Abstract
We report results using a microdevice for DNA sequencing using samples from chromosome 17, obtained from the Whitehead Institute Center for Genome Research (WICGR) production line. The device had an effective separation distance of 11.5 cm and a lithographically defined injection width of 150 μm. The four-color raw data were processed, base-called by the sequencing software Trout, and compared to the corresponding ABI 377 sequence from WICGR. With a criteria of 99% accuracy, we achieved average continuous reads of 505 bases in 27 min with 3% linear polyacrylamide (LPA) at 150 V/cm, and 460 bases in 22 min with 4% LPA at 200 V/cm at a temperature of 45°C. In the best case, up to 565 bases could be base-called with the same accuracy in <25 min. In some instances, Trout allowed for accurate base-calling down to a resolution R as low as R = 0.35. This may be due in part to the high signal-to-noise ratio of the microdevice. Unlike many results reported on capillary machines, no additional sample cleanup other than ethanol precipitation was required. In addition, DNA fragment biasing (i.e., discrimination against larger fragments) was reduced significantly through the unique sample injection mechanism of the microfabricated device. This led to increased signal strength for long fragments, which is of great importance for the high performance of the microdevice.
Significant advancement in the technology of DNA analysis is expected from the use of microfabricated electrophoretic devices for sequencing and genotyping. In this approach photolithography, combined with wet-etching and thermal wafer bonding, is used to construct enclosed intricate microchannel structures in glass and fused-silica substrates; these structures are then utilized for electrophoresis (Harrison et al. 1993). It has been speculated that these devices will allow DNA separations approaching the theoretical limits of electrophoresis and in a format that will reduce analysis time and extend parallelism and automation (Freemantle 1999), which might hence increase throughput well beyond current capillary array machines. For example, in recent experiments we have demonstrated genotyping at 10- to 100-fold reduced analysis times on microdevices when compared to capillaries and slab gels, respectively (Schmalzing et al. 1997, 1999). DNA sequencing of single-color pGEM and four-color M13 DNA standard sequencing samples has been demonstrated on 3.5-, 11.5-, and 7-cm-long microdevices (Woolley et al. 1995; Schmalzing et al. 1998; Liu et al. 1999). The feasibility of ultra-high sample throughput has been proven through still modest multiplexing up to 96 microchannels (Simpson et al. 1998; Koutny et al. 1999). However, to the best of our knowledge, all published studies on DNA sequencing by microdevices have been performed using DNA standard samples such as M13 or pGEM. Practical sequencing must deal with additional factors such as variable salt and template concentrations (Ruiz-Martinez et al. 1998; Salas-Solano et al. 1998), highly sample-specific compression regions, and the interplay between electrophoretic separation and base-calling software typical of production DNA sequencing samples. We report initial results on how microdevices perform under practical conditions using DNA sequencing samples as prepared for high throughput, cost-sensitive sequencing under the Human Genome Project. Our results suggest that much of the anticipated throughput improvement for microdevice sequencing is feasible.
RESULTS AND DISCUSSION
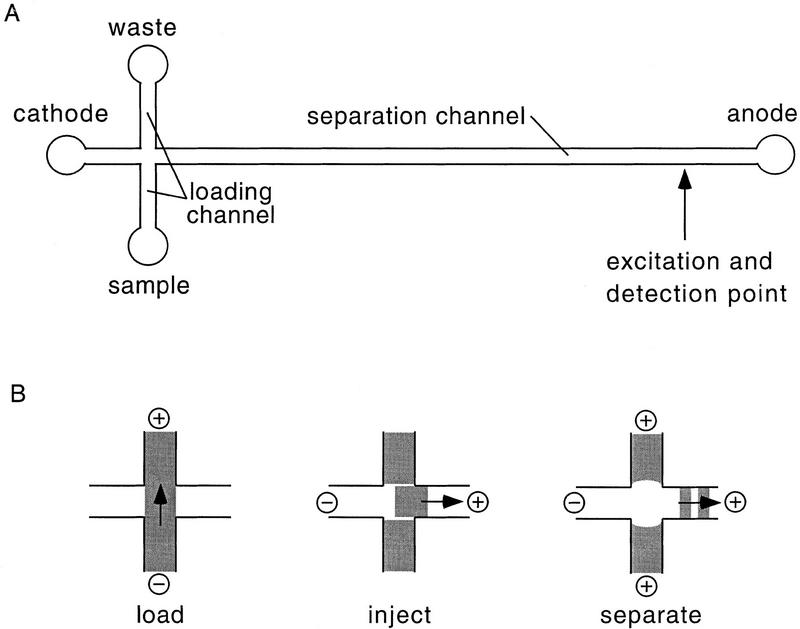
The electrophoretic microdevice used in this study consists of a 12.5-cm-long straight separation channel with an effective separation distance from injection to detection point of 11.5 cm (see Fig. 1A). The channel cross section is nearly semicircular with a 40-μm radius. A 1.0-cm-long loading channel intersects the separation channel at a distance 0.5 cm below the cathodic end and connects the sample and waste reservoirs. The intersection geometry defines a 150-μm-long injection plug of ∼0.5-nl volume. The loading and injection mechanism is illustrated in Figure 1B. The channel surfaces were passivated to neutralize electro-osmotic flow and minimize sample adsorption (Hjerten 1985). In this study we have used a replaceable high molecular weight linear polyacrylamide (LPA) separation matrix in 1× TBE with 3.5 m urea and 30% (wt/vol) formamide, chosen for its extremely high performance in DNA sequencing applications (Carrilho et al. 1996). The analysis temperature was 45°C.
Figure 1.
(A) Layout of the electrophoretic microdevice used in this study (not drawn to scale). The separation distance from injector to detector is 11.5 cm. The three short side channels are 0.5 cm long, 40 μm deep, and 90 μm wide. (B) Schematic of the injection port in load, inject, and run modes. (+, −) The polarity of the electric fields; (→) the direction of DNA movement; the dark regions indicate DNA.
We analyzed 12 different samples from the chromosome 17 with two separation conditions. The samples were taken randomly from the Whitehead Institute Center for Genome Research (WICGR) production line after ethanol precipitation. No additional sample treatment was performed. Six samples were sequenced under condition 1 [C1: 4% (wt/vol) LPA at 200 V/cm]. Another six were sequenced under condition 2 [C2: 3% (wt/vol) LPA at 150 V/cm]. Based on previous work, these conditions are expected to yield near optimum electrophoretic performance of the microdevice for DNA sequencing (Schmalzing et al. 1998). The four-color microdevice raw data were processed using manual editing of results generated by Trout sequencing software. Minor modifications of the Trout color matrix and temporal filters were used to adjust the software for the unusually high data rate and custom detector of the microelectrophoresis device. The final sequence was compared in blind tests with the sequence previously generated at the WICGR on an ABI 377 sequencer. Table 1 summarizes the results. For the data comparisons we defined read length as the contiguous length of sequence, measured in bases, which has a base-calling accuracy of ≥99%.
Table 1.
Microdevice Sequencing Error Rate as a Function of Base Number
| Sample | 35– 100 | 101– 150 | 151– 200 | 201– 250 | 251– 300 | 301– 350 | 351– 400 | 401– 450 | 451– 500 | 501– 550 | 551– 600 | Read length | Run time (min) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 11 | 25 | 415 | 22.9 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 21 | 515 | 22.7 |
| 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 17 | 520 | 22.8 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 19 | 515 | 21.1 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 10 | 27 | — | 425 | 20.5 |
| 6 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 4 | 8 | 21 | 365 | 20.5 |
| Average (RSD) | 460 (66} | 21.7 (1.2) | |||||||||||
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 565 | 25.7 |
| 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 7 | 465 | 27.3 |
| 9 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 7 | 515 | 28.8 |
| 10 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 7 | 500 | 26.1 |
| 11 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 5 | 10 | 470 | 26.1 |
| 12 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 4 | 515 | 26.6 |
| Average (RSD) | 505 (36} | 26.3 (1.2) | |||||||||||
Error rate after base-calling by Trout, manual editing, and comparison with ABI 377 data. Read length was defined as the length of sequence in bases that had a base-calling accuracy of at least 99%. Samples 1–6 were run at 200 V/ cm with 4% LPA (C1); samples 7–12 were run at 150 V/cm with 3% LPA (C2) at 45°C on a microdevice of 11.5 cm effective separation length. Samples were from the WICGR production line using DNA from chromosome 17.
The electrophoretic condition C1 produced an average read length of 460 bases with a root square deviation (RSD) of 66 bases in an average total run time of 21.7 min (RSD = 1.2 min). Condition C2 resulted in a somewhat longer average read length of 505 bases (RSD = 36 bases) in 26.7 min (RSD = 1.2 min). The extended read of C2 can be attributed to relative reductions in voltage and LPA concentration, which usually improve the electrophoretic separation of longer DNA fragments (Carrilho et al. 1996). In some cases, either condition generated runs with exceptionally long reads, for example, C1, 520 bases for sample 3; and C2, 565 bases for sample 7.
We evaluated several aspects of the errors in the called sequences. Most of the runs (9 of 12) were error-free between 100 and 450 bases. Errors clustered at the beginning and the end of the runs. Because of anomalous electrophoretic migration in the early part of the run, base-calling was unreliable below ∼35 bases for both conditions. However, there was no loss of valuable sequence information because most of the sequence in this region was M13 vector sequence. Some errors occurred between 35 and 100 bases; several of them (7 of 20) could be attributed to strong compressions. Less severe compression was also noticed in other parts of the runs but could usually be base-called by manual editing. This observation suggests that even stronger denaturing conditions should be used in the future. C1 had noticeably fewer errors than C2 in this region (5 vs. 15), in agreement with the general finding that both higher voltage (less time for diffusion) and higher concentration of sieving matrix (smaller pore size) increase the separation of small fragments (Carrilho et al. 1996).
The frequency of base-calling errors increased steadily for the late-migrating fragments. The increase was much more gradual for C2 than for C1 because of the superior electrophoretic performance of C2 in this region, which outweighed the higher error rate at the beginning of the C2 runs. Interestingly, the vast majority of base-calling errors (19 of 23) from 451 to 500 bases was associated with multiplets. Typically only n − 1 bases (n ≥ 2) were called when actually n bases were present in a given multiplet. Inspection of the raw data revealed that in this region multiplets started to become single broad peaks lacking the fine structure directly indicative of the number of bases constituting the multiplets. We speculate that further adaptation of the sequencing software for microdevice electrophoresis could reduce this type of error and lead to longer average read lengths. Beyond 500 bases, all types of base-calling errors could be seen. The sequences became unassignable at ∼600 bases. There was still DNA fragment sizing in this region, but single-base resolution was drastically below 0.5.
The question arises as to the extent that sequencing performance is limited by specific raw data patterns unique to the microdevice format, combined with our current degree of optimization of the base-calling software. One way to express electrophoretic performance without these confounding factors is through resolution, which is a function of the separation mechanism and the peak-broadening mechanisms alone. Resolution of 0.5 is defined as the point at which the migration time difference between two Gaussian peaks equals their average full widths at half maximum (Luckey et al. 1993). This resolution is commonly set as a minimum requirement for accurate robust sequencing (Best et al. 1994), although specialized base-calling software has been reported to operate to a resolution as small as 0.25 (B.L. Karger, unpubl.). In Figure 2 we plot average resolution as a function of DNA fragment size for the two electrophoretic conditions C1 and C2 (n = 6 in both cases). The two curves have the typical shape of resolution curves in DNA sequencing. Resolution was lower at the beginning of the runs when good selectivity was compromised by high diffusion and at the end when poor selectivity dominated over low diffusion. The best performance was found in the mid-range of fragment sizes, where selectivity and diffusion were balanced. The graph in Figure 2 shows total average read lengths of 415 bases for C1 (from base 35 to 450) and of 475 bases for C2 (from base 35 to 510), assuming a minimum resolution criteria of 0.5. Trout extended these read lengths on average by 45 bases for C1 and by 30 bases for C2. For sample 7, which gave the longest read with 565 bases, Trout increased the read length by 90 bases beyond what would be expected using the minimum resolution criteria of 0.5. Trout was reading this sequence with no errors up to base 600, where the resolution was only 0.35.
Figure 2.
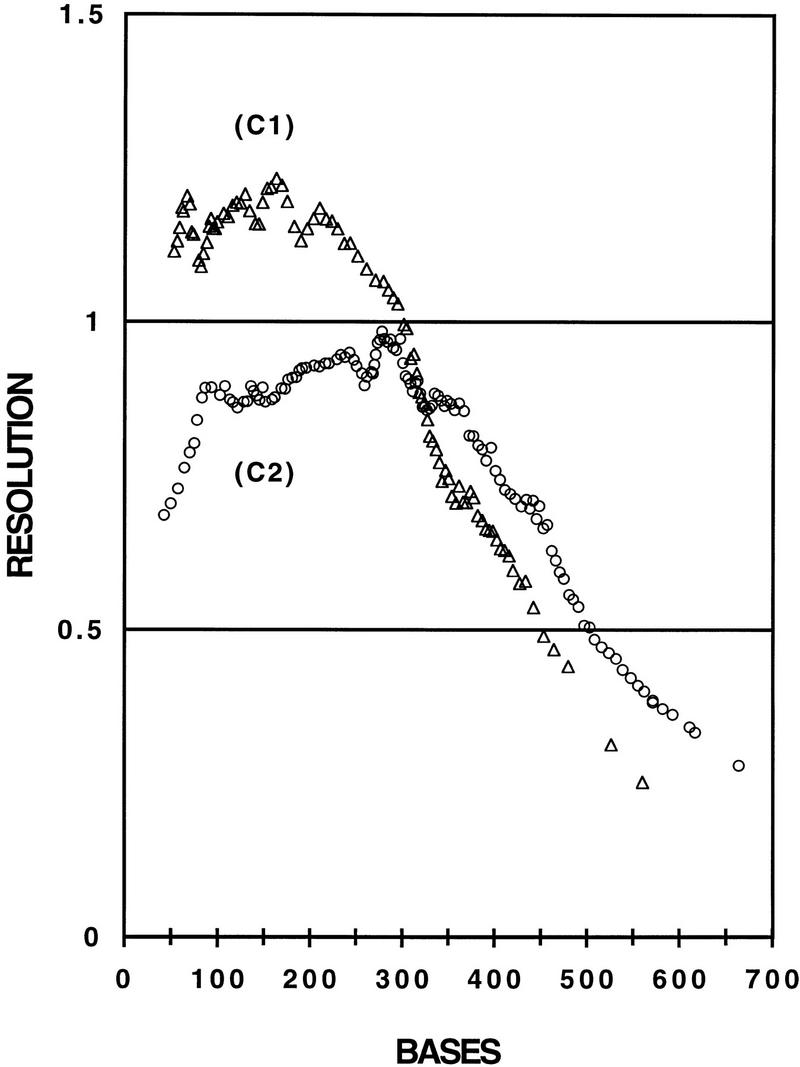
Averaged single base resolutions vs. base number calculated for samples 1–6 and 7–12 run at 200 V/cm with 4% LPA (C1) and at 150 V/cm with 3% LPA (C2) at 45°C, respectively, on a microdevice with 11.5 cm effective separation distance. Only the A traces were used for the calculation. (The base count starts with base number 35, as the sequences were not readable below 35 bases).
The signal-to-noise ratio varied from run to run in the range between 30 and 70. This may be a consequence of variability in the PCR amplifications or the inefficiency of the simple ethanol cleanup procedure to fully remove salt (Salas-Solano et al. 1998). Residual salt in the sample might influence the electrical resistance in the loading channel resulting in salt-dependent DNA velocities during the 2-min-long loading process and thereby introduce some variability of sample concentration during injection (Huang et al. 1988). The DNA velocities in the separation channel would not be affected, as only a miniscule amount of sample salt enters the separation channel during injection. In contrast to capillaries, the signals remained relatively stable in amplitude during the microdevice runs and did not decrease with increasing DNA fragment size, as is often observed for capillary electrophoresis. The microdevice cross-injector seems to permit representative DNA sample loading regardless of the composition of the sample. In addition, neither base-calling nor run times were found to be compromised.
Figure 3 shows the profile of the processed four-color sequencing run of sample 7 performed on a microfabricated device with an effective separation length of 11.5 cm filled with 3% LPA, 150 V/cm, at 45°C. The data presented have been processed and base-called by Trout, followed by manual editing. Errors are indicated as hyphens in the letter sequence. The primer region (eluting at ∼4 min) and the very end of the run are not shown. The presentation starts with the four-letter sequence TCCC (bases 32–35), which is the last section of the M13 vector sequence adjacent to the insert. Sequencing fragments of 400, 500, and 600 bases in length passed the detector after ∼16.5, 19.1, and 21.3 min, respectively. The total run time was 25.7 min, and the total read length was 565 bases.
Figure 3.
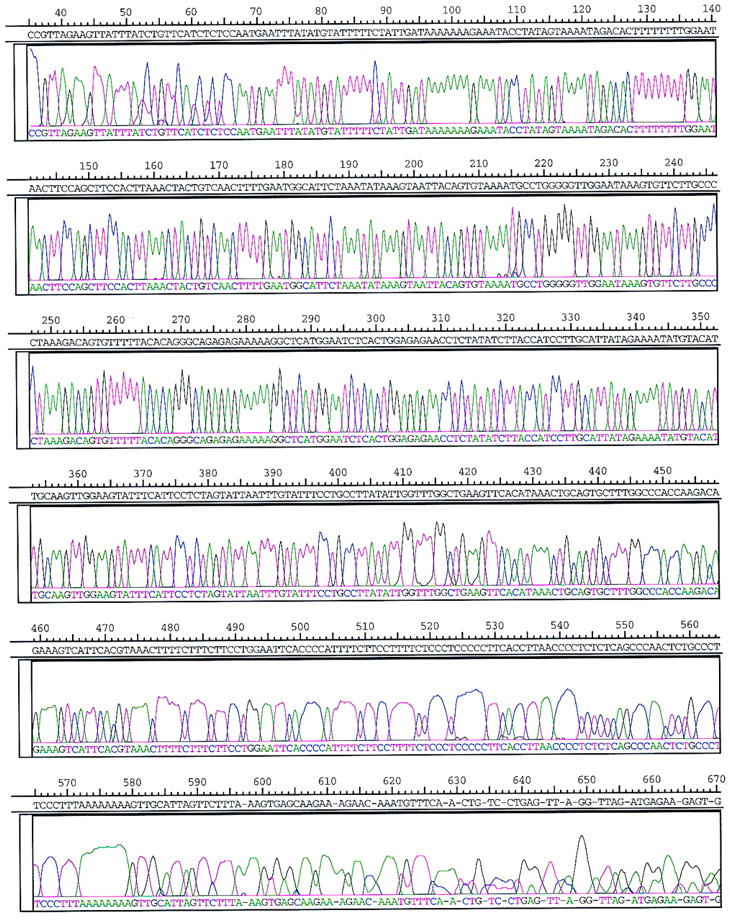
Four-color sequencing data of sample 7 after being processed, base called by Trout, and manually edited. Errors are indicated by hyphens in the letter sequence. The electrophoretic conditions were 3% LPA, 150 V/cm, 45°C, 11.5 cm effective length, and 150 μm geometrical width of cross-injector.
In conclusion, we have shown that microdevices are capable of high-quality DNA sequencing with practical de novo sequencing samples. Our data suggest that significantly less sample pretreatment might be required for consistent operation of microdevices when compared to capillaries. Analysis time is also reduced significantly in many cases. Robust continuous read lengths exceeded 500 bases in <30 min with limited optimization of base-calling software.
METHODS
Micromachining
Microdevices were built from 150-mm-diam. fused-silica wafers (Hoya, Tokyo, Japan) using photolithography, chemical wet-etching methods, laser drilling to form access holes, and thermal bonding (Koutny et al. 1996). Individual microdevices were cut from the bonded wafer pairs using a wafer saw (CHIPS, North Peabody, MA). Glass reservoirs (Ace Glass, Vineland, NJ) of 50 μl volume were affixed around the exit holes to hold sample and buffer.
Instrumentation
The apparatus for single-channel microdevice DNA analysis was described previously (Schmalzing et al. 1997).
Chemistry
The entire microchannel structure was chemically modified by grafting acrylamide to the channel surfaces according to the procedure of Hjerten (1985). High-molecular-weight LPA was used as replaceable sieving material. It was synthesized in-house by inverse emulsion polymerization (Goetzinger et al. 1998). Appropriate amounts of the LPA powder were dissolved in 1 × TBE buffer containing 3.5 m urea and 30% (vol/vol) formamide.
Sample Preparation
The samples were prepared at WICGR. The vector was M13mp18 with ∼2 kb human DNA inserts from Chromosome 17. The GenBank clone names were hRPK.1090_M_7 and hRPK.721_K_11. DYEnamic ET M13(21) primer chemistry (Amersham) was used to prepare the sequencing reaction mixtures. Template DNA (200 ng) was added to each of the four monomer reactions, which were thermocycled for 20 cycles using standard conditions, pooled together, and ethanol precipitated.
Slab Gel Electrophoresis
The standard samples for comparison were run on an ABI 377 sequencer at WICGR using 52-cm plates with a 48-cm well to read. They were run at 2.4 kV or ∼46 V/cm for 10 hr. The gel was 5% Long Ranger cross-linked acrylamide.
Microdevice Electrophoresis
Between each run the polymeric buffer solution present in the entire microchannel structure was replaced from the anodic end of the separation channel using a syringe attached to a mechanical fixture. The device was preelectrophoresed for 10 min at 200 V/cm and 45°C. The sequencing samples were dissolved in 20 μl of deionized water, heated to 95°C for 2 min, chilled on ice, and pipetted into the sample reservoir attached to one end of the loading channel. For representative sample loading, the samples were electrophoresed for 2 min at 200 V/cm across the separation channel. For injection and separation, the voltages were switched to create the desired field strength in the separation channel. Sequencing was performed at 45°C. To prevent leakage of excess sample into the separation channel, an electric field of ∼20 V/cm was applied to both side arms of the loading channel during electrophoresis.
Data Analysis
The ABI 377 data was signal processed using Plan package (Ewing and Green 1998) and base-called using Phred (Ewing et al. 1998). Plan is a Unix-based signal processing tool, similar in format to ABI processing software. It utilizes a mobility correction file (specific to the dye chemistry), a multicomponent matrix (specific to the sequencing machine) for color separation, amplitude normalization for the four channels, baseline substraction, and a smoothing algorithm. The microdevice data were collected using custom software written in HPVEE (Hewlett Packard). The microdevice data was processed further using the base-caller Trout. Trout is available on the WICGR ftp site (genome.wi.mit.edu) in the directory distribution/software/trout. Documentation is provided with the program.
Resolution Measurement
Single-base resolution R was calculated using the relationship R = [(2 ln 2)1/2 (t2 − t1)]/[(fwhm1 + fwhm2) Δb], where t is the migration time of the nth peak, fwhm is the full width at half-maximum of the nth peak, and Δb is the difference between the two peaks in base numbers (Δb > 1). C Grams software (Galactic Industries, Salem, NH) was used to measure t and fwhm of both isolated and partially resolved peaks in the A traces of the four-color runs of samples 1–12.
Acknowledgments
We thank WICGR for kindly providing access to the base-caller Trout. We also thank Mark Daly and Steve Rozen for valuable advice on Trout. This work was supported by the National Institutes of Health (grant HG01389) and by Air-force Office of Scientific Research (F49620-98-1-0235).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ehrlich@wi.mit.edu; FAX (617) 258-7663.
REFERENCES
- Best N, Arriaga E, Chen DY, Dovichi NJ. Separation of fragments up to 570 bases in length by use of 6% T non-crosslinked polyacrylamide for DNA sequencing in capillary electrophoresis. Anal Chem. 1994;66:4063–4067. doi: 10.1021/ac00094a031. [DOI] [PubMed] [Google Scholar]
- Carrilho E, Ruiz-Martinez MC, Berka J, Smirnov I, Goetzinger W, Miller AW, Brady D, Karger BL. Rapid DNA sequencing of more than 1000 bases per run by capillary electrophoresis using replaceable linear polyacrylamide solutions. Anal Chem. 1996;68:3305–3313. doi: 10.1021/ac960411r. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier LD, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Freemantle M. Downsizing chemistry. Chemical analysis and synthesis on microchips promise a variety of potential benefits. Chem Eng News. 1999;77:27–36. [Google Scholar]
- Goetzinger W, Kotler L, Carrilho E, Ruiz-Martinez MC, Salas-Solano O, Karger BL. Characterization of high molecular weight mass linear polyacrylamide powder prepared by emulsion polymerization as a replaceable polymer matrix for DNA sequencing by capillary electrophoresis. Electrophoresis. 1998;19:242–248. doi: 10.1002/elps.1150190217. [DOI] [PubMed] [Google Scholar]
- Harrison DJ, Fluri K, Seiler K, Fan Z, Effenhauser CS, Manz A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science. 1993;261:895–897. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- Hjerten S. High-performance electrophoresis elimination of electroendosmosis and solute adsorption. J Chromatogr. 1985;347:191–198. [Google Scholar]
- Huang X, Gordon MJ, Zare RN. Bias in quantitative capillary zone electrophoresis caused by electrokinetic sample injection. Anal Chem. 1988;60:375–377. [Google Scholar]
- Koutny LB, Schmalzing D, Taylor TA, Fuchs M. Microchip electrophoretic immunoassay for serum cortisol. Anal Chem. 1996;68:18–22. doi: 10.1021/ac9508311. [DOI] [PubMed] [Google Scholar]
- Koutny LB, Schmalzing D, Adourian A, Chisholm D, Matsudaira P, Ehrlich D. Proceedings from the Ninth International Symposium on Human Identification, 1998, Orlando, Florida. 1999. High speed STR analysis on microfabricated electrophoretic devices. Promega Corp., Madison, WI. (In press.) [Google Scholar]
- Liu S, Shi Y, Ja WW, Mathies RA. Optimization of high-speed DNA sequencing on microfabricated capillary electrophoresis channels. Anal Chem. 1999;71:566–573. doi: 10.1021/ac980783v. [DOI] [PubMed] [Google Scholar]
- Luckey JA, Norris TB, Smith LM. Analysis of resolution in DNA sequencing by capillary gel electrophoresis. J Phys Chem. 1993;97:3067–3075. [Google Scholar]
- Ruiz-Martinez MC, Salas-Solano O, Carrilho E, Kotler L, Karger BL. A sample purification method for rugged and high-performance DNA sequencing by capillary electrophoresis using replaceable polymer solutions. A. Development of the cleanup protocol. Anal Chem. 1998;70:1516–1527. doi: 10.1021/ac971143f. [DOI] [PubMed] [Google Scholar]
- Salas-Solano O, Ruiz-Martinez MC, Carrilho E, Kotler L, Karger BL. A sample purification method for rugged and high-performance DNA sequencing using replaceable polymer solutions. B. Quantitative determination of the role of sample matrix components on sequencing analysis. Anal Chem. 1998;70:1528–1535. doi: 10.1021/ac9711448. [DOI] [PubMed] [Google Scholar]
- Schmalzing D, Koutny L, Adourian A, Belgrader P, Matsudaira P, Ehrlich D. DNA typing in thirty seconds with a microfabricated device. Proc Natl Acad Sci. 1997;94:10273–10278. doi: 10.1073/pnas.94.19.10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzing D, Adourian A, Koutny L, Ziaugra L, Matsudaira P, Ehrlich D. DNA sequencing on microfabricated electrophoretic devices. Anal Chem. 1998;70:2303–2310. doi: 10.1021/ac971381a. [DOI] [PubMed] [Google Scholar]
- Schmalzing D, Koutny LB, Adourian A, Chisholm D, Matsudaira P, Ehrlich D. Two-color multiplexed analysis of eight short tandem repeat loci with an electrophoretic microdevice. Anal Biochem. 1999;270:148–152. doi: 10.1006/abio.1999.4067. [DOI] [PubMed] [Google Scholar]
- Simpson PC, Roach D, Woolley AT, Thorson T, Johnston R, Sensabaugh GF, Mathies RA. High-throughput genetic analysis using microfabricated 96-sample capillary array electrophoresis microplates. Proc Natl Acad Sci. 1998;95:2256–2261. doi: 10.1073/pnas.95.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley AT, Mathies RA. Ultra-high-speed DNA sequencing using capillary electrophoresis chips. Anal Chem. 1995;67:3676–3680. doi: 10.1021/ac00116a010. [DOI] [PubMed] [Google Scholar]