Abstract
The enediyne polyketides are secondary metabolites isolated from a variety of Actinomycetes. All members share very potent anticancer and antibiotic activity, and prospects for the clinical application of the enediynes has been validated with the recent marketing of two enediyne derivatives as anticancer agents. The biosynthesis of these compounds is of interest because of the numerous structural features that are unique to the enediyne family. The gene cluster for five enediynes has now been cloned and sequenced, providing the foundation to understand natures’ means to biosynthesize such complex, exotic molecules. Presented here is a review of the current progress in delineating the biosynthesis of the enediynes with an emphasis on the model enediyne, C-1027.
Keywords: Anticancer antibiotic, biosynthesis, C-1027, enediynes, maduropeptin, neocarzinostatin, polyketide synthase
INTRODUCTION
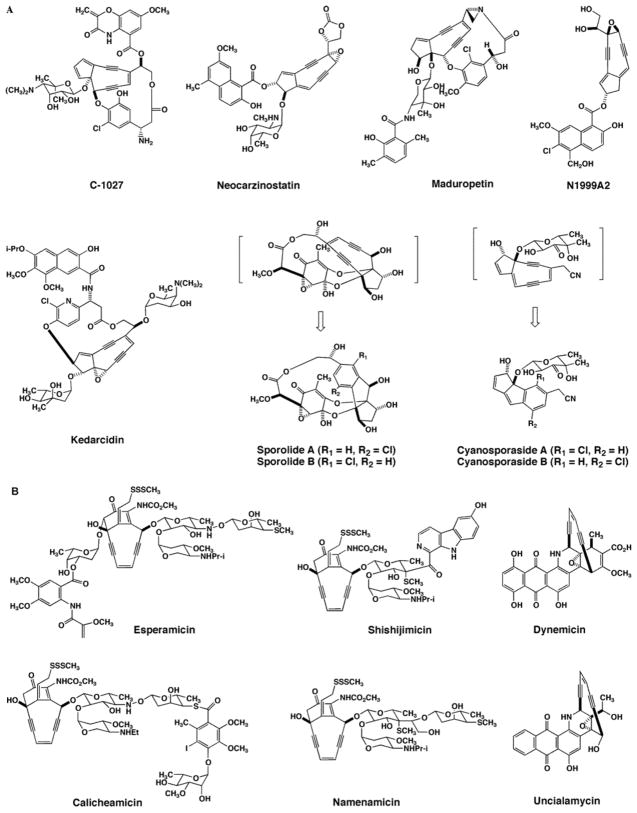
During the 1980s a new class of natural products named the enediynes was introduced with the structural elucidation of neocarzinostatin [1] and calicheamicin [2]. Since this time, thirteen enediynes have been structurally confirmed, which includes two probable enediynes isolated as inactive degradation products (Table 1; Fig. 1) [3–17]. All of the enediyne members share potent antibiotic and antitumor activities with cytotoxicity comparable to any known microbial metabolite [18]. Consequently, much effort has been put forth to develop the enediynes as anticancer agents.
Table 1.
Enediyne Natural Products
| Name | Producer | Yeara | Ref. |
|---|---|---|---|
| Nine-Membered Category | |||
| Auromomycin | Streptomyces macromomyceticus | 1968 | [3] |
| Largomycin | Streptomyces pluricolorescens | 1970 | [4] |
| Actinoxanthin | Actinomyces globisporus | 1976 | [5] |
| Sporamycin | Streptosporangium pseudovulgare | 1978 | [6] |
| Neocarzinostatin | Streptomyces carzinostaticus | 1985 | [1] |
| C-1027 | Streptomyces globisporus | 1991 | [7] |
| Maduropeptin | Actinomadura madurea | 1994 | [8] |
| Kedarcidin | Actinomycete L585-6 | 1997 | [9] |
| N1999A2 | Streptomyces sp. AJ9493 | 1998 | [10] |
| Sporolides A and Bb | Salinispora tropica | 2005 | [11] |
| Cyanosporasides A and Bb | Salinispora pacifica | 2006 | [12] |
| Ten-Membered Category | |||
| Esperamicin | Actinomadura verrucosospora | 1985 | [13] |
| Calicheamicin | Micromonospora echinospora ssp. calichensis | 1987 | [2] |
| Dynemycin | Micromonospora chersina | 1990 | [14] |
| Namenamicin | Polysyncraton lithostrotum | 1996 | [15] |
| Shishijimicin | Didemnum proliferum | 2003 | [16] |
| Uncialamycin | Unknownc | 2005 | [17] |
Year in which the endiyne metabolites were reported or structure determined.
Tricyclic core was proposed to be derived from an enediyne precursor based on chlorine substitution pattern after cycloaromatization.
Similar to Streptomyces cyanogenus from 16S RNA sequencing.
Fig. 1.
(A) Nine-membered enediynes and (B) ten-membered enediynes that have been structurally elucidated. The enediyne structures shown in brackets are hypothetical.
Regardless of their phenomenal biological activity, members of the enediyne family display structural rarities that have hitherto been unseen in other natural products. All of the members share a characteristic unsaturated core containing two acetylenic groups conjugated to a double bond, harbored within a nine- or ten-membered ring termed the enediyne core. The majority of the ten-membered enediynes contain an allylic trisulfide that is directly implicated in triggering the generation of the reactive chemical species for their biological activity. Several of the ten-membered enediynes also contain unusual thiol-sugars. Finally, while mono-chlorinated aromatic moieties are common in enediyne structures, unique to the ten-membered enediyne calicheamicin is an iodinated orsellinic acid component that has been shown to directly aid in DNA-binding [19].
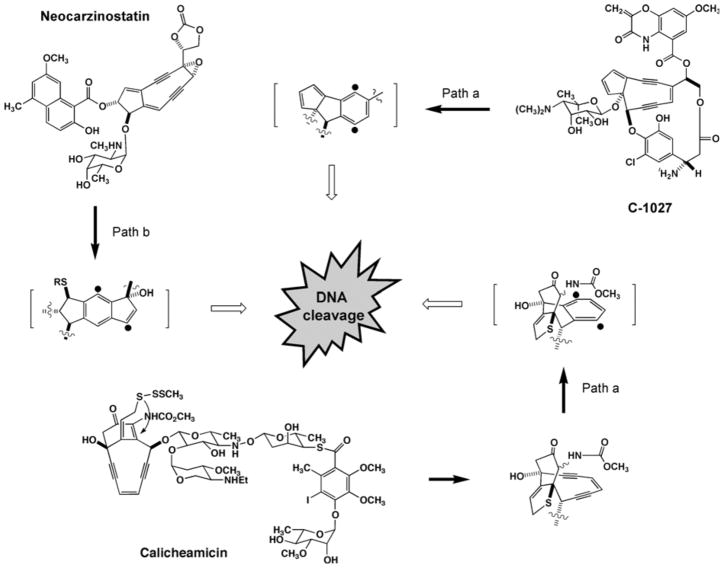
While the peripheral chemistry displayed for each enediyne fine-tunes the biological activity, it is the enediyne core that is the hallmark feature essential for their remarkably potent cytotoxicity. Upon an environmental trigger such as thiol activation or ultraviolet light, the enediyne core undergoes Bergman or Myers-Saito cyclization to yield a benzenoid diradical (Fig. 2). The diradical species is capable of abstracting hydrogen atoms from DNA, resulting in adverse cellular effects such as mutations and environmental trigger. C-1027 [20] and maduropeptin [21] are exceptionally reactive and have been shown to cycloaromatize without external activation. If they are indeed enediynes, the sporolides [11] and cyanospora-sides [12] represent the extreme case since neither is isolated in the enediyne form.
Fig. 2.
Enediyne activation via Bergman (path a) or Myers-Saito (path b) cyclization to yield a benzenoid diradical that causes the eventual DNA cleavage.
The biosynthesis of the enediynes is intriguing because of the plethora of unique chemical features incorporated into each molecule. The origin of the enediyne core was initially studied using isotope-labeling experiments by monitoring the production of neocarzinostatin [22], dynemicin [23], and esperamicin [24]. While the data unequivocally established acetate as a precursor unit, the results did not distinguish whether the enediyne core was constructed by the degradation of fatty acids or by de novo biosynthesis with a dedicated fatty acid or polyketide synthase (PKS). Since the time of the feeding experiments, the gene cluster for 5 enediynes, C-1027 [25], neocarzinostatin (NCS) [26], calicheamicin [27], dynemicin [28,29], and maduropeptin [63], have been cloned and characterized providing the foundation to investigate enediyne biosynthesis. Presented here are highlights of recent molecular and biochemical studies deciphering enediyne biosynthesis [30]. The reader is referred to other articles in this issue and a number of excellent reviews for information regarding total synthetic accomplishments [31–34], mechanisms of resistance to enediynes [35,36], and the biological activity of the enediyne family [37,38].
FINDING THE GENE CLUSTER
A distinguishing characteristic of the nine-membered enediyne family is the isolation of the molecule as a chromoprotein complex consisting of a binding protein, also known as the apo-protein, and a dissociable enediyne chromophore. The binding protein directs transport of the reactive enediyne chromophore to the extracellular environment [35,39], and is also essential for self-resistance by stabilizing the reactive enediyne chromophore. The latter function has been suggested from observations that the gene for the binding protein of C-1027 [40] and macromomycin [41] are constitutively expressed and, as previously mentioned, the 9-membered enediynes C-1027 and maduropeptin readily undergo cycloaromatization in the absence of the binding protein. Interestingly, no binding protein was found for N1999A2, which is the only 9-membered enediyne that has been isolated in the enediyne form without a binding protein [10].
Pioneering work on the isolation of C-1027 and NCS chromoprotein complexes established the amino acid sequence of the homologous binding proteins [35], which ultimately led to the cloning and the sequencing of the gene (cagA for C-1027 [42] and ncsA for NCS [43]). This locus was subsequently used, in combination with degenerate primers for a predicted dNDP-glucose-4,6-dehydratase gene ubiquitously found in pathways for the production of 6-deoxy aminosugars, as probes to identify and localize the entire gene cluster for C-1027 [25,44]. A different strategy was used to clone the gene cluster for the ten-membered enediyne calicheamicin since there was no luxury of a binding protein and its sequence [27]. By screening clones capable of conferring calicheamicin resistance and using PCR-based screens, followed up by DNA-shotgun sequencing, the calicheamicin gene cluster was localized and cloned. While numerous differences between the clusters exist, a unified biosynthetic scheme for the nine- and ten-membered enediynes was evident with the uncovering of a shared iterative type I PKS that is unique to the enediyne family, and many shared open reading frames are apparent [28]. The conserved architecture of enediyne biosynthetic gene clusters, featuring the enediyne PKS, formed the basis of several expedient strategies to clone additional enediyne biosynthetic gene clusters as exemplified by the NCS, maduropeptin, and dynemicin gene clusters via a PCR approach [28] and numerous yet-to-be characterized enediyne gene clusters from previously unknown producers via a genome scanning method [29].
GENERAL BIOSYNTHETIC STRATEGY
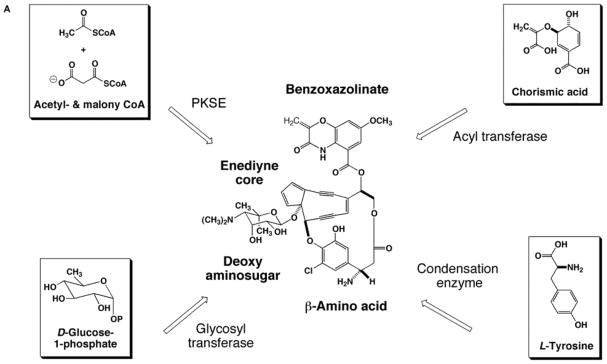
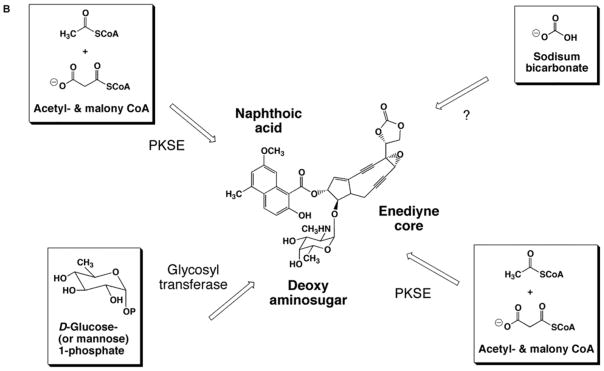
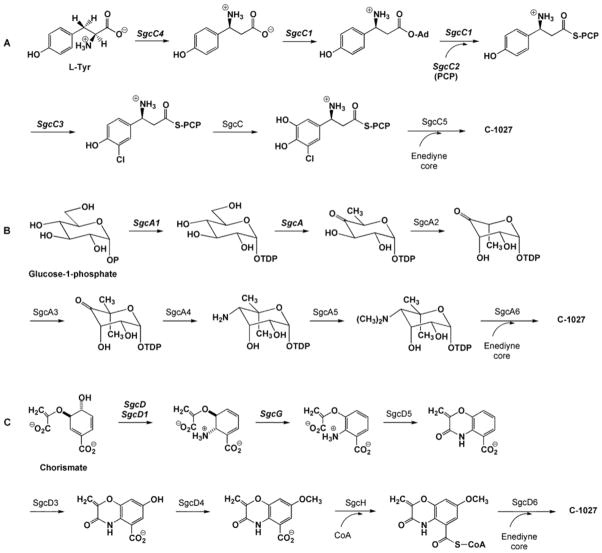
The C-1027 chromophore can be dissected into 4 biosynthetic building blocks: an enediyne core, a β-amino acid, a deoxy aminosugar, and a benzoxazolinate moiety (Fig. 3A). Sequencing of the gene cluster revealed homologs for a glycosyl transferase, an acyltransferase, and condensation enzymes, suggesting a convergent biosynthetic approach is used in enediyne assembly [25]. Similarly, the calicheamicin [27], NCS [26], and maduropeptin [63] gene clusters also contain open reading frames with analogous predictions, as shown in Fig. 3B for NCS. As discussed below, the initial enzymatic step(s) in every pathway for C-1027 have now been analyzed using a combination of in vivo gene inactivation and in vitro characterization of recombinant enzymes, unambiguously establishing the starting metabolite for each moiety and providing substantial evidence for a convergent approach in enediyne biosynthesis.
Fig. 3.
A model of convergent biosynthesis for the enediynes as exemplified by (A) C-1027 and (B) neocarzinostatin.
PKSE: AN ITERATIVE TYPE I PKS
Bacterial polyketide biosynthesis typically follows one of three paradigms: biosynthesis by a noniterative, modular PKS (type I), a multienzyme complex of iteratively acting PKS activities (type II), or homodimeric, iteratively acting condensing PKS without an acyl carrier protein (ACP) (type III) [45,46]. While these three PKS archetype have provided the molecular rationale to explain structural diversity among polyketides as well as have provided the basis for the discovery of novel natural products by genetic engineering, data from numerous genome-sequencing and secondary metabolite gene cluster-sequencing projects have now revealed alternative means to polyketide biosynthesis that extend beyond the classical paradigms. The enediyne PKS family represents one such extension.
To date, among the five gene clusters cloned only one type of PKS has been found, an iterative type I PKS. The gene cluster for C-1027 contains a single PKS that is shared by sequence homology and domain architecture among the enediyne family (Fig. 4A). Disruption of this PKS (SgcE) in the C-1027 producer abolished production of C-1027, which was restored by the introduction of a complementation plasmid containing sgcE under control of the constitutive ermE* promoter [25]. Identical results have been obtained with the homologous neocarzinostatin PKS, NcsE [26], and in total the results provided definitive evidence that the enediyne core is produced by a polyketide pathway.
Fig. 4.
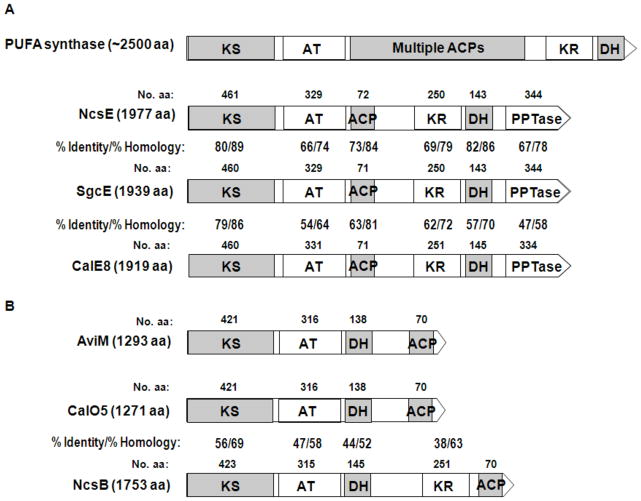
Architecture and domain organization of ierative type I PKSs found in enediyne gene clusters: (A) the enediyne PKSE and its relationship with PUFA synthase and (B) PKS for the aromatic polyketide moieties of enediynes and its comparison with orsellinic acid synthase AviM. ACP, acyl carrier protein, AT, acyl transferase; DH, dehydratase; KR, ketoreductase; KS, ketosynthase; PPTase, phosphopantetheinyl transferase.
Bioinformatic analysis of this PKS family, PKSE, identified 4 domains in the following order (N- to C-terminus): a ketosynthase (KS), an acyltransferase (AT), a ketoreductase (KS), and dehydrogenase (DH), with closest sequence homology to polyunsaturated fatty acid (PUFA) synthases involved in the biosynthesis of docosahexaenoic acid in Moritella marina and eicosapentaenoic acid in Shewanella [47] (Fig. 4A). An internal region located between the AT and KR was initially proposed to be an ACP based on the exact architecture to PUFA synthases, although this region has no homology to any proteins in public databanks. The C-terminal domain based on structural modeling predictions, was proposed to be a phosphopantetheinyl transferase (PPTase) responsible for loading the phosphopantetheine (Ppant) cofactor to the ACP to initiate polyketide biosynthesis [29].
Amino acids essential for the function of individual domains were tested using the established in vivo system for the C-1027 producer and the ΔsgcE mutant by preparing point mutations of SgcE within the complementation construct [64]. Of the multiple mutations prepared, the results have established that S974, the serine originally hypothesized to be the site of Ppant modification within the ACP domain, and D1827, an aspartic acid proposed to be critical for PPT activity within the C-terminal PPT domain, both abolished the production of C-1027, consistent with the designation of the ACP and PPTase domains.
The PKSE was also analyzed using recombinant protein to further probe domain predictions. SgcE and NcsE are produced as soluble proteins in E.coli, and production of PKSE results in a yellow pigment associated with the protein and cell debris suggesting the protein is in the active form. The purified ~210 kDa protein was subjected to trypsin digestion and peptide mapping using liquid chromatography-fourier transform mass spectroscopy. As predicted S974 and the analogous residue of NcsE had +340 amu shifts consistent with modification by a Ppant group. Mutation of this serine eliminated the mass shift, as did mutations within the predicted PPTase domain. In contrast, all mutations outside the ACP and PPTase domains did not alter the observed Ppant mass shift. As expected, the S974A and D1827A point mutations also eliminated the production of the yellow pigment associated with the sgcE or ncsE gene product. In total the data is consistent with self-phosphopanetheinylation of a unique ACP, unambiguously establishing an ACP-dependent PKSE-catalyzed pathway for enediyne biosynthesis (Fig. 5A). While self-phosphopantetheinylation has been observed for a yeast fatty acid synthase [48], this is the first example of a bacterial PKS with such catalytic activity and domain organization [49], representing a new PKS paradigm.
Fig. 5.
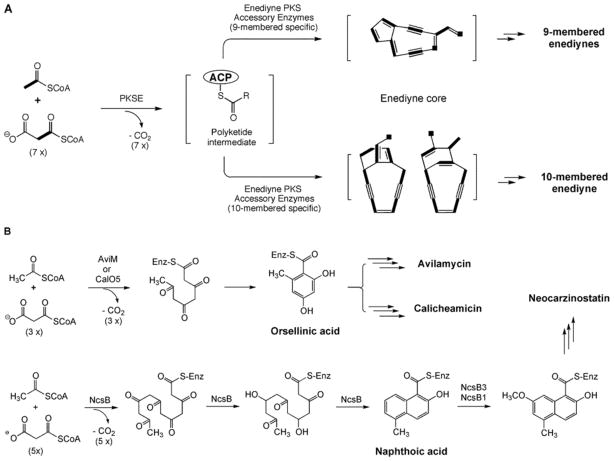
(A) Proposed pathway for biosynthesis of a polyunsaturated intermediate (structure unknown) from acyl CoA by PKSE and the subsequent transformation by enediyne PKS associated enzymes into putative nine- or ten-membered enediyne cores that are finally tailored to individual enediyne natural product. Atoms that were incorporated intact from acyl CoA precursors to the enediyne cores are shown in bold. (B) Proposed pathway for the biosynthesis of aromatic polyketides by iterative type I PKS as exemplified by AviM or CalO5 for orsellinic acid and by NcsB for a naphthoic acid.
OTHER ITERATIVE TYPE I PKS
In addition to the shared iterative type I PKS, PKSE, a second, separate family of iterative type I PKS is also found in the calicheamicin [27], NCS [26], and maduropeptin [62] gene clusters. The 3 PKSs have a minimum of 4 domains in the following order: KS, AT, DH, and ACP, and the domain organization is similar to AviM involved in the biosynthesis of the orsellinic acid of avilamycin in S. viridochromogenes Tü57 (Fig. 4B) [50]. The calicheamicin gene cluster contains CalO5, presumed to be responsible for the production of the orsellinic acid moiety [27]. Within the maduropeptin gene cluster, a PKS (MadB) was discovered that is likely responsible for the biosynthesis of the 6-methylsalicyclic acid moiety [63]. Finally, the neocarzinostatin gene cluster contains a second iterative PKS, NcsB [28], and this PKS has already been shown to heterologously produce the precursor naphthoic acid of NCS [51]. Together, the family represents a separate class of iterative type I PKS that provides an alternative biosynthetic approach to monocyclic or bicyclic aromatic polyketides (Fig. 5B).
PATHWAYS OF C-1027 BIOSYNTHESIS
The majority of the biosynthetic investigations have focused on elucidating the initial steps of C-1027 biogenesis. An attractive feature of this enediyne is the robust genetic system that has been established for the producing organism Streptomyces globisporus [25,44]. Of the 56 open reading frames contained within the boundaries of the cluster, 30 have been successfully inactivated confirming their involvement in C-1027 production [Ben Shen, unpublished data]. While the majority solely abolished C-1027 production, several mutants accumulated new intermediates, and, in total, the gene inactivations have provided the preliminary rationale for in vitro examination of specific biosynthetic enzymes.
Sequence analysis of a free-standing adenylation enzyme, SgcC1, suggests the β-amino acid moiety originates from L-α-tyrosine, which, through 5 steps, is converted to the final product (Fig. 6A). While the biosynthesis does indeed begin with L-α-tyrosine, the initial step is not catalyzed by SgcC1 but has been shown to be the conversion of L-α-tyrosine to (S)-β-tyrosine by an unprecedented aminomutase, SgcC4 [52,53]. It is the SgcC4 product that is the preferred substrate of SgcC1 that only loads β-tyrosine and other β-analogs, as opposed to any α-amino acid, to a free-standing peptidyl carrier protein (PCP), SgcC2 [54]. After covalent tethering to the PCP, a flavin-dependent halogenase SgcC3 incorporates chlorine followed by hydroxylation catalyzed by SgcC. While inactivation of sgcC3 [54] and sgcC [25] resulted in the isolation of the deschloro- and deshydroxy-analogs of C-1027, suggesting these two steps may occur after incorporation to the enediyne core (i.e., the final steps in C-1027 maturation), we now have in vitro data supporting halogenation and hydroxylation occurring on the SgcC2 PCP-tethered β-tyrosine substrate. Finally, a type II condensation enzyme, SgcC5, incorporates the fully modified β-tyrosyl moiety into the enediyne core, although the precise timing of the coupling step relative to the final assembly of other moieties remains to be resolved.
Fig. 6.
Proposed biosynthetic pathways of the peripheral moieties of C-1027: (A) β-amino acid, (B) deoxy aminosugar, and (C) benzoxazolinate. Recombinant enzymes that have been characterized experimentally in vitro are shown in bold-italics.
The initial step in the biosynthesis of the deoxy aminosugar of C-1027 has been confirmed and the recombinant enzyme, SgcA1, characterized (Fig. 6B) [55]. SgcA1, similar to the large family of NDP-sugar synthases, utilizes dTTP and α-D-glucose to form dTDP-glucose. Inspired by the biochemical data, a significant yield improvement in C-1027 isolation was accomplished by overexpressing sgcA1 and it’s flanking region in the producer S. globisporus [55]. Subsequent steps in the deoxy aminosugar biosynthesis involve 5 additional enzymes, a dTDP-glucose-4,6-dehydratase (SgcA), a dTDP-4-keto-6-deoxyglucose epimerase (SgcA2), a C-methyl transferase (SgcA3), an amino transferase (SgcA4), an N-methyl transferase, and a glycosyl transferase, catalyzing conversions common for deoxy aminosugar biosynthesis in secondary metabolism [56].
The remaining moiety of C-1027 is the benzoxazolinate, an uncommon structural feature in natural products. A subcluster within the C-1027 locus contains homologs to anthranilate synthase components I and II (SgcD and SgcD1), suggesting the pathway begins from chorismate via the conversion to anthranilate. Production and purification of recombinant SgcD did not result in anthranilate synthase activity, but instead SgcD converts chorismate to 2-amino-2-deoxyisochorismate [65], which is the first half-reaction of anthranilate synthase I (Fig. 6C) [57]. Subsequently, an iron-sulfur, flavin-dependent dehy-drogenase, SgcG, has been shown to convert the SgcD product into 3-O-enolpyruvalan-thranilate. Final reactions include amide bond formation (SgcD5), hydroxylation (SgcD3), O-methylation (SgcD4), and CoA-activation (SgcH) and covalent attachment to the enediyne core (SgcD6), although the final steps await in vitro characterization.
Although differences will undoubtedly exist in the maturation process for assembly of other enediynes, the general biosynthetic strategy of C-1027 is likely shared among the family. Findings from studying the C-1027 biosynthetic machinery as a model system, therefore, should greatly facilitate the characterization of other enediyne biosynthetic pathways.
NEW PATHWAYS AND UNUSUAL CHEMISTRY
As expected from the exotic structures, many of the enzymatic steps in C-1027 and enediyne biosynthesis represent new types of chemical conversions or contain unexpected transformations and pathways. Firstly, PKSE represents a new architecture for a bacterial PKS family, and contains ACP and PPTase domains that do not have homology to proteins with the respective functions. The ability to heterologously express sgcE in E. coli in an active form has now set the stage to identify the first intermediate produced by PKSE and explore the activity and mechanism of iterative type I PKS, both of which are currently in progress. Furthermore, the results also laid the groundwork to explore the mechanism of triple bond formation, of which little is currently known [58].
Secondly, during the first step of the biosynthesis of the modified-β-amino acid, an aminomutase SgcC4 catalyzes amine migration. Unlike other aminomutases described to date, SgcC4 does not employ any of the following cofactors: cobalamin (or AdoMet) and an iron-sulfur cluster, pyridoxamine phosphate, or ATP [53]. Instead, SgcC4 is post-translationally modified to contain a 4-methylidenei-midazole-5-one (MIO), a cofactor that is necessary for phenylalanine or histidine ammonia lyase activity. However, SgcC4 extends ammonia lyase activity by catalyzing Michael addition of the released ammonia to the bound p-hydroxycinnamic acid intermediate to form (S)-β-tyrosine, representing a novel class of aminomutases [59].
Processing of (S)-β-tyrosine is carried out by a free-standing adenylation domain, SgcC1. While SgcC1 was predicted to activate L-α-tyrosine from sequence comparisons, activity tests with α-amino acids were to no avail [54]. After discovering the novel aminomutase, it became evident that SgcC1 activity was specific for S-β-tyrosine as we have now demonstrated [54]. Interestingly, while absolutely no α-amino acids are turned over by SgcC1, several β-tyrosine analogs are recognized and processed setting the stage to engineer new C-1027 analogs using a chemoenzymatic approach [66].
Finally, the discovery of SgcD activity represents the challenges that remain in analyzing sequence data for the purpose of pathway predictions. SgcD has lost the 2-amino-2-deoxyisochorismate lyase activity inherent in anthranilate synthase, a prediction not readily observed from sequence comparisons [65]. At least 4 different families of enzymes with sequence homology to anthranilate synthase are now known, suggesting the anthranilate synthase template is readily amenable to mutations that evolve new functions [60,61]. Furthermore, the tandem conversion by SgcD and SgcG represents yet another biosynthetic pathway that originates from chorismate, of which a minimum of 6 is currently known [62].
During the cloning and sequencing of the maduropeptin gene cluster, a new type of enediyne binding protein was identified [63]. The yellow pigment associated with expression of SgcE and NcsE was identified as a linear polyene; when sgcE or ncsE was coexpressed with the gene encoding a putative type II thioesterase found within all enediyne gene clusters, 1,3,5,7,9,11,13-pentadecaheptaene was isolated and spectroscopically characterized [64]. The X-ray crystal structure of the aminomutase SgcC4 was recently reported [68]. While SgcC1 adenylates several β-tyrosine analogs, kinetic analysis revealed SgcC1 is most efficient with (S)-β-tyrosine, suggesting (S)-β-tyrosine is the in vivo substrate [66]. The halogenase SgcC3 has been characterized as an FAD-dependent halogenase that regiospecifically chlorinates (S)-β-tyrosyl-S-SgcC2 [69]. Genome sequencing of the marine actinomycete Salinispora tropica revealed two independent enediyne PKS homologs suggesting this organism produces enediyne natural products [70]. Genetic engineering with the C-1027 producer has led to the biosynthesis of new enediyne analogs that have an altered ability to produce double-strand breaks and interstrand cross-links in cellular DNA and an altered cellular response to damage [71].
PROSPECTS FOR CLINICAL APPLICATIONS
Despite their remarkable potency as a family, only two second generation of enediynes have seen use in clinical settings: a polymer derivative of NCS (SMANCS®) marketed in Japan and an antibody conjugate of calicheamicin (Mylotarg®) used in the United States. So how can the information gathered from the biosynthetic studies presented here be applied to discovering enediynes with a better therapeutic index?
First and foremost, new enediynes are continually being discovered (Table 1). We have used a comparative genomics approach based on the knowledge of a few known enediyne gene clusters to discover new potential enediynes [29]. Furthermore, even without using a genomics-guided approach, standard screens for bioactive metabolites have yielded shishijimicins A-C [16], uncialamycin [17], sporolides A and B [11], and cyanosporasides A and B [12]. Interestingly, these new enediynes were isolated from marine organisms, a diverse population that has generated substantial interest due to its rich, untapped source of natural products. Secondly, the producers for C-1027 [25] and NCS [26] are readily manipulated using genetic engineering, which has allowed the isolation of new enediyne analogues with altered reactivity that have improved bioactivity traits [67]. Finally, the biosynthetic studies discussed here have paved the way to rationally engineer new enediynes that are of great challenge to prepare by total synthesis. Our biosynthetic studies have also revealed that heterologous hosts such as E. coli are suitable for enediyne engineering since the prerequisite building blocks are abundant and because of the self-activating nature of the enediyne PKS, PKSE.
In total, the foundation for the discovery of the next generation of enediynes is now in place. Using a combination of synthetic and biosynthetic studies, large libraries of enediynes can be prepared using combinatorial biosynthesis and chemoenzymatic approaches. It is anticipated a fraction of these compounds will have the appropriate compromise of potency and cytotoxicity to be utilized as anticancer agents.
Acknowledgments
We acknowledge and are grateful for the work of Dr. Pieter C. Dorrestein and Prof. Niel L. Kelleher, University of Illinois at Urbana-Champaign, who performed FT-MS experiments with recombinant SgcE, NcsE, and SgcC2. We also acknowledge other contributors from the Shen Lab for the unpublished data, including Wen Liu, Jian Zhang, Shuangjun Lin, Wenli Li, and Jianhua Ju, other members of the Shen Lab whose contribution were cited in the references, and Prof. Y. Li, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences, Beijin, China for the wild-type C-1027 producing strain of Streptomyces globisporus. Enediyne biosynthesis described from the Shen Lab was supported in part by National Institute of Health grant CA78747. S.V.L is a recipient of an NIH postdoctoral fellowship CA1059845, and B.S is the recipient of an NIH Independent Scientist Award AI51689.
ABBREVIATIONS
- ACP
Acyl carrier protein
- AT
Acyltransferase
- DH
Dehydratase
- KR
Ketoreductase
- KS
Ketosynthase
- NCS
Neocarzinostatin
- PCP
Peptidyl carrier protein
- PKS
Polyketide synthase
- Ppant
Phosphopantetheine
- PPTase
Phosphopantetheinyl transferase
- PUFA
Polyunsaturated fatty acid
References
- 1.Edo K, Mizugaki M, Koide Y, Seto H, Furihata K, Otake N, Ishida N. The Structure of Neocarzinostatin Chromophore Possessing a Novel Bicyclo[7.3.0]dodecadiyne System. Tetrahedron Lett. 1985;26:331–340. [Google Scholar]
- 2.Lee MD, Dunne TS, Siegel MM, Chang CC, Morton GO, Borders DB. Calichemicins, a Novel Family of Antitumor Antibiotics. 1. Chemistry and Partial Structure of Calichemicin. Gamma.1I. J Am Chem Soc. 1987;109:3464–3466. [Google Scholar]
- 3.Chimura H, Ishizuka M, Hamada M, Hori S, Kimura K. A New Antibiotic, Macromomycin, Exhibiting Antitumor and Antimicrobial Activity. J Antibiot. 1968;21:44–49. doi: 10.7164/antibiotics.21.44. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguch T, Furumai T, Sato M, Okuda T, Ishida N. Studies on a New Antitumor Antibiotic, Largomycin. I. Taxonomy of the Largomycin-Producing Strain and Production of the Antibiotic. J Antibiot. 1970;23:369–372. doi: 10.7164/antibiotics.23.369. [DOI] [PubMed] [Google Scholar]
- 5.Khokhlov AS, Reshetov PD, Chupova LA, Cherches BZ, Zhigis LS, Stoyachenko IA. Chemical Studies on Actinoxanthin. J Antibiot. 1976;29:1026–1034. doi: 10.7164/antibiotics.29.1026. [DOI] [PubMed] [Google Scholar]
- 6.Komiyama K, Umezawa I. Tissue Pharmacokinetics and Inhibition of DNA Synthesis in Mice Treated with Sporamycin. J Antibiot. 1978;31:473–476. doi: 10.7164/antibiotics.31.473. [DOI] [PubMed] [Google Scholar]
- 7.Otani T, Minami Y, Sakawa K, Yoshida K. Isolation and Characterization of Non-Protein Chromophore and Its Degradation Product from Antibiotic C-1027. J Antibiot. 1991;44:564–568. doi: 10.7164/antibiotics.44.564. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder DR, Colson KL, Klohr SE, Zein N, Langley DR, Lee MS, Matson JA, Doyle TW. Isolation, Structure Determination, and Proposed Mechanism of Action for Artifacts of Maduropeptin Chromophore. J Am Chem Soc. 1994;116:9351–9352. [Google Scholar]
- 9.Kawata S, Ashizawa S, Hirama M. Synthetic Study of Kedarcidin Chromophore: Revised Structure. J Am Chem Soc. 1997;119:12012–12013. [Google Scholar]
- 10.Ando T, Ishii M, Kajiura T, Kameyama T, Miwa K, Sugiura Y. A New Non-Protein Enediyne Antibiotic N11999A2: Unique Enediyne Chromophore Similar to Neocarzinostatin and DNA Cleavage Feature. Tetrahedron Lett. 1998;39:6495–6498. [Google Scholar]
- 11.Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Sporolides A and B: Structurally Unprecedented Halogenated Macrolides from the Marine Actinomycete Salinispora tropica. Org Lett. 2005;7:2731–2734. doi: 10.1021/ol050901i. [DOI] [PubMed] [Google Scholar]
- 12.Oh D, Williams PG, Kauffman CA, Jensen PR, Fenical W. Cyanosporasides A and B, Chloro- and Cyano-cyclopenta [a]indene Glycosides from the Marine Actinomycete “Salinispora pacifica”. Org Lett. 2006;8:1021–1024. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 13.Konishi M, Ohkuma H, Saitoh K, Kawaguchi H, Golik J, Dubay G, Groenewold G, Krishnan B, Doyle TW. Esperamicins, a Novel Class of Potent Antitumor Antibiotics. I. Physico-Chemical Data and Partial Structure. J Antibiot. 1985;38:1605–1609. doi: 10.7164/antibiotics.38.1605. [DOI] [PubMed] [Google Scholar]
- 14.Konishi M, Ohkuma H, Tsuno T, Oki T, Vanduyne GD, Clardy J. Crystal and Molecular Structure of Dynemicin A: a Novel 1,5-Diyn-3-ene Antitumor Antibiotic. J Am Chem Soc. 1990;112:3715–3716. [Google Scholar]
- 15.McDonald LA, Capson TL, Krishnamurthy G, Ding WD, Ellestad GA, Bernan VS, Maiese WM, Lassota P, Discafani C, Kramer RA, Ireland CM. Namenamicin, a New Enediyne Antitumor Antibiotic from the Marine Ascidian Polysyncraton lithostrotum. J Am Chem Soc. 1996;118:10898–10899. [Google Scholar]
- 16.Oku N, Matsunaga S, Fusetani N. Shishijimicins A–C, Novel Enediyne Antitumor Antibiotics from the Ascidian Didemnum proliferum. J Am Chem Soc. 2003;125:2044–2045. doi: 10.1021/ja0296780. [DOI] [PubMed] [Google Scholar]
- 17.Davies J, Wang H, Taylor T, Warabi K, Huang X, Andersen RJ. Uncialamycin, A New Enediyne Antibiotic. Org Lett. 2005;7:5233–5236. doi: 10.1021/ol052081f. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Xie H. C-1027. Drugs of the Future. 1999;24:847–852. [Google Scholar]
- 19.Ikemoto N, Kumar RA, LT, Ellestad GA, Danishefsky SJ, Patel DJ. Calicheamicin -DNA Complexes: Warhead Alignment and Saccharide Recognition of the Minor Groove. Proc Natl Acad Sci US A. 1995;92:10506–10510. doi: 10.1073/pnas.92.23.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu YJ, Zhen YS, Goldberg IH. C1027 Chromophore, a Potent New Enediyne Antitumor Antibiotic, Induces Sequence-Specific Double-Strand DNA Cleavage. Biochemistry. 1994;33:5947–5954. doi: 10.1021/bi00185a036. [DOI] [PubMed] [Google Scholar]
- 21.Zein N, Schroeder DR. Kedarcidin and Maduropeptin: Two Novel Antitumor Chromoproteins with Selective Protease Activity and DNA Cleaving Properties. Adv DNA Seq -Spec Agents. 1998;3:201–225. [Google Scholar]
- 22.Hensens OD, Giner J, Goldberg IH. Biosynthesis of NCS Chrom A, the Chromophore of the Antitumor Antibiotic Neocarzinostatin. J Am Chem Soc. 1989;111:3295–3299. [Google Scholar]
- 23.Tokiwa Y, Miyoshi-Saitoh M, Kobayashi H, Sunaga R, Konishi M, Oki T, Iwasaki S. Biosynthesis of Dynemicin A, a 3-Ene-1,5-diyne Antitumor Antibiotic. J Am Chem Soc. 1992;114:4107–4110. [Google Scholar]
- 24.Lam KS, Veitch JA, Golik J, Krishnan B, Klohr SE, Volk KJ, Forenza S, Doyle TW. Biosynthesis of Esperamicin A1, an Enediyne Antitumor Antibiotic. J Am Chem Soc. 1993;115:12340–12345. [Google Scholar]
- 25.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the Enediyne Antitumor Antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. The Neocarzinostatin Biosynthetic Gene Cluster from Streptomyces carzinostaticus ATCC 15944 Involving Two Iterative Type I Polyketide Synthases. Chem Biol. 2005;12:1–10. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The Calicheamicin Gene Cluster and Its Iterative Type I Enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, Ahlert J, Gao Q, Wendt-Pienkowski E, Shen B, Thorson JS. Rapid PCR Amplification of Minimal Enediyne Polyketide Synthase Cassettes Leads to a Predictive Familial Classification Model. Proc Natl Acad Sci USA. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. A Genomics-Guided Approach for Discovering and Expressing Cryptic Metabolic Pathways. Nat Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 30.Shen B, Liu W, Nonaka K. Enediyne Natural Products: Biosynthesis and Prospects towards Engineering Novel Antitumor Agents. Curr Med Chem. 2003;10:2317–2325. doi: 10.2174/0929867033456701. [DOI] [PubMed] [Google Scholar]
- 31.Jones GB, Fouad FS. Designed Enediyne Antitumor Agents. Curr Pharm Des. 2002;8:2415–2440. doi: 10.2174/1381612023392810. [DOI] [PubMed] [Google Scholar]
- 32.Danishefsky SJ, Shair MD. Observations in the Chemistry and Biology of Cyclic Enediyne Antibiotics: Total Syntheses of Calicheamicin γ1I and Dynemicin A. J Org Chem. 1996;61:16–44. [Google Scholar]
- 33.Smith AL, Nicolaou KC. The Enediyne Antibiotics. J Med Chem. 1996;39:2103–2117. doi: 10.1021/jm9600398. [DOI] [PubMed] [Google Scholar]
- 34.Thorson JS, Sievers EL, Ahlert J, Shepard E, Whitwam RE, Onwueme KC, Ruppen M. Understanding and Exploiting Nature’s Chemical Arsenal: The Past, Present, and Future of Calicheamicin Research. Curr Pharm Des. 2000;6:1841–1879. doi: 10.2174/1381612003398564. [DOI] [PubMed] [Google Scholar]
- 35.Thorson JS, Shen B, Whitwam RE, Liu W, Li Y, Ahlert J. Enediyne Biosynthesis and Self-Resistance: A Progress Report. Bioorg Chem. 1999;27:172–188. [Google Scholar]
- 36.Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Antitumor Antibiotics: Bleomycin, Enediynes, and Mitomycin. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 37.Brukner I. C-1027 Taiho Pharmaceutical Co Ltd. Curr Opinion Oncol Endoc Met Invest Drugs. 2000;2:344–352. [Google Scholar]
- 38.Schor NF. The Enediynes. In: Teicher BA, editor. Cancer Therapeutics: Experimental and Clinical Agents; Humana Press Inc; Totowa, NJ: 1997. pp. 229–239. [Google Scholar]
- 39.Xi Z, Goldberg IH. DNA-Damaging Enediyne Compounds. In: Kool ET, editor. Comprehensive Natural Products Chemistry. Vol. 7. Elsevier; New York: 1999. p. 533. [Google Scholar]
- 40.Sakata N, Kanbe T, Tanabe M, Hayashi H, Hori M, Hotta K, Hamada MJ. Nucleotide Sequence of the Macromomycin Apoprotein Gene and Its Expression in Streptomyces macromomyceticus. J Antibiot. 1989;42:1704–1712. doi: 10.7164/antibiotics.42.1704. [DOI] [PubMed] [Google Scholar]
- 41.Sakata N, Ikeno S, Hori M, Hamada M, Otani T. Cloning and Nucleotide Sequencing of the Antitumor Antibiotic C-1027 Apoprotein Gene. Biosci Biotechnol Biochem. 1992;56:1592–1595. doi: 10.1271/bbb.56.1592. [DOI] [PubMed] [Google Scholar]
- 42.Otani T, Yasuhara T, Minami Y, Shimazu T, Zhang R, Xie MY. Purification and Primary Structure of C-1027-AG, a Selective Antagonist of Antitumor Antibiotic C-1027, from Streptomyces globisporus. Agric Biol Chem. 1991;55:407–417. [PubMed] [Google Scholar]
- 43.Gibson BW, Herlihy WC, Samy TS, Hahm KS, Maeda H, Meienhofer J, Biemann K. A Revised Primary Structure for Neocarzinostatin Based on Fast Atom Bombardment and Gas Chromatographic-Mass Spectrometry. J Biol Chem. 1984;259:10801–10806. [PubMed] [Google Scholar]
- 44.Liu W, Shen B. Genes for Production of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus are Clustered with the cagA Gene that Encodes the C-1027 Apoprotein. Antimicrobio Agents Chemother. 2000;44:382–392. doi: 10.1128/aac.44.2.382-392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wenzel SC, Müller R. Formation of Novel Secondary Metabolites by Bacterial Multimodular Assembly Lines: Deviations from Textbook Synthetic Logic. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Shen B. Polyketide Biosynthesis Beyond the Type I, II, and III Polyketide Synthase Paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 47.Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science. 2001;293:290–293. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- 48.Fichtscherer F, Wellein C, Mittag M, Schweizer E. A Novel Function of Yeast Fatty Acid Synthase. Subunit α is Capable of Self-Pantetheinylation. Eur J Biochem. 2000;267:2666–2671. doi: 10.1046/j.1432-1327.2000.01282.x. [DOI] [PubMed] [Google Scholar]
- 49.Kasahara K, Fujii I, Oikawa H, Ebizuka Y. Expression of Alternaria solani PKSF Generates a Set of Complex Reduced-Type Polyketides with Different Carbon-Lengths and Cyclization. ChemBioChem. 2006;7:1–5. doi: 10.1002/cbic.200600034. [DOI] [PubMed] [Google Scholar]
- 50.Weitnauer G, Mühlenweg A, Trefzer A, Hoffmeister D, Süßmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Biosynthesis of the orthosomomycin antibiotic avilamycin A: Deductions from the Molecular Analysis of the avi Biosynthetic Gene Cluster of Streptomyces viridochromogenes Tü57 and Production of New Antibiotics. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- 51.Sthapit B, Oh T, Lamichhane R, Liou K, Lee HC, Kim C, Sohng JK. Neocarzinostatin Naphthoate Synthase: An Unique Iterative Type I PKS from Neocarzinostatin Producer Streptomyces carzinostaticus. FEBS Lett. 2004;566:201–206. doi: 10.1016/j.febslet.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 52.Christenson SD, Liu W, Toney MD, Shen B. A Novel 4-Methylideneimidazole-5-one-Containing Tyrosine Aminomutase in Enediyne Antitumor Antibiotic C-1027 Biosynthesis. J Am Chem Soc. 2003;125:6062–6063. doi: 10.1021/ja034609m. [DOI] [PubMed] [Google Scholar]
- 53.Christenson SD, Wu W, Spies MA, Shen B, Toney MD. Kinetic Analysis of the 4-Methylideneimidazole-5-one-Containing Tyrosine Aminomutase in Enediyne Antitumor Antibiotic C-1027 Biosynthesis. Biochemistry. 2003;42:12708–12718. doi: 10.1021/bi035223r. [DOI] [PubMed] [Google Scholar]
- 54.Van Lanen SG, Dorrestein PC, Christenson SD, Liu W, Ju J, Kelleher NL, Shen B. Biosynthesis of the β-Amino Acid Moiety of the Enediyne Antitumor Antibiotic C-1027 Featuring β-Amino Acyl-S-Carrier Protein Interrmediates. J Am Chem Soc. 2005;127:11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- 55.Murrell JM, Liu W, Shen B. Biochemical Characterization of the SgcA1 α-D-Glucopyranosyl-1-phosphate Thymidylyl-transferase from the Enediyne Antitumor Antibiotic C-1027 Biosynthetic Pathway and Overexpression of sgcA1 in Streptomcyes globisporus to Improve C-1027 Production. J Nat Prod. 2004;67:206–213. doi: 10.1021/np0340403. [DOI] [PubMed] [Google Scholar]
- 56.He XM, Liu HW. Formation of unusual Sugars: Mechanistic Studies and Biosynthetic Applications. Annu Rev Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]
- 57.Morollo AA, Bauerle R. Characterization of Composite Aminodeoxyisochorismate Synthase and Aminodeoxyisochorismate Lyase Activities of Anthranilate Synthase. Proc Natl Acad Sci US A. 1993;90:9983–9987. doi: 10.1073/pnas.90.21.9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee M, Lenman M, Banas A, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson P, Sjodahl S, Green A, Stymne S. Identification of Non-Heme Diiron Proteins That Catalyze Triple Bond and Epoxy Group Formation. Science. 1998;280:915–918. doi: 10.1126/science.280.5365.915. [DOI] [PubMed] [Google Scholar]
- 59.Walker KD, Klettke K, Akiyama T, Croteau R. Cloning, Heterologous Expression, and Characterization of a Phenylalanine Aminomutase Involved in Taxol Biosynthesis. J Biol Chem. 2004;279:53947–53954. doi: 10.1074/jbc.M411215200. [DOI] [PubMed] [Google Scholar]
- 60.He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of Mechanism in Three Chorismate-Utilizing Enzymes. J Amer Chem Soc. 2004;126:2378–2385. doi: 10.1021/ja0389927. [DOI] [PubMed] [Google Scholar]
- 61.Parsons JF, Jensen PY, Pachikara AS, Howard AJ, Eisenstein E, Ladner JE. Structure of Escherichia coli Aminodeoxychorismate Synthase: Architectural Conservation and Diversity in Chorismate-Utilizing Enzymes. Biochemistry. 2002;41:2198–2208. doi: 10.1021/bi015791b. [DOI] [PubMed] [Google Scholar]
- 62.Walsh CT, Liu J, Rusnak F, Sakaitani M. Molecular Studies on Enzymes in Chorismate Metabolism and the Enterobactin Biosynthetic Pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 63.Van Lanen SG, Oh T-J, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the Maduropeptin Biosynthetic Gene Cluster from Actinomadura madurae ATCC 39144 Supporting a Unifying Paradigm for Enediyne Biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A Self-Phosphopantetheinylating Polyketide Synthase Producing a Linear Polyene to Initiate the Biosynthesis of the Enediyne Antitumor Antibiotics. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0711625105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Lanen SG, Lin S, Shen B. The Biosynthesis of the Antitumor Antibiotic C-1027 Involves a New Branching Point in Chorismate Metabolism. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0708750105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Lanen SG, Lin S, Dorrestein PC, Kelleher NL, Shen B. Substrate Specificity of the Adenylation Enzyme SgcC1 Involved in the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027. J Biol Chem. 2006;281:29633–29640. doi: 10.1074/jbc.M605887200. [DOI] [PubMed] [Google Scholar]
- 67.Kennedy DR, Gawron LS, Ju J, Liu W, Shen B, Beerman TA. Single Chemical Modifications of the Enediyne Core of C-1027, a Radiomimetic Antitumor Drug, Result in Markedly Varied Cellular Responses to DNA Double Strand Breaks. Cancer Res. 2007;67:773–781. doi: 10.1158/0008-5472.CAN-06-2893. [DOI] [PubMed] [Google Scholar]
- 68.Christianson CV, Montavon TJ, Van Lanen SG, Shen B, Bruner SD. The Structure of L-Tyrosine 2,3-aminomutase from C-1027 Enediyne Antitumor Antibiotic Biosynthetic Pathway. Biochemistry. 2007;46:7025–7214. doi: 10.1021/bi7003685. [DOI] [PubMed] [Google Scholar]
- 69.Lin S, Van Lanen SG, Shen B. Regiospecific Chlorination of (S)-β-Tyrosyl-S-Carrier Protein Catalyzed by SgcC3 in the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027. J Am Chem Soc. 2007;129:12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Udwary DW, Zeigler L, Asolkar N, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome Sequencing Reveals Complex Secondary Metabolome in the Marine Actinomycete Salinispora tropica. Proc Natl Acad Sci US A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kennedy DR, Ju J, Shen B, Beerman TA. Designer Enediynes Generate DNA Breaks, Interstrand Cross-links, or Both, with Concomitant Changes in the Regulation of DNA Damage Responses. Proc Natl Acad Sci US A. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]