable-mobility allele and electrophoresed the PCR products on formamide-containing acrylamide gels. Overall, the variable-mobility alleles demonstrated slightly decreased mobility on the formamide-containing gels, although the range of mobility on formamide and standard gels overlapped extensively (see Table 1). Thus, use of formamide in the gels reduced, but did not eliminate, the variation in mobility of the alleles containing G(CG)7–8C. These alleles demonstrate extreme levels of variation in mobility (see Table 1), possibly depending on undetectably minor variations in experimental conditions.
Identification of (CG)≥5 Repeats Through Database Searches
The published sequence of D18S58 contains five contiguous CG dinucleotides, and we have shown that some alleles of this marker contain eight contiguous CG’s. Such runs of CG are unusual in vertebrate genomes (Tautz et al. 1986). We examined the prevalence of human sequences containing (CG)≥5 in sequence databases. Database searches identified 56 sequence tagged sites (STSs), of which 54 are CA-repeat-containing microsatellites. Seven extended regions of genomic sequence and 18 CpG island sequences containing (CG)≥5 were also identified; in four and five of these sequences, respectively, the (CG)≥5 repeat was separated by 10 bp or less from (CA)≥5. These results suggest that (CG)m repeats tend to be adjacent to (CA)n repeats. Furthermore, we noticed a striking pattern in the position of (CG)m repeats in relation to adjacent (CA)n repeats. In 55 of 63 (87%) of these (CG)m–(CA)n compound repeat sequences, the run of CGs is 5′ to the longest perfect run of CAs. This proportion is significantly different from the 50% expected if the order of the two repeats were random (P < 0.000001). In only 8 of the 63 (13%) sequences, the (CG)m repeat lies 3′ to the longest perfect run of CAs or between two equally long runs.
Odd-Sized Alleles at D18S58 and at Other (CG)m–(CA)n Microsatellites
Examination of the published allele distribution for D18S58 (Gyapay et al. 1994) indicates that, although most alleles fall into a ladder of sizes that differ from each other by an even number of base pairs, as expected for a dinucleotide repeat, two alleles, with frequencies of 15% and 2% in the Centre d’Etudes du Polymorphisme Humain (CEPH) pedigree collection (CEPH Genotype Database), differ by an odd number of base pairs from the other alleles. The reported sizes of these two “odd-sized” alleles are within the apparent size range of the variable-mobility alleles that we identified.
Additionally, we examined the sizes of alleles reported for other (CG)m–(CA)n compound repeats. For 40 of the 54 (CG)m–(CA)n STSs identified in the databases, we were able to obtain information on allele sizes and frequencies from the CEPH, the Genome Database, or other sources. Eighteen of these 40 microsatellites (45%) had one or more odd-sized alleles. In contrast, odd-sized alleles were reported for only one of 40 markers (2.5%) randomly chosen from among the 473 markers used in the previously mentioned linkage study (McInnes et al. 1996). Additionally, only 7.9% (5 of 63) of the (CA)n repeat markers on chromosome 18 in the 1994 Genethon genetic map (Gyapay et al. 1994) had one or more odd-sized alleles. These differences in the frequency of odd-sized alleles between the (CG)m–containing markers and both the markers randomly chosen from the entire genome, and the markers from chromosome 18, are statistically significant (P < 0.0001).
Typing of Additional (CG)m–(CA)n Compound Microsatellites Possessing Odd-Sized Alleles
We identified a subset of the markers possessing odd-sized alleles, in which at least one odd-sized allele was present in one or more of three CEPH individuals (1347-2, 1331-1, or 1331-2). These three CEPH individuals were genotyped for three of these markers, D2S2351, D9S1799, and D10S1789, on a standard gel. Additionally, D2S2351 and D9S1799 reactions were run on a formamide gel. In contrast to the data obtained in our laboratory with hundreds of other markers, for each of these three markers the data were not entirely consistent with the results reported in the CEPH pedigree collection. Also, for the two markers for which reactions were run on both standard and formamide gels, apparent allele sizes differed between gels. These results indicate that it may be difficult to obtain reproducible results from many (CG)m–(CA)n repeats and that our findings with D18S58 are indicative of a general phenomenon observed when using (CG)m–(CA)n repeats for genotyping studies.
Sequence Characterization of Human Compound (CA)n Microsatellites in General
We determined how the frequency and patterns of sequence and variation observed for (CG)m–(CA)n repeats compare with those of other classes of compound (CA)n repeats, using a slightly different database searching strategy than was used in the initial examination of (CG)m–(CA)n repeats described above. Because other dinucleotide repeats are more common in the human genome than are (CG)m repeats, we focused our database searches by searching for (CA)n repeats directly adjacent to other dinucleotide repeats, for example, (CT)5(CA)5. This approach identifies compound microsatellite repeats in which the two repeats are directly adjacent to each other, with no region of imperfect repeat sequence between them [i.e., it identifies a subset of those repeats that would be found with a search performed just for the non-(CA) repeat]. We searched the STS database for five classes of compound (CA)n repeats; those in which (CT)m, (TA)m, (GA)m, (CG)m, or (GT)m repeats were also present. We performed this search with the eight different permutations of sequence possible for each class of compound repeat (see Table 3, columns A and B). The sequences obtained with these eight different searches for a single class of repeat are likely to overlap extensively [e.g., the same microsatellite is often identified by searching for (CT)5(CA)5 or by searching for (TC)5(AC)5]; therefore, the results of these searches do not indicate the total number of unique sequences in each general class but provide an estimate of the relative number of repeats in each general class, as well as the specific number of repeats within each subclass of a single class of repeat.
Table 3.
Compound (CA)n Repeats
| A compound CA class | B Repeat | C No. | D No. (%) ± 1 bp | E % (CA)n 5′ | F % (CA)n 3′ | G % imp. period. |
|---|---|---|---|---|---|---|
| (CT) or (TC) | (CT)5(CA)5 | 199 | 1/40 (2.5) | 29.4 | 70.6 | 2.7 |
| (CA)5(CT)5 | 85 | |||||
| (TC)5(CA)5 | 4 | |||||
| (CA)5(TC)5 | 0 | |||||
| (CT)5(AC)5 | 12 | |||||
| (AC)5(CT)5 | 0 | |||||
| (TC)5(AC)5 | 200 | |||||
| (AC)5(TC)5 | 88 | |||||
| (TA) or (AT) | (TA)5(CA)5 | 121 | 7/40 (17.5) | 49.3 | 50.7 | 0.9 |
| (CA)5(TA)5 | 99 | |||||
| (AT)5(CA)5 | 0 | |||||
| (CA)5(AT)5 | 1 | |||||
| (TA)5(AC)5 | 0 | |||||
| (AC)5(TA)5 | 3 | |||||
| (AT)5(AC)5 | 108 | |||||
| (AC)5(AT)5 | 120 | |||||
| (GA) or (AG) | (GA)5(CA)5 | 2 | 2/40 (5) | 98.1 | 1.9 | 0.5 |
| (CA)5(GA)5 | 101 | |||||
| (AG)5(CA)5 | 0 | |||||
| (CA)5(AG)5 | 0 | |||||
| (GA)5(AC)5 | 0 | |||||
| (AC)5(GA)5 | 1 | |||||
| (AG)5(AC)5 | 2 | |||||
| (AC)5(AG)5 | 100 | |||||
| (CG) or (GC) | (CG)5(CA)5 | 30 | 13/36 (36.1) | 16.5 | 83.5 | 2.2 |
| (CA)5(CG)5 | 8 | |||||
| (GC)5(CA)5 | 1 | |||||
| (CA)5(GC)5 | 0 | |||||
| (CG)5(AC)5 | 1 | |||||
| (AC)5(CG)5 | 0 | |||||
| (GC)5(AC)5 | 44 | |||||
| (AC)5(GC)5 | 7 | |||||
| (GT) or (TG) | (GT)5(CA)5 | 0 | N.A. | N.A. | N.A. | N.A. |
| (CA)5(GT)5 | 0 | |||||
| (TG)5(CA)5 | 0 | |||||
| (CA)5(TG)5 | 1 | |||||
| (GT)5(AC)5 | 0 | |||||
| (AC)5(GT)5 | 0 | |||||
| (TG)5(AC)5 | 0 | |||||
| (AC)5(TG)5 | 0 | |||||
Column A indicates the general classes of compound (CA)n repeat. Column B indicates the different subclasses of each type of repeat. Column C shows the number of repeats identified in each subclass. The number and percentage of examined repeats in each general class for which odd-sized alleles were reported is shown in column D. Columns E and F indicate the percentage of repeats in each general class for which the (CA)n repeat was located 5′ or 3′ of the other repeat, respectively. Column G indicates the proportion of repeats in each general class that demonstrates imperfect dinucleotide periodicity. (N.A.) not available.
Results of these database searches are shown in Table 3, in which the classes of compound human (CA)n repeats are listed from most to least frequently identified, and the numbers of human repeats identified in each subclass are shown. Interestingly, those repeats containing (CT)m appear to be almost threefold more common than those repeats containing (GA)m, which is the same repeat, only present on the other DNA strand.
We randomly chose 40 different markers in each compound repeat class and examined their reported allele sizes, to determine whether any odd-sized alleles had been observed (see Table 3, column D). [For the compound (CA)5 repeats containing (GT)m on the same strand as the (CA)5 repeat, this analysis was not possible, because only one repeat was found; also, allele frequency distributions could only be found for 36 of the repeats containing (CG)m. This number of repeats is lower than the number for which allele information could be found above (i.e., 40), because only those repeats without any imperfect repeat sequence between the two repeats are studied here.] In this particular search, those repeats containing (CG)m possessed one or more reported odd-sized alleles with the highest frequency, 36.1% (see Table 3, column D). This number is not significantly different from the frequency (45%) with which odd-sized alleles were reported for (CG)m–(CA)n repeats in the previous search, discussed above (P = 0.57, N.S.).
The compound (CA)n repeat class with the next highest level of reported odd-sized alleles is the class containing (TA)m repeats, in which 17.5% of the repeats had odd-sized alleles reported. This proportion is not statistically different from the proportion of (CG)m–(CA)n repeats for which odd-sized alleles were reported (P = 0.11, N.S.). The other two compound repeat categories for which the proportion of repeats with odd-sized alleles could be calculated had negligible numbers of them. Five percent of repeats containing (GA)m had reported odd-sized alleles, as did 2.5% of repeats containing (CT)m. These proportions do not differ from each other (P= 0.55, N.S.) or from the 17.5% of (TA)m-containing repeats with odd-sized alleles (P = 0.16, N.S. and P = 0.0624, N.S., respectively) but are statistically different from the 36.1% of (CG)m–(CA)n repeats with odd-sized alleles [P = 0.0018 and P = 0.0005, respectively (still significant after correction for performance of six tests to compare the proportion of odd-sized alleles in the four categories)].
We also examined the relative positions of the two repeats in the four classes of repeat for which sufficient data was available (see Table 3, columns F and G). This was done by tallying the number of repeats in a given class that were identified by those searches using sequences of the type (XY)5(CA)5 or (XY)5(AC)5 versus those identified using sequences of the type (CA)5(XY)5 or (AC)5(XY)5. Whereas data for the repeat class containing (AT)m or (TA)m indicated no bias in position of the (AT)m or (TA)m repeat, relative to that of the (CA)n or (AC)n repeat (P = 0.81, N.S.), compound repeats in the other three classes do appear to have a bias in the relative position of the two repeats, and interestingly, this bias is not the same for the different repeat types (see Table 3, columns E and F). In 83.5% of the compound (CA)n repeats containing (CG)m or (GC)m, the (CA)n or (AC)n is the more 3′ of the two repeats. [This number differs from 50% (P < 0.0001) and is similar to the 87% found in the initial analysis discussed above.] Similarly, in 70.6% of the repeats containing (CT)n or (TC)n, the (CA)n or (AC)n repeat is located 3′ to the other repeat. (Again, this number differs from 50%, with P < 0.0001.) In contrast, the repeats containing (GA)n or (AG)n demonstrate very strikingly the opposite pattern; in 98.1% of cases, the (CA)n or (AC)n repeat is 5′ of the other repeat. (This number differs from 50%, with P < 0.0001).
We also examined how frequently the two repeats demonstrated perfect dinucleotide periodicity, defined to mean that the two neighboring repeats directly abut each other such that the base present in both repeats occurs every other base pair [i.e., (CT)5(CA)5 or (TC)5(AC)5 repeats, as opposed to (CT)5(AC)5 or (TC)5(CA)5 repeats]. For all four general classes of compound (CA)n repeats examined, the proportion of repeats that did not demonstrate perfect dinucleotide periodicity was negligible (0.5%–2.7%) (see Table 3, column G).
DISCUSSION
In this paper we used detailed analysis of a single (CG)m(CA)n compound microsatellite, D18S58, as well as genotyping of three additional similar markers and an examination of sequence databases, to evaluate compound (CA)n microsatellite repeats, particularly those containing (CG)m, as a source of complexity among human microsatellites. Through direct sequencing of several D18S58 alleles, we demonstrated that variation in the number of (CG)m repeats was responsible for the observation that certain alleles of this marker differed in migration patterns between experiments. Those alleles that demonstrated variable mobility possessed longer (CG)m repeats than those that did not. Given that (CG)m is the dinucleotide repeat predicted to form the most stable hairpin structure (Gacy et al. 1995), the variable mobility is probably due to altered secondary structure in single-stranded PCR products of those alleles with a longer run of (CG)m. We also showed that similar variation in mobility occurs at other (CG)m–(CA)n repeats. The unusual properties of these compound microsatellites stimulated us to evaluate the characteristics of (CG)m–(CA)n repeats and other compound (CA)n repeats, in the human genome.
CG dinucleotides are under-represented in vertebrate DNA, because of C → T mutation due to methylation of cytosine in CG dinucleotides, followed by deamination. However, runs of (CG)m are observed more rarely than expected, given the frequency of CG dinucleotides (Tautz et al. 1986; Jurka and Pethiyagoda 1995). Additionally, (CG)m repeats are rare, even in Drosophila, which does not have C methylation or under-representation of CG dinucleotides (Lowenhaupt et al. 1989; Stallings 1992). Our results indicate that (CA)n-repeat-containing regions appear enriched for contiguous runs of (CG)m, in comparison with other genomic regions. There are several possible (not mutually exclusive) explanations for this observation. (CG)m runs may have been the original nuclei for the formation of some (CA)n repeats through a process of expansion and mutation (Levinson and Gutman 1987), or compound (CG)m–(CA)n repeats may have analogously arisen from simple (CA)n repeats. Given the presence of adjacent runs of CG and CA, replication slippage, the mechanism most likely to account for expansion of simple sequence repeats (Tautz and Schlötterer 1994), may have caused expansion of both repeats. It is possible that the current representation of (CG)m–(CA)n repeats in the databases provides an underestimate of the frequency with which ancestral (CG)m and (CA)n repeats co-occurred and coexpanded; as C → T mutation commonly occurs at CG dinucleotides, many ancestral CG dinucleotides have probably been converted to TG (Levinson and Gutman 1987). Our finding that (TG)m–(CA)n-type repeats appear very rare in the human genome suggests that if (CG)m–(CA)n repeats mutate through the methylation–deamination process, there may be a strong preference for the strand on which this occurs; methylation and mutation on the (CA)n strand would lead to (TG)m–(CA)n-type repeats, which appear rare, whereas methylation on the (TG)n strand would lead to simple (CA)n repeats. Alternatively, perhaps (CG)m repeats directly adjacent to (CA)n repeats are relatively protected from this mutation process, due to undermethylation.
Because of their under-representation in genomic DNA, it has been hypothesized that (CG)m repeats assume a conformation deleterious to chromosome structure (Stallings 1992). Several studies have suggested that C + G-rich repeats are particularly prone to forming unusual structures, such as hairpins and quadruplexes (e.g., see Chen et al. 1995; Darlow and Leach 1995; Gacy et al. 1995; Kettani et al. 1995; Nadel et al. 1995; Smith et al. 1995; Warren 1996; Petruska et al. 1996; for review, see Pearson and Sinden 1998). Although many of these studies focus on C + G-rich trinucleotide repeats that, when expanded, lead to human diseases, it has been suggested that CG dinucleotide repeats may be even more prone than trinucleotide repeats to form hairpins (Gacy et al. 1995). Our observation that the length of the D18S58 (CG)m repeat affects its conformation suggests that the tendency of (CG)m repeats to be located near (CA)n repeats should be considered in relationship to alternative DNA structures. With respect to the co-occurrence of (CA)n and (CG)m repeats, it is noteworthy that Z-DNA is a candidate alternative structure for (CG)m and (CA)n repeats. Z-DNA is most easily formed by certain sequences of altering purines and pyrimidines; both (CG)m/(CG)m and (CA)n/(TG)n runs have high potential to form Z-DNA, in contrast to (AT)n/(AT)n (Jovin et al. 1983). It has been proposed that (CA)n repeats may assume a Z-DNA structure under certain in vivo conditions, and available data suggest that Z-DNA formation may play a role in cellular processes including transcription and recombination (Hamada and Kakunaga 1982; Berger et al. 1998; for review, see Herbert and Rich 1996). If (CA)n repeats have a function for which they assume Z-DNA structure, the relatively high frequency with which CG runs occur next to (CA)n repeats might be explained by the fact that (CG)m repeats have an even stronger Z-DNA-forming potential.
Over one-third of the (CG)m–(CA)n microsatellites for which genotyping information was available have odd-sized alleles, whereas few other markers that we examined for comparison do, suggesting that (CG)m repeats may commonly vary in length and that alleles with longer CG runs may demonstrate variable mobility, producing apparently odd-sized alleles. The only other class of repeat we examined for which a sizable proportion of members possessed odd-sized alleles were (TA)m–(CA)n compound repeats; (AT)n repeats are predicted to form hairpins second in stability only to those formed by (CG)n repeats (Gacy et al. 1995). Perhaps these odd-sized alleles are also caused by secondary structure formation that alters gel mobility. Of course, some odd-sized alleles result from other mechanisms, such as insertion–deletion mutations (Grimaldi and Crouau-Roy 1997; Brinkmann et al. 1998b).
Our examination of the five different classes of compound (CA)n microsatellites yielded additional findings. This set of analyses was done on sequences present in the STS database, which is likely to be enriched for microsatellites with high levels of polymorphism, relative to those with low genetic informativeness. If the different types of compound (CA)n repeat have differing levels of polymorphism, then the relative frequencies of these repeats found in the STS database may differ from those in the genome as a whole. This seems unlikely, but confirmation of lack of bias requires analysis of random genomic sequence from a large proportion of the human genome and is beyond the scope of this paper. Other than this risk of bias and the possibility that different compound (CA)n repeats could differ in clonability, the compound (CA)n repeat sequences in the STS database should be fairly representative of those in the genome, because (CA)n repeats are generally identified using techniques that focus only on identification of the (CA)n repeat and not on inclusion or exclusion of other adjacent repeats.
The relative frequency of different classes of compound (CA)n microsatellites can be estimated from our search results. The relative frequency of the different compound repeats cannot be completely explained by the average base composition of the human genome nor by the relatively higher frequency of transition compared with transversion mutations seen in the human genome (Cooper and Krawczak 1993); if these compound repeats frequently form by mutation from simple CA repeats, one might expect (TA)m–(CA)n microsatellites to be most common, because a TA dinucleotide can form by one transition mutation from a CA dinucleotide. However, (CT)m–(CA)n repeats appear most common; although, for a CT dinucleotide to form from a CA dinucleotide, a transversion mutation is required. A logical explanation for the dearth of (GT)m–(CA)n-type repeats is that, to generate such a repeat from a simple (CA)n repeat would require a mutation event in both base pairs of the repeat. However, one might expect that such repeats would sometimes form through expansion of a GT dinucleotide present by chance immediately adjacent to the CA repeat.
The apparent tendency of CG and CT runs to lie 5′, and of GA runs to lie 3′, of (CA)n repeats is also intriguing, because it suggests that the formation or retention of runs of CG, CT, and GA near (CA)n repeats has polarity that differs depending on the repeat type. [Runs of TA do not appear to demonstrate polarity in their position relative to (CA)n repeats]. A clear example of polarity in microsatellite mutation is seen in the fragile X trinucleotide repeat, in which variation in repeat length occurs mainly at the 3′ end of the repeat (Eichler et al. 1994; Hirst et al. 1994; Kunst and Warren 1994; Snow et al. 1994).
In our search, as well as in a previously reported analysis of some compound repeats (Epplen et al. 1997), the proportion of compound microsatellites demonstrating imperfect dinucleotide periodicity was small, supporting the hypothesis that such repeats generally arise through a process of mutation and replication slippage (Levinson and Gutman 1987). A fuller examination of the characteristics of compound (CA)n microsatellite periodicity would require examination of those repeats in which the (CA)n and other repeats were not directly adjacent to each other and is beyond the scope of this paper.
Screening of available sequence databases indicates that numerous (CG)m–(CA)n compound microsatellites exist and are in use as genetic markers. Devising a method for obtaining consistent and accurate genotyping results for (CG)m–(CA)n microsatellites would increase their reliability as genetic markers. Perhaps substitution during PCR of 7-deaza-dGTP or dITP (an analog with even less tendency to form secondary structures) for dGTP or of N4-methyl-dCTP (Li et al. 1993; McCrea et al. 1993) for dCTP would induce the alleles carrying long CG runs to have the mobility expected given their length. However, PCR conditions used might need to be modified, because these nucleotide analogs are generally incorporated less efficiently than dGTP and dCTP (Li et al. 1993; McCrea et al. 1993).
Relatively little sequencing of microsatellite alleles has been done. However, such sequencing can be particularly useful for population studies, and for increasing understanding of the origin, mutational processes, and structure of microsatellites, in addition to allowing more informed interpretation of genotyping data, as discussed above. In genetic studies that involve comparison of genotypes across a population, rather than within a family, it is particularly important to be able to distinguish alleles that are identical by state (IBS) from those identical by descent (IBD). Such differentiation is difficult to do with microsatellites; because they have a relatively high mutation rate (Weber and Wong 1993), an allele of a given size may arise independently multiple times. By sequencing compound microsatellites, such as D18S58, one may distinguish alleles of the same length that have differing sequences, that is, are IBS, from those that have identical sequence, that is, are probably IBD (Garza and Freimer 1996). Sequencing of such alleles may be helpful for fine mapping of disease genes using association and linkage disequilibrium approaches and may increase the amount of information about populations obtainable through study of microsatellites.
METHODS
Genotyping
Genotyping was performed as described previously (Bull et al. 1999), except as follows: Where indicated, gels containing 40% formamide were used. For these gels, the protocol in Ausubel et al. (1995) was followed, except that Long Ranger Gel Solution (J.T. Baker) was used. Also, prior to loading onto a gel, samples were sometimes denatured by heating at 94°C for 3 min in a PCR machine, rather than as described previously (Bull et al. 1999).
Sequencing
PCR was performed on the DNA samples using standard conditions, and PCR products were cloned using the pGem-T Vector System (Promega) and sequenced using the fmol DNA Sequencing System (Promega) and radioactively end-labeled primers. Reactions were run on gels containing 40% formamide, to resolve sequence compression seen in the (CG)m repeat region. We focused on sequencing each allele on the strand on which the (CG)m repeat lay 5′ to the (CA)n repeat, because sequence of the (CG)m repeat was easier to read on this strand.
Database Searches
Searches of sequence databases were performed using BLAST and FASTA. Searches focusing specifically on (CG)m repeats were performed July–September 1996, and the following procedure was used: BLAST searches were performed on the STS division of the GenBank database without filtering, using all possible combinations of (CG)mN, as 11 bp of sequence was the minimum required to allow identification of homologies. The score cutoff for the BLAST searches was 55, which corresponded to a perfect match at all 11 bases of the query sequence. FASTA searches were performed on the STS and human divisions of the EMBL database, also using all possible combinations of (CG)mN and a score cutoff (44) corresponding to a perfect match with the query sequence. [Because there was a large number of repeats with the sequence (CG)mC, searches of the human division were performed using FASTA and all possible combinations of (CG)mCN, with a score cutoff of 48]. We did not extensively evaluate (CG)m repeats present in characterized human genes and ESTs, although some do exist. The large size and/or redundancy of the relevant databases and the necessity for knowledge of exon–intron boundaries make this a complicated task. The searches for the five classes of compound (CA)n repeats were performed in February and March 1998 using BLAST and the STS database. Sequences of the form (XY)5(CA)5 (see Table 3) were used and a score cutoff of 100, which corresponded to a perfect match at all 20 bases of the query sequence.
Statistics
Statistical comparisons were done using the χ2 test.
Acknowledgments
We thank L. Alison McInnes for providing the initial observations that led us to perform this work, and Victoria Carlton, Christopher Vulpe, Susan Service, and Carlos Garza for helpful comments on the manuscript. This work was supported by a National Institutes of Health R01 grant (MH49499) and an National Institute of Mental Health (NIMH) Research Scientist Development Award to N.B.F. L.N.B. was supported by a postdoctoral National Research Service Award from the NIMH and by a Young Investigator Award from the National Alliance for Research on Schizophrenia and Depression. C.P.-P. was supported by a Research Project Grant from the Minority Research Resources branch of the NIMH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Nelson@ngl.ucsf.edu; FAX (415) 476-7389.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology, Vol. 1. New York, NY: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- Berger I, Winston W, Manoharan R, Schwartz T, Alfken J, Kim Y-G, Lowenhaupt K, Herbert A, Rich A. Spectroscopic characterization of a DNA-binding domain, Z-alpha, from the editing enzyme, dsRNA adenosine deaminase: Evidence for left-handed Z-DNA in the Z-alpha-DNA complex. Biochem. 1998;37:13313–13321. doi: 10.1021/bi9813126. [DOI] [PubMed] [Google Scholar]
- Blanquer-Maumont A, Crouau-Roy B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J Mol Evol. 1995;41:492–497. doi: 10.1007/BF00160321. [DOI] [PubMed] [Google Scholar]
- Brinkmann B, Junge A, Meyer E, Wiegand P. Population genetic diversity in relation to microsatellite heterogeneity. Hum Mutat. 1998a;11:135–144. doi: 10.1002/(SICI)1098-1004(1998)11:2<135::AID-HUMU6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: Influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998b;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull LN, Juijn JA, Liao M, van Eijk MJT, Sinke RJ, Stricker NL, DeYoung JA, Carlton VEH, Baharloo S, Klomp LWJ, et al. Fine-resolution mapping by haplotype evaluation: The examples of PFIC1 and BRIC. Hum Genet. 1999;104:241–248. doi: 10.1007/pl00008714. [DOI] [PubMed] [Google Scholar]
- Chen X, Mariappan SVS, Catasti P, Ratliff R, Moyzis RK, Laayoun A, Smith SS, Bradbury EM, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: Structure and biological implications. Proc Natl Acad Sci. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Human gene mutation. Oxford, UK: Bios Scientific; 1993. [Google Scholar]
- Darlow JM, Leach DRF. The effects of trinucleotide repeats found in human inherited disorders on palindrome inviability in Escherichia coli suggest hairpin folding preferences in vivo. Genetics. 1995;141:825–832. doi: 10.1093/genetics/141.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Holden JJA, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Epplen C, Santos EJM, Mäueler W, van Helden P, Epplen JT. On simple repetitive DNA sequences and complex diseases. Electrophoresis. 1997;18:1577–1585. doi: 10.1002/elps.1150180916. [DOI] [PubMed] [Google Scholar]
- Estoup A, Tailliez C, Cornuet JM, Solignac M. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae) Mol Biol Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- Garza JC, Freimer NB. Homoplasy for size at microsatellite loci in humans and chimpanzees. Genome Res. 1996;6:211–217. doi: 10.1101/gr.6.3.211. [DOI] [PubMed] [Google Scholar]
- Grimaldi MC, Crouau-Roy B. Microsatellite allelic homoplasy due to variable flanking sequences. J Mol Evol. 1997;44(3):336–340. doi: 10.1007/pl00006151. [DOI] [PubMed] [Google Scholar]
- Guyer MS, Collins FS. The Human Genome Project and the future of medicine. Am J Dis Child. 1993;147:1145–1152. doi: 10.1001/archpedi.1993.02160350019003. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. The 1993-94 Généthon human genetic linkage map. Nat Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- Hamada H, Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982;298:396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Herbert A, Rich A. The biology of left-handed Z-DNA. J Biol Chem. 1996;271(20):11595–11598. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- Hirst MC, Grewal PK, Davies KE. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet. 1994;3:1553–1560. doi: 10.1093/hmg/3.9.1553. [DOI] [PubMed] [Google Scholar]
- Jovin TM, McIntosh LP, Arndt-Jovin DJ, Zarling DA, Robert-Nicoud M, van de Sande JH, Jorgenson KF, Eckstein F. Left-handed DNA: From synthetic polymers to chromosomes. J Biomol Struct Dyn. 1983;1:21–57. doi: 10.1080/07391102.1983.10507425. [DOI] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: Compilation and analysis. J Mol Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Kettani A, Kumar RA, Patel DJ. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J Mol Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- Kunst CB, Warren ST. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li S, Haces A, Stupar L, Gebeyehu G, Pless RC. Elimination of band compression in sequencing gels by the use of N4-methyl-2′-deoxycytidine 5′-triphosphate. Nucleic Acids Res. 1993;21:2709–2714. doi: 10.1093/nar/21.11.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Jin L, Lin X, Voros A, Underhill P, Mignot E. Microsatellite single nucleotide polymorphisms in the HLA-DQ region. Tissue Antigens. 1998;52:9–18. doi: 10.1111/j.1399-0039.1998.tb03018.x. [DOI] [PubMed] [Google Scholar]
- Lowenhaupt K, Rich A, Pardue ML. Nonrandom distribution of long mono- and dinucleotide repeats in Drosophila chromosomes: Correlations with dosage compensation, heterochromatin, and recombination. Mol Cell Biol. 1989;9:1173–1182. doi: 10.1128/mcb.9.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea KW, Marrs CF, Gilsdorf JR. Gel compressions and artifact banding can be resolved in the same DNA sequence reaction. Biotechniques. 1993;15:843–844. [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, Rojas E, Spesny M, Baharloo S, Blankenship K, et al. A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci. 1996;93:13060–13065. doi: 10.1073/pnas.93.23.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel Y, Weisman-Shomer P, Fry M. The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J Biol Chem. 1995;270:28970–28977. doi: 10.1074/jbc.270.48.28970. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Sinden RR. Trinucleotide repeat DNA structures: Dynamic mutations from dynamic DNA. Curr Opin Struct Biol. 1998;8:321–330. doi: 10.1016/s0959-440x(98)80065-1. [DOI] [PubMed] [Google Scholar]
- Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GK, Jie J, Fox GE, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 1995;23:4303–4311. doi: 10.1093/nar/23.21.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow K, Tester DJ, Kruckeberg KE, Schaid DJ, Thibodeau SN. Sequence analysis of the fragile X trinucleotide repeat: Implications for the origin of the fragile X mutation. Hum Mol Genet. 1994;3:1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Stallings RL. CpG suppression in vertebrate genomes does not account for the rarity of (CpG)n microsatellite repeats. Genomics. 1992;17:890–891. doi: 10.1016/0888-7543(92)90178-u. [DOI] [PubMed] [Google Scholar]
- Tautz D. Notes on the definition and nomenclature of tandemly repetitive DNA sequences. Exs. 1993;67:21–28. doi: 10.1007/978-3-0348-8583-6_2. [DOI] [PubMed] [Google Scholar]
- Tautz D, Schlötterer C. Simple sequences. Curr Opin Genet Devel. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Tautz D, Trick M, Dover GA. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986;322:652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Urquhart A, Kimpton CP, Downes TJ, Gill P. Variation in short tandem repeat sequences—A survey of twelve microsatellite loci for use as forensic identification markers. Int J Leg Med. 1994;107(1):13–20. doi: 10.1007/BF01247268. [DOI] [PubMed] [Google Scholar]
- Warren ST. The expanding world of trinucleotide repeats. Science. 1996;271:1374–1375. doi: 10.1126/science.271.5254.1374. [DOI] [PubMed] [Google Scholar]
- Weber JL. Informativeness of human (dC-dA)n · (dG-dT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]