Abstract
With the genomic sequencing of Arabidopsis nearing completion and rice sequencing very much in its infancy, a key question is whether we can exploit the Arabidopsis sequence to identify candidate genes for traits in cereal crops using a map-based approach. This requires the existence of colinearity between the Arabidopsis and cereal genomes, represented by rice, which is readily detectable using currently available resources, that is, Arabidopsis genomic sequence, rice ESTs, and genetic and physical maps. A detailed study of the colinearity remaining between two small regions of Arabidopsis chromosome 1 and rice suggests that at least in these regions of the Arabidopsis genome, conservation of gene orders with rice has been eroded to the point that it is no longer identifiable using comparative mapping. Although our analysis does not preclude that tracts of colinear gene orders may be identified using sequence comparisons or may exist in other regions of the rice and Arabidopsis genomes, it is unlikely that the extent of colinearity will be sufficient to allow map-based cross-species gene prediction and isolation. Our research also highlights the difficulties encountered in identifying orthologs using BLAST searches in incomplete sequence databases. This complicates the interpretation of comparative data among highly divergent species and limits the exploitation of Arabidopsis sequence in monocot studies.
Comparative genome analyses have shown the existence of conserved gene orders (colinearity) in the genomes of different plant and mammal species. In plants, this is best documented in the grass family, where colinearity has been maintained over evolutionary periods as long as 60 million years (Devos and Gale 1997; Gale and Devos 1998). In mammals, the most comprehensive comparative maps are available for human and mouse, which diverged ∼70 million years ago (Carver and Stubbs 1997). Although short-range conserved synteny has been demonstrated between the genomes of human and chicken (Klein et al. 1996) and human and pufferfish (Elgar et al. 1996), which diverged some 300 and 400 million years ago, respectively, conserved synteny does not imply conservation of gene orders. Paterson et al. (1996) predicted that 43%–58% of chromosomal tracts ≤3 cM should have remained colinear over the evolutionary time period [130–240 million years (Wolfe et al. 1989; Crane et al. 1995)] separating the monocots and eudicots and provided some empirical mapping data to support this hypothesis. With a large part of the Arabidopsis sequence available, we aimed to investigate whether Arabidopsis–rice colinearity can be identified and thus exploited using currently available data and tools. The key issue is not the existence of colinearity at the sequence level. It is clear that any colinearity that can be detected only when the genomic sequence is available for both rice and Arabidopsis will have limited applications. Once the rice sequence is available, the exploitation of rice, and not Arabidopsis, sequence will be the priority in cereal research. In the absence of rice genomic sequence, a comparative genetic study of the location in the rice genome of expressed sequence tags (ESTs) with homology to genes from the top of Arabidopsis chromosome 1 failed to demonstrate that gene orders had remained conserved over the monocot-eudicot divide to the extent that Arabidopsis sequence could be exploited for the map-based identification and isolation of genes underlying cereal traits. The study also highlights the practical problems involved in establishing relationships between highly divergent genomes.
RESULTS AND DISCUSSION
To establish whether gene orders remained conserved over smaller genetic distances, genes belonging to the same BAC and to BACs spaced over two regions of maximum 3 cM of Arabidopsis chromosome 1 were selected for an Arabidopsis–rice comparative study (Fig. 1a). BLAST searches identified one or more rice ESTs (n ≥ 200) for 53 of 128 annotated Arabidopsis genes from five BAC clones. A further 47 hits were obtained when querying the rice EST database with the complete sequence of five nonannotated BACs (data available at http://pgec-genome.pw.usda.gov/sequencing.html in January 1998). Initially, Arabidopsis genes for which no putative rice homolog could be identified at the nucleotide level, were rescreened at the amino acid level. However, levels of homology between Arabidopsis genes and rice ESTs that were identified at the amino acid level only were generally too low for the hits to be considered orthologs. BLAST searches at the amino acid level were therefore not pursued further. Because it was expected that a number of the rice hits obtained at the nucleotide level were also not orthologous to the Arabidopsis query sequence, the rice ESTs were rescreened against the Arabidopsis database. About 30% of the identified rice ESTs displayed a higher degree of homology to Arabidopsis BACs other than those originally used to select them, indicating that they do not correspond to the Arabidopsis genes in the target region.
Figure 1.
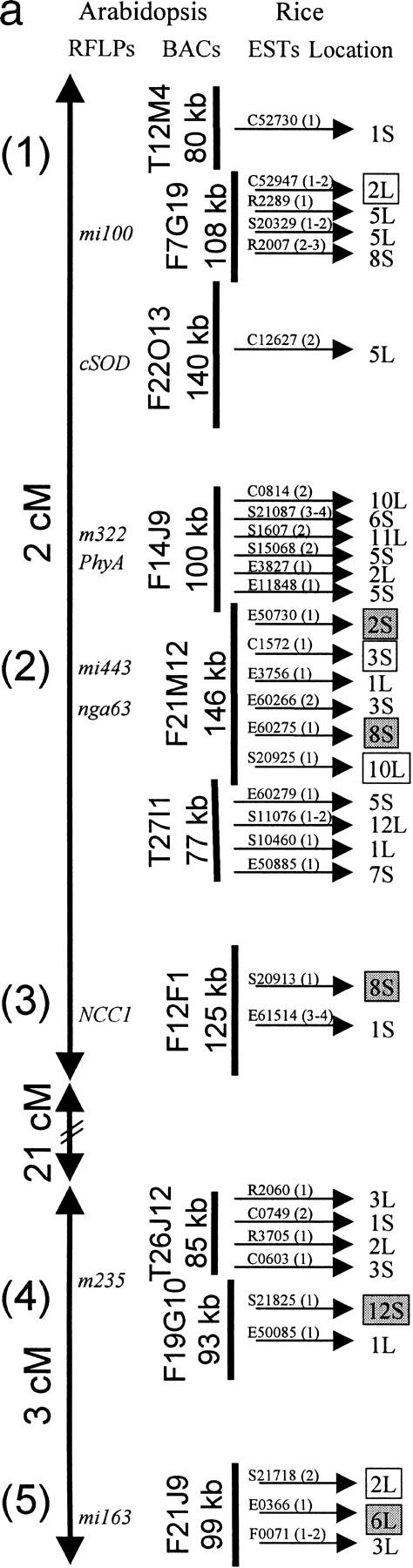
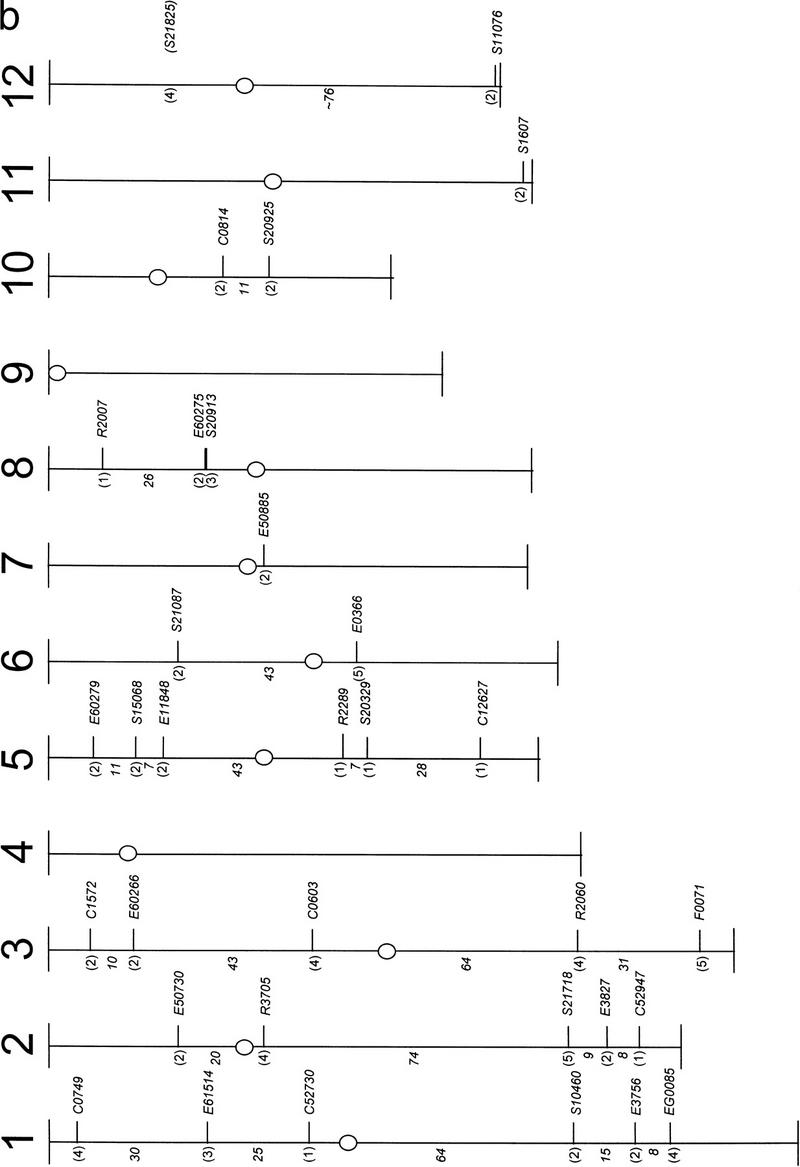
Comparative analysis of the location of homologous sequences in Arabidopsis and rice. (a) Rice ESTs with putative homology to Arabidopsis genes from 10 BAC clones organized in five contigs, and their chromosomal location and copy number in rice. Copy numbers were estimated from the number of hybridizing fragments. Probes detecting a strong hybridizing fragment in addition to weak fragments were assigned a copy number of 1. Boxed locations indicate that the rice ESTs displayed homology at both 5′ and 3′ ends with the BAC sequence. Shaded boxes indicate that homology of loci was established based on cross-hybridization data. Genetic distances are shown but not drawn to scale. Contig numbers are indicated in parenthesis. (b) Map position in rice for the identified rice ESTs. The contig number is given in parenthesis. Centimorgan distances between the loci are in italics. (○) Centromeres.
Of the remaining rice ESTs, 33 were mapped to 33 loci on 10 of the 12 rice chromosomes (Fig. 1b). Their estimated copy number in rice, based on the number of hybridizing fragments, is given in Figure 1a. For probes that detected weak fragments in addition to a strongly hybridizing fragment, only the map position of the latter was included in the analysis. Sixty-seven percent of these loci mapped to rice chromosomes 1, 2, 3, and 5. This could be defined as synteny strictu sensu, that is, genes lying on the same chromosomes, without making assumptions about genetic linkage. Potential regions of conserved synteny could be observed, for example, between contigs 1 and 2 of Arabidopsis chromosome 1 and rice chromosome 5 (Fig. 1b). Nevertheless, within these regions, little evidence was found for conserved gene orders.
Elsewhere on rice chromosome arm 8S, two markers from contigs 2 (E60275) and 3 (S20913), which spanned a genetic distance of 1.2 cM in Arabidopsis, detected closely linked (0.3 cM) loci (Fig. 1). No conserved positions in rice were found, however, for the other Arabidopsis genes located in this apparently conserved region. Assuming that this linkage is a remnant of ancestral gene associations, our data suggest that the conservation between monocot and eudicot species of ∼50 % of 3-cM intervals, as suggested by Paterson et al. (1996), may be an overestimate. A level of conserved synteny, as identified between Arabidopsis chromosome 1 and rice, has limited applications in map-based gene prediction.
It thus appears that the 130–240 million years that separate Arabidopsis and rice have largely eroded close linkages, at least in the area under investigation. One can argue that although a number of nonorthologs were discarded based on the results of a reciprocal BLAST search, no evidence is available to suggest that the remaining rice ESTs are orthologous to the Arabidopsis target genes on chromosome 1. Rice ESTs were initially selected mainly on the basis of 5′ homology. To estimate the extent of homology between the rice and the corresponding Arabidopsis genes more accurately, 17 of the mapped rice ESTs were also sequenced from the 3′ end, and these 3′ ends were subjected to BLAST searches against the Arabidopsis database. Nine clones showed a level of homology with both the 5′ and 3′ ends to the BAC originally used to select the rice ESTs (Fig. 1a). For one rice EST, different BACs were identified with the 3′ and 5′ end sequences, whereas the 3′ ends of the remaining clones displayed no homology to Arabidopsis BAC sequences. Lack of homology, especially when the 3′ end sequence consists of <400 bp, may be explained by the presence of a 3′-untranslated region, which is unlikely to have remained conserved between Arabidopsis and rice. However, it is possible that a number of these BLAST hits represent domain homologies rather than gene homologies.
To determine the level of error introduced in BLAST-based comparative analyses, correspondence of the hybridization patterns of 11 Arabidopsis exons and their putative rice homologs were used to evaluate orthology between the identified Arabidopsis and rice sequences. From the seven genes that cross-hybridized, two produced a pattern different than that of the corresponding rice ESTs. Good correspondence between the hybridization patterns was obtained for the remaining five Arabidopsis sequences and rice ESTs, all of which displayed homology with Arabidopsis at both the 5′ and 3′ ends. However, for genes belonging to multigene families or displaying different copy numbers in rice and Arabidopsis, paralogous loci could have been mapped in the two species. From the nine mapped rice ESTs that displayed good homology at both the 5′ and 3′ ends with Arabidopsis, only three produced single copy patterns in both rice and Arabidopsis. For two further ESTs, a comparison of the relative signal strengths of the hybridizing fragments in Columbia (the ecotype used to construct the Arabidopsis BAC library), in the target BAC, and in rice confirmed that orthologous loci had been mapped in Arabidopsis and rice (Fig. 1a). No conclusions could be drawn for the other four genes. Although this stringent selection retained the two loci that were linked in both Arabidopsis and rice, colinearity was not maintained for an additional locus from the same BAC (Fig. 1a). Two loci from two other BACs that mapped <3 cM apart were also not linked in rice (Fig. 1a).
In addition to demonstrating a lack of identifiable genome conservation between the top of Arabidopsis chromosome 1 and rice using currently available comparative mapping tools, our study highlighted the need for careful interpretation of comparative data between species as divergent as monocots and eudicots. The alignment of conserved domains in nonorthologous genes in BLAST queries and the identification of different members of multigene families may confound relationships. Although this would most likely lead to an underestimation of the extent of synteny, the alignment of nonorthologous genes following a BLAST search in a database biased toward genes with nonrandom genome distribution could provide apparent support for conserved relationships. Based on the stringency of the parameters used in defining genome conservation, different investigators may therefore come to varying conclusions.
A fuller picture of the precise relationship between the Arabidopsis and rice genomes will emerge as more sequence data become available. However, even if tracts of conserved gene orders do exist between the two model species at the DNA sequence level, the fact that they are not readily identifiable using currently available comparative mapping tools will greatly diminish the impact of comparative knowledge between Arabidopsis and rice on grass genome analyses. The exploitation of rice rather than Arabidopsis genomic sequence will then be the priority in cereal research. Nevertheless, for certain applications such as the identification of orthologous relationships between different members of a gene family and the study of positional effects on gene function, the relative position of genes in the Arabidopsis and rice genomes will continue to be important.
METHODS
Homology Searches
A BLAST search was conducted at the nucleotide level with the Arabidopsis gene sequences from five annotated BAC clones from the top of Arabidopsis chromosome 1 (F21M12, F7G19, F19G10, T26J12, and F21J9; Fig. 1a) against the rice EST database of the Japanese Rice Genome Programme (RGP), which contains mainly 5′ end sequences. For the remaining BACs (Fig. 1a), the complete sequence was used in BLAST searches. Reciprocal BLAST searches using selected rice ESTs as query against the Arabidopsis thaliana Database (http://genome-www.stanford.edu/Arabidopsis/) were also carried out at the nucleotide level.
DNA Sequencing
Rice ESTs were sequenced from the 3′ end by the dideoxy termination method using fluorescent primers on an Applied Biosystems ABI 377 automated DNA sequencer.
Plant Material
An F2 population of 186 plants from the Oryza sativa cross Nipponbare (japonica) × Kasalath (indica) was used for mapping in rice (Kurata et al. 1994; Harushima et al. 1998). Additional mapping was carried out in a population of 155 F2 progeny or their F3 families from the cross IR20 (indica) × 6383 (japonica) (Quarrie et al. 1997). The A. thaliana ecotype Columbia was used for cross-hybridization experiments.
Primers and Probes
Primers to rice and Arabidopsis sequences were designed using the program “Primer” (Whitehead Institute for Biomedical Research, Cambridge, MA). Rice ESTs were obtained from RGP, and Arabidopsis BACs were from The Arabidopsis Biological Resource Center (Columbus, OH). Arabidopsis probes were prepared by PCR amplification of exon sequences from BAC F21M12 using standard conditions.
Marker Analyses
DNA isolation and digestion, electrophoresis, and Southern blot transfers were performed as described by Kurata et al. (1994) and Sharp et al. (1988), for use with chemiluminescent and radioisotope labeling and detection systems, respectively. For chemiluminescent labeling and detection of probes, the ECL-Direct kit (Amersham) was used according to the supplier’s instructions. Hybridization conditions following radioactive labeling with 32P were performed as described by Laurie et al. (1993). Restriction fragment length polymorphism (RFLP) markers were added to the existing Nipponbare × Kasalath and IR20 × 6383 genetic maps using the “try” command of the program Mapmaker version 3.0 (Whitehead Institute for Biomedical Research).
Physical mapping of rice ESTs to YACs by PCR was carried out as described by Umehara et al. (1996).
Acknowledgments
K.M.D. was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) through a David Phillips Research Fellowship. Part of the work was carried out at RGP under a STAFF Visiting Research Fellowship Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL katrien.devos@bbsrc.ac.uk; FAX 44 1603 502 241.
REFERENCES
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Crane PR, Friis EM, Raunsgaard-Pedersen K. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- Devos KM, Gale MD. Comparative genetics in the grasses. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, Brenner S. Small is beautiful: Comparative genomics with the pufferfish (Fugu rubripes) Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamota T, Lin SY, Antonio BA, Parco A, et al. A high density-rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Morrice DR, Sang H, Crittenden LB, Burt DW. Genetic and physical mapping of the chicken IGF1 gene to chromosome 1 and conservation of synteny with other vertebrate genomes. J Hered. 1996;87:10–14. doi: 10.1093/oxfordjournals.jhered.a022946. [DOI] [PubMed] [Google Scholar]
- Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin S-Y, et al. A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Devos KM, Leitch IJ, Gale MD. The distribution of RFLP markers on chromosome 2(2H) of barley in relation to the physical and genetic location of 5S rDNA. Theor Appl Genet. 1993;87:177–183. doi: 10.1007/BF00223762. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lan T-H, Reischmann KP, Chang C, Lin Y-R, Liu S-C, Burow MD, Kowalski SP, Katsar CS, DelMonte TA, et al. Toward a unified genetic map of higher plants transcending the monocot-dicot divergence. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Laurie DA, Zhu J, Lebreton C, Semikhodskii A, Steed A, Witsenboer H, Calestani C. QTL analysis to study the association between leaf size and abscissic acid accumulation in droughted rice leaves and comparisons across cereals. Plant Mol Biol. 1997;35:155–165. [PubMed] [Google Scholar]
- Sharp PJ, Kreis M, Shewry PR, Gale MD. Location of beta-amylase sequences in wheat and its relatives. Theor Appl Genet. 1988;75:286–290. [Google Scholar]
- Umehara Y, Tanoue H, Kurata N, Ashikawa I, Minobe Y, Sasaki T. An ordered yeast artificial chromosome library covering over half of rice chromosome 6. Genome Res. 1996;6:935–942. doi: 10.1101/gr.6.10.935. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]


