Abstract
The retina contains ganglion cells (RGCs) that respond selectively to objects moving in particular directions. Individual members of a group of ON-OFF direction-selective RGCs (ooDSGCs) detect stimuli moving in one of four directions: ventral, dorsal, nasal, or temporal. Despite this physiological diversity, little is known about subtype-specific differences in structure, molecular identity, and projections. To seek such differences, we characterized mouse transgenic lines that selectively mark ooDSGCs preferring ventral or nasal motion as well as a line that marks both ventral- and dorsal-preferring subsets. We then used the lines to identify cell surface molecules, including Cadherin 6, CollagenXXVα1, and Matrix metalloprotease 17, that are selectively expressed by distinct subsets of ooDSGCs. We also identify a neuropeptide, CART (cocaine- and amphetamine-regulated transcript), that distinguishes all ooDSGCs from other RGCs. Together, this panel of endogenous and transgenic markers distinguishes the four ooDSGC subsets. Patterns of molecular diversification occur before eye opening and are therefore experience independent. They may help to explain how the four subsets obtain distinct inputs. We also demonstrate differences among subsets in their dendritic patterns within the retina and their axonal projections to the brain. Differences in projections indicate that information about motion in different directions is sent to different destinations.
Introduction
The brain is remarkable not only for its staggering number of neurons, but also for its panoply of neuron types. To understand how brain circuits work, it will be essential to define these classes, recognize them in vivo, and match them with the functions they serve. Numerous molecular markers of neuronal subtypes have been identified using immunohistochemistry, gene expression analysis, and transgenic technology (Lein et al., 2007; Wässle et al., 2009; Siegert et al., 2009). It remains unclear, however, whether expression of one or a few genes will suffice to distinguish closely related neuronal subtypes.
To address this issue, we sought differences among four closely related neuronal subtypes in mouse retina. The retina contains five main neuronal types, which are divided into 60–100 subtypes (Masland, 2001; Sanes and Zipursky, 2010), a number probably typical for any brain area. The subtypes on which we focus here are ON-OFF direction-selective retinal ganglion cells (ooDSGCs). ooDSGCs respond to both the onset and termination of a flashed spot of light. If the spot moves through the receptive field of an ooDSCG, it fires most strongly for one (preferred) direction of motion and remains silent for the opposite (null) direction (Barlow et al., 1964). The dendrites of ooDSGCs arborize in two sublaminae of the inner plexiform layer (IPL), where they receive synapses from bipolar and starburst amacrine cells in circuits that produce the direction selectivity (Barlow and Levick, 1965; Demb, 2007; Zhou and Lee, 2008; Lee et al., 2010).
Directional preferences of ooDSGCs are not uniformly distributed; instead, discrete groups of ooDSGCs prefer ventral, dorsal, temporal, or nasal motion on the retina (Oyster and Barlow, 1967; Elstrott et al., 2008). These distinct preferences suggest that the four subtypes receive different inputs and might have distinct synaptic targets in the brain. Furthermore, it is clear that the four subtypes recognize themselves as distinct: gap junctions selectively couple members of the same subtype (DeBoer and Vaney, 2005) and dendritic fields of a given subtype overlap minimally, whereas dendrites of ooDSGCs with different preferred directions overlap freely (Amthor and Oyster, 1995).
Despite intensive study of ooDSCGs generally, and the availability of mouse transgenes that label subsets of ooDSGCs (Huberman et al., 2009; Kim et al., 2010), no differences in morphology, endogenous gene expression, or targets have been described among the four directionally distinct subtypes. Here, we used these previously described mice together with novel transgenic lines to seek distinctions among ooDSGCs. Using gene expression profiling, we identify markers that, in combination, label all four subtypes with ∼90% accuracy. Subtype identity is specified molecularly before eye opening, suggesting that differences among ooDSGCs arise independent of visual experience. We also document differences in dendritic arbors and axonal projections that distinguish the ooDSGCs selective for ventral motion from those selective for nasal motion. In addition to providing reagents for mechanistic studies of ooDSGC subtype diversification and synaptic specificity, our results provide support for the idea that molecular markers can be used to distinguish closely related neuronal subtypes.
Materials and Methods
Mice.
FSTL4-CreER transgenic mice, used to label bistratified dendrite-retinal ganglion cells (BD-RGCs), were described by Kim et al. (2010). Briefly, the transgene was generated by insertion of a tamoxifen-responsive cre recombinase (CreER) cDNA at the initiation codon of the Fstl4 coding sequence in a bacterial artificial chromosome. These animals were crossed to mice that express a fluorescent protein following Cre-mediated excision of “stop” sequences. The following three reporter lines were used: Thy1-STOP-YFP mice line 15 (Buffelli et al., 2003), and mice that conditionally express tdTomato (Madisen et al., 2010) (obtained from Jackson Laboratories, catalog #007909) or green fluorescent protein (GFP) (Kim et al., 2009) (kindly provided by S. Dymecki, Harvard Medical School, Boston, MA) from the Rosa26 locus. Tamoxifen (100 μg, Sigma) was injected intraperitoneally into double transgenics at postnatal day (P) 0–1 to activate CreER and thereby initiate expression of reporter.
The W9 mouse line was generated in parallel with the W3 and W7 lines described by Kim et al. (2010). In this transgene, Thy1 regulatory elements drive expression of YFP. YFP was expressed in distinct and nonoverlapping subsets of RGCs in the W3, W7, and W9 lines, presumably due to effects of sequences near the site of transgene integration in the genome (Feng et al., 2000).
To mark cdh6-expressing cells, CreER was inserted into the initiation codon of the cdh6 gene by homologous recombination in embryonic stem cells. The targeting vector was generated by lambda phage-mediated recombineering (Chan et al., 2007). Chimeric mice with the targeted embryonic stem cells were generated and mated to obtain germ line transmission. Mice were mated to reporter lines as described above for FSTL4-CreER.
Thy1-YFP-H mice were generated and characterized by Feng et al. (2000). Dopamine receptor D4-GFP (DRD4-GFP) mice were obtained from Mutant Mouse Regional Resource Center-University of North Carolina (www.mmrrc.org).
Electrophysiology.
Methods for electrophysiological analysis have been described by Kim et al. (2008, 2010). Briefly, dark-adapted retinas were isolated in Ringer's solution and targeted for cell-attached recording with patch microelectrodes. Light stimuli were delivered from a computer-driven video projector through a custom-made substage lens. Receptive field centers were determined with small flashing spots, then direction selectivity was assessed with stimuli moving through the receptive field center in eight different directions.
Histology.
Methods for histological analysis of retina and brain are described by Yamagata et al. (2006) and Kim et al. (2010). Antibodies used were as follows: rabbit anti-GFP, goat anti-choline acetyltransferase, mouse anti-Brn3a (all from Millipore), goat anti-vesicular acetylcholine transporter (Promega), rabbit anti-CART (Phoenix), and rabbit anti-MMP17 (Epitomics). Secondary antibodies were from Invitrogen or Jackson ImmunoResearch.
Cell isolation and expression profiling.
To isolate BD-RGCs, retinas from P6 mice were dissociated using papain (Worthington). The cell suspension was incubated with anti-Thy1 antibodies conjugated to magnetic beads (Miltenyi Biotec), and passed over a magnetic column, according to the manufacturer's instructions. This RGC-enriched cell fraction was then passed through a MoFlo cytometer (Dako) to select YFP-positive cells, which were sorted directly into RNA-stabilizing lysis buffer from the PicoPure RNA Isolation Kit (MDS).
To isolate starburst amacrines, we used a Thy1 transgenic line in which all starbursts and a subset of RGCs are labeled with the Kusabira Orange fluorescent protein (OFP; line Thy1-OFP3) (J. Livet and J. R. Sanes, unpublished observation). Purification of these cells used the same procedure as above, except that the Thy1-negative fraction (cells not retained on the magnetic column) was used for sorting. We confirmed that all OFP-positive cells recovered in this manner were starbursts by plating the sorted cells and staining for starburst and RGC markers.
For microarray hybridization, RNA was isolated using the PicoPure kit. Two rounds of amplification were performed with the MessageAmpII system (Ambion/Applied Biosystems), the second resulting in biotin-labeled samples. These were hybridized to Affymetrix Mouse 430 2.0 arrays according to the manufacturer's instructions. We hybridized two replicates per transgenic line, each representing a sample of ∼200 cells collected from different litters. Data were analyzed, and cell-type-specific genes identified using Resolver (Rosetta) and dChip software (Li and Wong, 2001).
Results
Transgenic lines marking ooDSGCs that prefer ventral or nasal motion
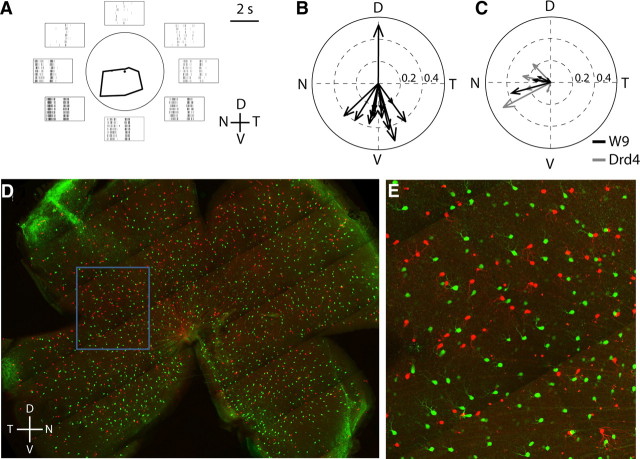
We recently generated a transgenic line in which a group of ooDSGCs, which we call BD-RGCs, are labeled with a fluorescent protein (Kim et al., 2010). To begin this study, we assessed the directional preference of BD-RGCs in adult retina (eccentricity of <60% measured from the optic nerve head to the retinal periphery). Each cell was stimulated with bars moving in eight directions. Over 90% of BD-RGCs responded best to bars moving from dorsal to ventral on the retina (i.e., to ventral motion) (Fig. 1A,B).
Figure 1.
Transgenic markers of ooDSGCs that prefer different directions. A, Responses of a BD-RGC to a white rectangle moving across the receptive field center in eight different directions at 575 μm/s. Average responses are displayed in a polar plot and surrounding traces show raster plots for seven repeats. Note ON and OFF phases of spiking in rasters. V, Ventral; D, dorsal; N, nasal; T, temporal. B, Preferred directions of BD-RGCs recorded from <60% eccentricity. Arrow length indicates the extent of direction selectivity, calculated as in Kim et al. (2010). C, Preferred directions of W9-RGCs (black arrows) and DRD4-RGCs (gray arrows). D, E, Retina whole mount showing BD-RGCs (red) and DRD4-RGCs (green), in triple transgenic mouse (DRD4-GFP × FSTL4-CreER × ROSA-CAGS-STOP-tdTomato). Blue box in D shows region enlarged in E. RGCs express either green or red fluorescent proteins, but not both, demonstrating that DRD4- and BD-RGCs are distinct populations.
We also recorded from RGCs in a transgenic line called W9, which was generated in parallel with the W3 and W7 lines described previously (Kim et al., 2010). The ventral retina of the W9 line contained a nearly homogeneous population of labeled bistratified RGCs. These cells were ooDSGCs that responded preferentially to nasal motion (Fig. 1C). This preference was similar to that reported previously for ooDSGCs labeled in a transgenic line generated by the GENSAT project and called DRD4-GFP (Huberman et al., 2009). We obtained this line and confirmed that DRD4-RGCs prefer nasal motion (Fig. 1C).
These results imply that BD-RGCs are distinct from W9 and DRD4-RGCs. To test this idea, we generated BD+DRD4 double-transgenic mice in which BD-RGCs were labeled with a red fluorophore and could thus be distinguished from GFP-positive DRD4-RGCs. As expected, few fluorescent cells in these retinas (∼1%) were both RFP and GFP positive (Fig. 1D,E). We also expect that W9-RGCs and DRD4-RGCs are overlapping subsets, but because they are labeled with indistinguishable fluorophores (YFP and GFP), we could not test this idea directly.
Based on these results, we used the BD, W9, and DRD4 lines to compare the structural and functional properties of ooDSGCs selective for ventral and nasal motion. We used both DRD4 and W9 lines in most studies but, unless otherwise noted, present results from DRD4 here.
Different patterns of asymmetry in cells preferring ventral and nasal motion
BD-, DRD4-, and W9-RGCs all had bistratified dendritic arbors that, when viewed en face, were generally displaced asymmetrically around the somata (Fig. 2A–C,E). The degree of asymmetry varied among cells but in no case was as dramatic as that documented previously for OFF-DSGCs (Kim et al., 2008). Consistent with previous reports (Oyster et al., 1993; Huberman et al., 2009), there was no obvious relationship between the structural asymmetry and directional preference of W9- or DRD4-RGCs. For example, the W9-RGC in Figure 2A and the DRD4-RGC shown in Figure 2C had dendrites displaced dorsonasally and dorsotemporally from their somata, respectively, but preferred nasal motion.
Figure 2.
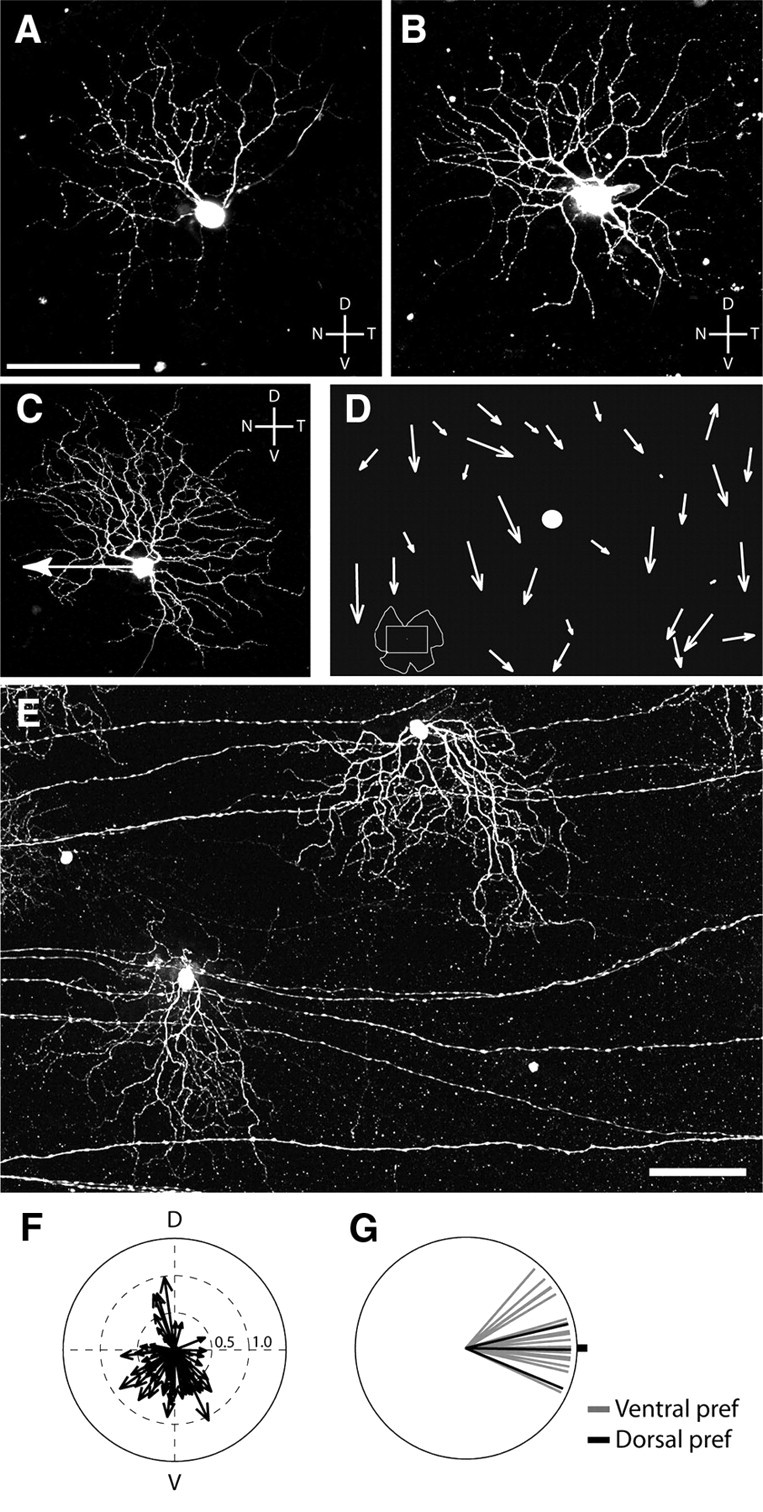
Relationship between structure and function of ooDSGCs. A, B, Morphology of W9 RGCs does not correlate with their preferred direction. Confocal stack z-projection showing two W9 cells that were injected with Lucifer yellow following recording. Scale bar, 100 μm. C, DRD-4-RGC filled with Lucifer yellow following recording. Arrow indicates preferred direction, which is distinct from the orientation of its dendritic asymmetry. D, Sketch of part of a whole mounted retina (see inset at bottom left) showing dendritic asymmetry of the BD-RGCs. Arrows originate from the somas and point in the direction of dendritic asymmetry. Length of arrow is proportional to degree of dendritic asymmetry. Dot, Optic disc. E, Micrograph of two BD-RGCs from the retina. Scale bar: (in E) C, E, 100 μm. F, Polar plot summarizing the dendritic asymmetry of BD-RGCs from a retina similar to that shown in D and E. G, Relationship between dendritic asymmetry and direction selectivity of 22 BD-RGCs. The preferred direction is plotted relative to the direction of the dendritic arbor (dot). Black lines indicate dorsal-preferring cells from the dorsal margin of the retina.
In striking contrast, most of the asymmetric BD-RGCs had dendrites that were displaced vertically from the soma. In most of the retina, the predominant direction was ventral, but a small number of BD-RGCs near the dorsal pole of the retina had arbors directed dorsally (Fig. 2D–F). Together with the physiological results presented above, this finding suggested a correlation between the structure and function of BD-RGCs. To test this relationship stringently, we recorded from 13 BD-RGCs in central retina and 3 BD-RGCs at the dorsal pole, then imaged their dendrites following recording. In 15, including all 3 of the dorsal cells, the preferred direction of the cell, determined physiologically, corresponded to the direction in which the dendritic arbor was displaced from the soma (Fig. 2G); the 16th had a symmetrical dendritic arbor. Thus, not only the majority of BD-RGCs that prefer ventral motion but also the minority in dorsal retina that prefer dorsal motion exhibit an association of structural and functional asymmetries.
Distinct molecular signatures of RGCs that prefer vertical and nasal motion
To identify genes selectively expressed in BD-RGCs cells, we isolated them based on their fluorescence and profiled gene expression using microarrays (see Experimental procedures). In parallel, we isolated and profiled several other RGC and amacrine subtypes (Kay, 2011), including starburst amacrines, which provide the main inhibitory input to ooDSGCs. We filtered the microarray data to identify genes expressed at several fold higher levels in BD-RGCs and/or starburst amacrines compared with other cell types in the dataset, then used in situ hybridization (ISH) to ask they whether they were expressed by BD-, DRD4-, or W9-RGCs.
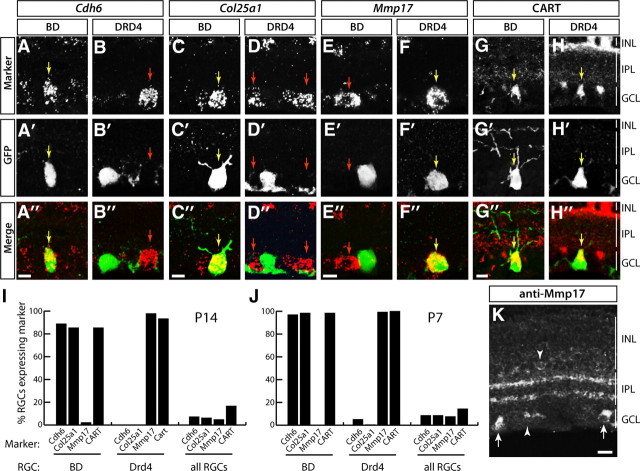
Two genes, cadherin 6 (Cdh6) and collagen 25a1 (Col25a1), were expressed by BD-RGCs but not by DRD4-RGCs (Fig. 3A–D,I). Cdh6, a type II cadherin, was previously shown to be expressed in mouse retina (Honjo et al., 2000); Col25a1 encodes a transmembrane collagen that is expressed in the nervous system and accumulates in amyloid plaques in Alzheimer's disease (Hashimoto et al., 2002). Cdh6 and Col25a1 were also expressed by starburst amacrines but not by other retinal cells.
Figure 3.
Molecular markers for subsets of ooDSGCs. A, B, In situ hybridization for Cdh6 RNA (red) combined with anti-GFP antibody staining (green) reveals expression of Cdh6 in BD- but not DRD4-RGCs. Yellow arrows in A–H indicate double-labeled cells. Red arrows in A–H indicate marker-positive cells that do not express GFP. C, D, In situ hybridization for Col25a1 RNA (red) shows expression in GFP-labeled (green) BD-RGCs but not DRD4-RGCs. E, F, In situ hybridization for Mmp17 RNA (red) shows expression in GFP-labeled (green) DRD4- but not BD-RGCs. G, H, CART is a marker of both BD- and DRD4-RGCs as shown by immunostaining with anti-CART (red) and anti-GFP (green). Laminae marked in H (INL, inner nuclear layer; IPL, inner plexiform layer) apply to A–H, and K. Scale bars: 10 μm. I, J, Quantification of the fraction of RGCs expressing Cdh6, Col25a1, or Mmp17 at P14 (I) and at P7 (J) (n ≥ 44 for BD-RGCs, ≥104 for DRD4-RGCs, ≥1198 for all RGCs). K, MMP17 immunoreactivity in section from P14 retina. Antibody labels the putative starburst layers of the IPL, suggesting immunoreactivity in the dendrites of starburst amacrines and/or ooDSGCs. A subset of RGC cell bodies (arrows) as well as putative starburst amacrines (arrowheads) are labeled in the GCL and INL. Scale bar, 25 μm.
We were also interested in identifying markers of ooDSGCs that prefer nasal motion. The most obvious candidate was Drd4, since regulatory elements from this gene drive expression of GFP in the DRD4 transgenic line. In some cases, however, including the line that labels BD-RGCs, transgenes are expressed in cells that do not express the endogenous gene, because of omission of critical regulatory elements from the transgene or to influences at the genomic site of integration (Feng et al., 2000; Haverkamp et al., 2009; Kim et al., 2010). Indeed, consistent with data from the rat (Klitten et al., 2008), in situ hybridization showed that Drd4 was expressed prominently by photoreceptors and at low, uniform levels in the ganglion cell layer (GCL) of wild-type mice (Fig. 4A). In contrast, the Drd4 probe strongly labeled GFP-positive RGCs, along with photoreceptors, in the DRD4-GFP transgenic retina (Fig. 4B,B′). This result is not unexpected, in that the genomic fragment used to generate the transgene contains sequences that could be transcribed and recognized by the ISH probe. Thus, Drd4 mRNA in DRD4-RGCs is derived from the transgene, and it is unlikely that GFP-positive cells in the DRD4 line express significant levels of endogenous Drd4.
Figure 4.
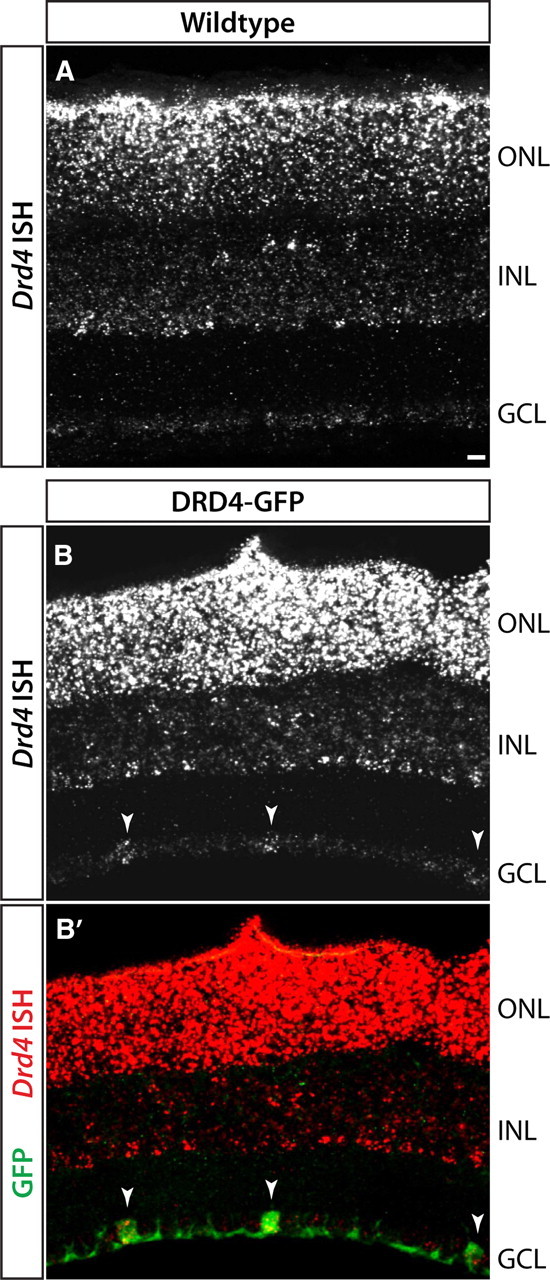
ISH for Drd4 RNA in P14 retina of wild-type or DRD4-GFP transgenic mice. A, Wild-type mice show Drd4 expression in photoreceptors in the outer nuclear layer (ONL) and in a subset of inner nuclear layer (INL) cells. Very low levels of transcript are detected in the GCL. B, DRD4-GFP transgenic mice show the same pattern of transcripts in photoreceptors and INL as in A but also exhibit Drd4 signals in DRD4-GFP-positive RGCs (arrowheads). The difference in labeling between wild-type and transgenic mice suggests that Drd4 transcripts in DRD4-RGCs of DRD4-GFP mice arise from the transgene, not the endogenous Drd4 gene. Scale bar, 25 μm.
As an alternative approach, we tested genes that we had found by in situ hybridization to be expressed by starburst amacrines and a small subset of RGCs but not by BD-RGCs. One of these, matrix metalloprotease 17 (Mmp17), was expressed by ≤5% of all RGCs and by few if any BD-RGCs (≤1%) or Col25a1-positive RGCs (1/179) but by >90% of DRD4 RGCs (Fig. 3E,F,I). MMP17 is a cell surface-associated enzyme (Sohail et al., 2008) expressed by some neurons in brain (Rikimaru et al., 2007). Immunostaining confirmed the selective association of MMP17 with DRD4-RGCs. Immunoreactivity was localized in somata of RGCs and starburst amacrines, and in the ON and OFF sublaminae of the IPL, where their processes arborize (Fig. 3K). Approximately 70% of MMP17-positive RGCs were DRD4-RGCs as assessed by ISH and immunostaining. MMP17-positive, GFP-negative RGCs might be ooDSGCs that prefer nasal motion but are not labeled by the DRD4-GFP transgene or, as discussed below, RGCs of other types.
Expression patterns of Cdh6, Col25a1, and Mmp17 at P7, before eye opening (∼P12), were similar to those at P14, after eye opening (Fig. 3I,J). The correspondence of endogenous gene and transgene expression at this early stage indicates that visual experience is not required for ooDSGC subtypes to acquire their distinct molecular identities.
CART selectively labels ooDSGCs
Another gene shown by microarray analysis to be expressed by BD-RGCs was “cocaine- and amphetamine-regulated transcript” (CART; gene symbol Cartpt). This gene encodes several neuropeptides, including CART (Rogge et al., 2008). Antibodies to CART labeled ∼15% of all RGCs as well as a small group of nonstarburst amacrines in the inner nuclear layer that we did not characterize further. CART was present in nearly all BD-, W9-, and DRD4-RGCs (Fig. 3G–J) and in nearly all RGCs that expressed Col25a1 or Mmp17 (data not shown). Moreover, the fraction of RGCs that was CART positive corresponded approximately to the fraction of RGCs that are ooDSGCs (Sun et al., 2002; Badea and Nathans, 2004; Coombs et al., 2006; Völgyi et al., 2009). We therefore considered the possibility that CART might be expressed by all and only ooDSGCs.
To test this idea, we used a transgenic line, Thy1-YFP-H (Feng et al., 2000). Approximately 200 RGCs are labeled per retina in this line, so that RGCs are sufficiently well separated to permit imaging of the complete dendritic arbor of a single cell. Labeled cells include all known RGC subtypes (Coombs et al., 2006). Whole mounts of retinas from Thy1-YFP-H mice were stained with anti-CART and then imaged (Fig. 5A,C–E). All of the CART-positive, YFP-positive RGCs imaged (22 in 3 retinas) were morphologically similar and had bistratified dendritic arbors, consistent with their being ooDSGCs (Fig. 5B). Seven of these cells were reconstructed at high resolution; all had dendrites that cofasciculated with the processes of starburst amacrines, confirming their identity as ooDSGCs (Fig. 5D–F). In contrast, none of 140 CART-negative, YFP-positive RGCs imaged in these retinas resembled ooDSGCs. Thus, CART is a specific marker of ooDSGCs.
Figure 5.
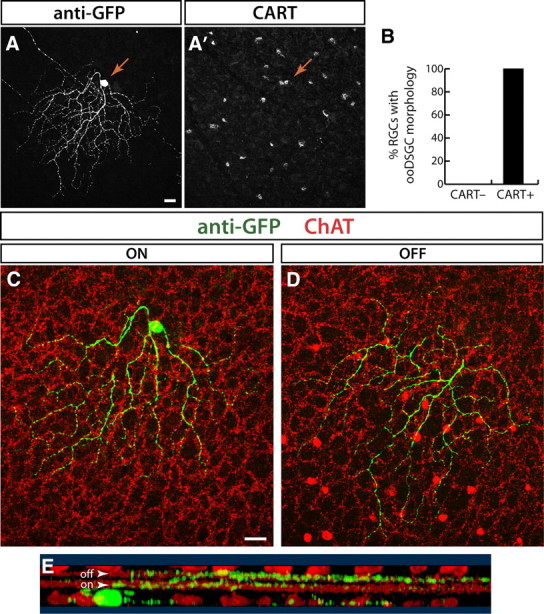
CART antibody labels ooDSGCs. A, CART and anti-GFP immunostaining identify double-positive RGCs in a retinal whole mount from line YFP-H. GFP channel shows the morphology of a YFP+ CART immunoreactive RGC (arrow; z-projection of confocal stack). CART channel (A′) shows CART+ RGCs in a single confocal plane through the GCL. Arrow indicates the soma of the YFP+CART+ double-positive cell. B, Morphological analysis of RGCs in line YFP-H that were CART+ and CART− (n = 140). All CART-immunoreactive RGCs (n = 22) have the bistratified morphology of ooDSGCs. By contrast, none of the CART− cells showed this morphology. C, D, Single confocal planes through the cell shown in A reveal morphological features that identify it as an ooDSGC. The cell dendrites (green) are bistratified, with both the ON (C) and OFF arbors (D) cofasciculating with the choline acetyltransferase (ChAT)-positive processes of starburst amacrines (red). E, Rotation of a 3-D reconstruction of the dendrites of this cell show that it has bistratified projections to the OFF and ON starburst IPL sublaminae (arowheads). GFP (green) and ChAT (red) channels are shown. Scale bars: 20 μm.
Cadherin 6 is selectively expressed by ooDSGCs responsive to vertical motion
Of genes selectively expressed by BD-RGCs, we focused on Cdh6 because previous work suggested that cadherins are expressed by RGC subsets (Yamagata et al., 2006) and involved in retinotectal patterning (Inoue and Sanes, 1997). We generated a targeted allele in which tamoxifen-activated CreER was inserted in place of the first coding exon of the Cdh6 gene. These mice were mated to mice in which expression of YFP was Cre dependent.
Tamoxifen was administered to neonates, and retinas were examined at ≥P21. Two sets of retinal cells were YFP positive: starburst amacrine cells (identified by double labeling with antibodies to ChAT) and RGCs (Fig. 6A,B). Most (≥99%) of the Cdh6-positive RGCs had bistratified dendrites that arborized with starburst amacrine processes (Fig. 6A), suggesting, along with microarray results presented above, that they were ooDSGCs. (Using other reporters described in Materials and Methods, only ∼95% of labeled RGCs were ooDSGCs; most of the remainder had dendrites in the outermost sublamina of the IPL.) Physiological analysis confirmed that the YFP-labeled Cdh6-RGCs were direction selective. Over 90% (22/23) were ooDSGCs; one was an ON-DSGC. Approximately 90% preferred vertical motion (10 dorsal and 11 ventral) (Fig. 6C). Thus, Cdh6 is a selective marker of RGCs that respond to vertical motion.
Figure 6.
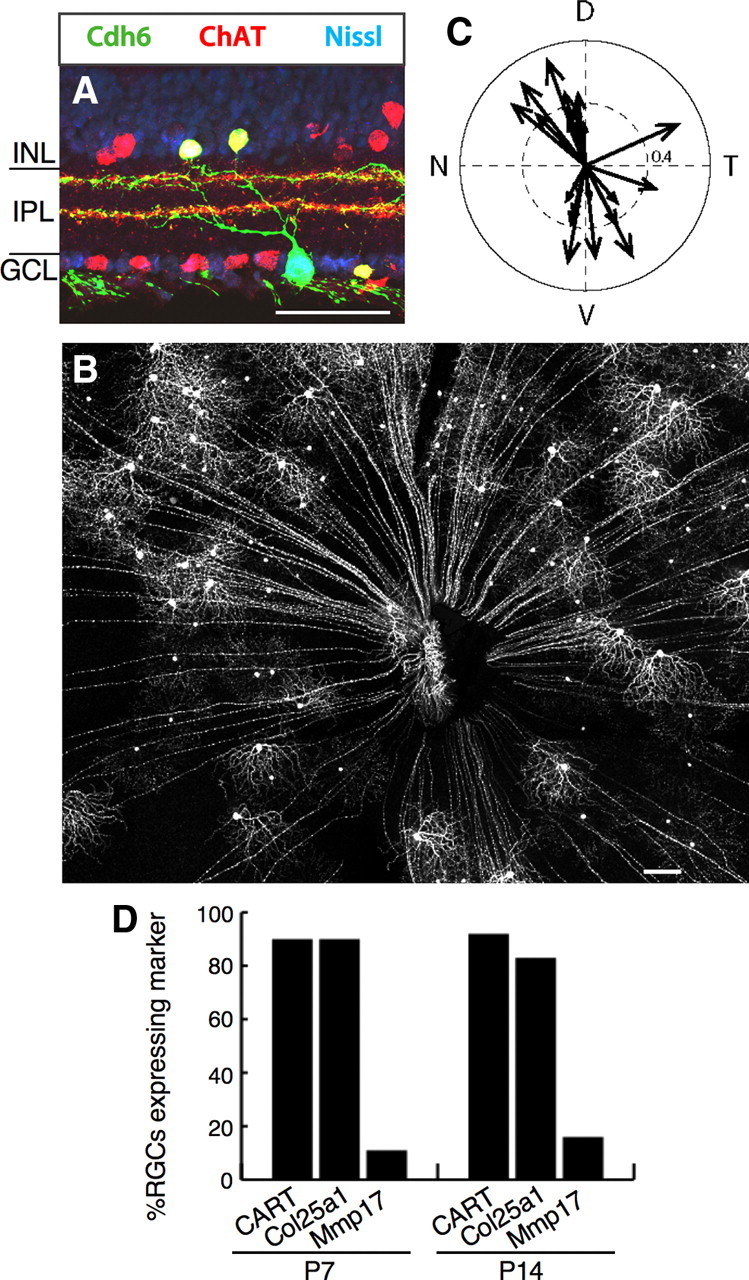
Cadherin 6-positive ooDSGCs prefer vertical motion. A, Cdh6-RGCs and starburst amacrine cells labeled with YFP (green) in retina sections from a cdh6 knock-in heterozygote (Cdh6-CreER × Thy1-stop-YFP). Choline acetyltransferase (ChAT) (red) labels starburst amacrine cell somas and dendrites. Cdh6-RGC dendrites project to starburst IPL layers. Blue, Fluorescent Nissl stain. B, Retina whole mount from a cdh6 knock-in heterozygote. C, Responses of Cdh6-RGCs, showing that the vast majority prefer dorsal or ventral motion (n = 23 cells from 7 retinas, 7 mice). D, Molecular markers expressed by Cdh6-RGCs. Scale bars: A, 50 μm; B, 200 μm.
Most BD-RGCs are selective for ventral motion, whereas similar numbers of Cdh6-RGCs prefer dorsal and ventral motion. Thus, molecular differences between these two populations would provide clues to the molecular identity of DSGCs that prefer dorsal motion. We used the panel of markers defined above to assess gene expression in Cdh6-RGCs. At P7, nearly all Cdh6-RGCs (∼90%) were CART and Col25a1 positive, but only ∼10% were Mmp17 positive (Fig. 6D). Thus, expression of endogenous genes assessed to date does not distinguish ooDSGCs that prefer ventral and dorsal motion. It is possible that the Mmp17-positive Cdh6-RGCs (Fig. 6D) correspond to the small fraction of Cdh6-RGCs that prefer temporal motion.
Central projections of ooDSGCs
To visualize the central projections of ooDSGCs, we stained coronal brain sections with antibodies to CART. The full extent of all retinal projections was revealed by intraocular injection of a tracer, cholera toxin B, and the retinal origin of CART-labeled fibers was assessed by their loss following monocular enucleation. Retinally derived CART-positive fibers were abundant in the dorsal lateral geniculate nucleus (dLGN) and superior colliculus (Fig. 7A–C). Within the superior colliculus, CART-positive fibers were concentrated in the superficial half of the retinorecipient zone (Fig. 7C), the region previously shown to be innervated by BD-RGC axons (Kim et al., 2010). Small numbers of retinally derived CART-positive fibers were present in the ventral LGN, the medial temporal nucleus (MTN), and the nucleus of the optic tract (NOT). No retinally derived CART-positive fibers were found in the suprachiasmatic nucleus, or in accessory optic or pretectal nuclei other than MTN and NOT (data not shown).
Figure 7.
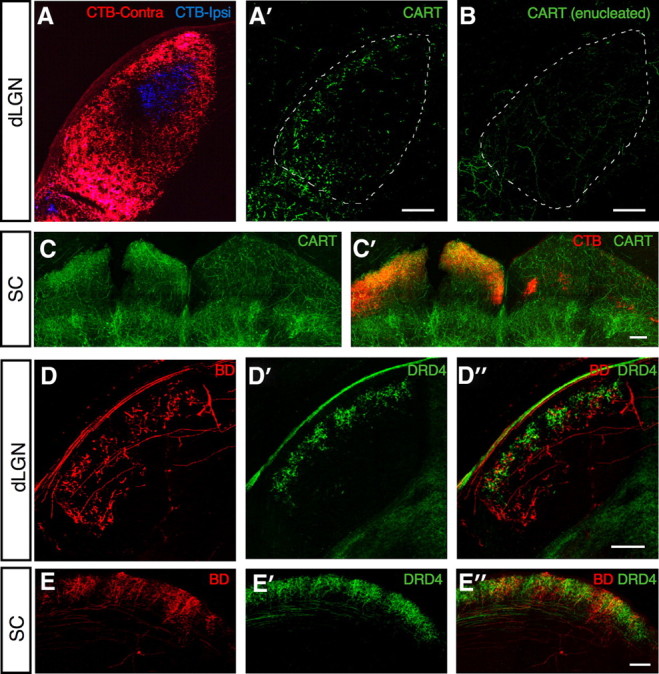
Lamina-specific projections of different ooDSGC functional subtypes. A, Cholera toxin subunit B (CTB)-labeled axons in the dLGN arising from the contralateral (red) and ipsilateral (blue) eyes. A′, CART+ axons (green) overlap with CTB-labeled axons from the contralateral eye (red). B, After monocular enucleation, the CART+ axon terminals (green) disappear from the dLGN contralateral to the eye removed, indicating that these terminals originate from RGCs. C, Projections of CART+ RGCs to superior colliculus (SC). After monocular enucleation CART+ axons (green) are present only in the SC contralateral to the remaining eye. CTB-labeled axons in the contralateral (left) SC (red) overlap with CART+ axon terminals. No CART+ RGC terminals are seen in the CTB-labeled ipsilateral projection (right). D, BD- (red) and DRD4-RGC (green) axons in the dLGN. Overlay in D″ shows distinct laminar targeting of the two RGC subsets. E, BD- (red) and DRD4-RGC (green) axons in the SC. Scale bars: A–D, 200 μm; E, 400 μm.
Inputs from the contralateral and ipsilateral eyes are spatially separated in both the dLGN and the superior colliculus (Godement et al., 1984). In the dLGN, ipsilateral input is confined to a central core, surrounded by a contralaterally innervated shell. In the superior colliculus, ipsilateral input is confined to a narrow sublamina in the deepest portion of the retinorecipient zone. Remarkably, in both structures few if any retinally derived CART-positive fibers were present in ipsilaterally innervated regions (Fig. 7A–C). Thus, central projections of ooDSGCs are predominantly if not entirely contralateral.
Quina et al. (2005) have shown that RGCs expressing the transcription factor Brn3a are excluded from the ipsilateral pathway. Consistent with this result, we found that >95% of BD-RGCs, DRD4-RGCs, Cdh6-RGCs, and CART-positive RGCs are Brn3a-positive (data not shown).
Distinct projections of ooDSGCs that prefer vertical and nasal motion
We used the transgenic lines described above to ask whether ooDSGC subsets projected to distinct regions within their target areas. BD-, DRD4-, and Cad6-RGCs all projected to the dLGN, the ventral lateral geniculate nucleus (vLGN), and the superior colliculus (Figs. 7D,E, 8A,B). In the dLGN, however, the laminar position of their terminal arbors differed. DRD4-RGC arbors occupied a narrow superficial lamina, directly beneath the optic tract. In contrast, arbors of BD- and Cad6-RGCs extended deeper (Fig. 7D). In the superior colliculus, DRD4-RGC arbors started branching at a more superficial point than those of BD- and Cad6-RGCs, but the difference was not striking (Fig. 7E).
Figure 8.
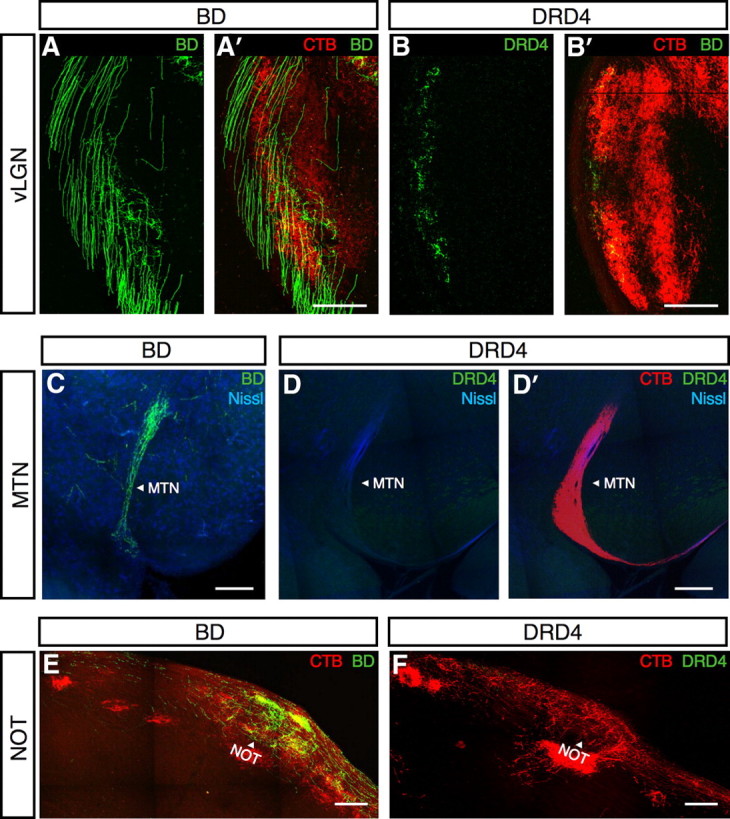
ooDSGCs project to the vLGN and the pretectal system. A, BD-RGC axons (green) arborize in the vLGN. A′, Merge of CTB (red) and BD-RGC (green) axons. B, DRD4-RGC axons (green) arborize in the vLGN. B′, Merge of CTB (red) and DRD4-RGC (green) axons. C, BD-RGC axons (green) project to the MTN. Blue, fluorescent Nissl stain. D, DRD4-RGC axons (green) are not present in the MTN. D′, Merge of DRD4-RGC (green) and CTB (red) axons in the MTN. Blue, Fluorescent Nissl stain. E, BD-RGC axons (green) arborize in the nucleus of the optic tract (NOT; arrowhead). CTB (red) labels the optic tract and the nucleus. F, DRD4 axons (green) are absent from the NOT (arrowhead), labeled with CTB (red). Scale bars: A–E, 200 μm.
In contrast to these structures, which were innervated by BD-, Cdh6-, and DRD4-RGCs, only the vertically preferring ooDSGCs (BD- and Cdh6-RGC) projected to NOT (Fig. 8E,F). Likewise, the MTN was innervated by BD- and Cad6-RGCs but not by nasal-preferring DRD4-RGCs (Fig. 8C,D). Previous studies in mice have shown that the MTN receives input from RGCs that respond to vertical but not horizontal motion (Yonehara et al., 2009). Our results are consistent with this pattern. Although most RGCs that project to mouse MTN are ON-DSGCs rather than ooDSGCs (Yonehara et al., 2009), rat MTN does receive input from ooDSGCs (Dann and Buhl, 1987).
Discussion
In many respects, ooDSGCs appear to be a single major RGC subtype: they are similar in dendritic morphology (the main criterion by which RGCs are classified), in response properties and in the biophysical mechanisms that underlie their direction selectivity (Demb, 2007; Zhou and Lee, 2008). Yet in one respect they are readily divisible: they can be grouped into four distinct sets, each responsive to one of the four cardinal directions (Oyster and Barlow, 1967). Thus, ooDSGCs are more properly viewed as comprising four closely related subtypes that, despite many common features, are likely to be molecularly distinct and might send their information to different central targets. However, despite intensive study of ooDSGCs in the aggregate, differences among them have not been explored until very recently (Huberman et al., 2009). Here, we used a set of four transgenic lines to mark ooDSGCs with preferences for ventral (BD), nasal (DRD4 and W9), and vertical (dorsal and ventral; Cdh6) motion. By comparing the lines, we identified molecular and structural differences among ooDSGC subsets and documented differences in their central targets.
Molecular markers for ooDSGCs with distinct directional preferences
The fact that transgenic lines mark subsets of ooDSGCs with distinct directional preferences implies that these subsets are molecularly distinct. Unfortunately, the three lines with which we began our study (BD, W9, and DRD4) provided no information on what these differences might be. This is because transgene expression patterns reflect influences from the chromosomal region at which the transgene is integrated, rather than or in addition to regulatory sequences within the transgene itself. Such ectopic expression is not uncommon. For example, Haverkamp et al. (2009) recently characterized marked retinal subsets in four BAC transgenic lines and showed that none of the marked cells expressed the corresponding endogenous gene.
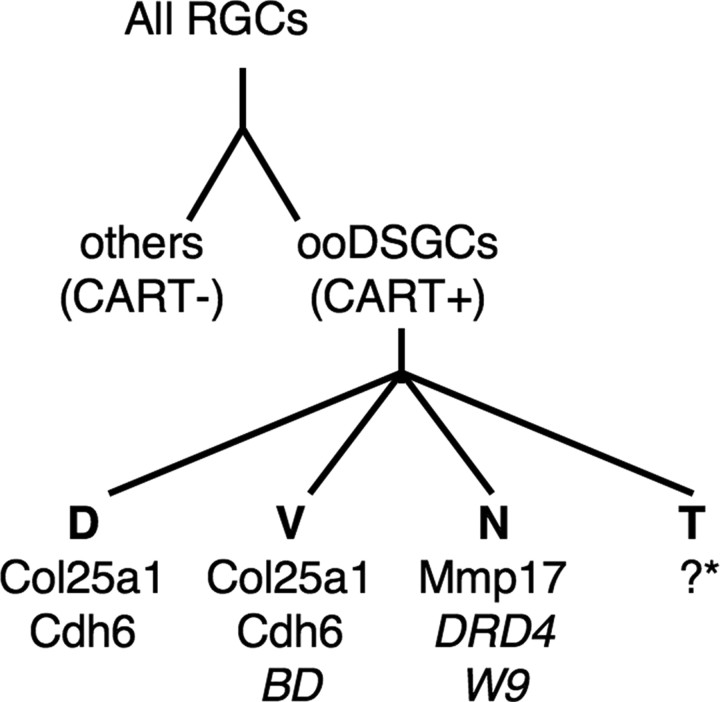
As an alternative, we isolated marked RGCs from several transgenic lines, based on their fluorescence, and used expression profiling to seek endogenous genes selectively expressed by one line. This strategy led to identification of Cdh6 and Col25a1, which are expressed selectively (though not exclusively) by BD-RGCs. Further screening led to identification of Mmp17 as a gene expressed by DRD4- and W9- but not BD-RGCs. Another gene identified in this screen, Cartpt, is expressed by most if not all ooDSGCs and by few if any other RGCs. Together, the panel of markers we describe here provides a molecular signature for each of the four ooDSGC subtypes (Fig. 9).
Figure 9.
Markers that identify the four ooDSGC subsets and distinguish ooDSGCs from other RGCs. The schematic includes genes identified in the microarray analysis (CART, Col25a1, Cdh6, and Mmp17) as well as the transgenes, which do not correspond to endogenous genes(BD, DRD4, and W9). *Some of the Cdh6-RGCs ooDSGCs that prefer temporal motion may be Mmp17 positive.
Of the many genes identified using our sorting and microarray strategy, we chose these genes for investigation because their products are cell surface or secreted proteins that might be involved in intercellular interactions required for development or function of ooDSGCs. Initial analysis of Cdh6-null mutants has so far not revealed severe defects in RGCs, but further studies of these and other mutants are now in progress.
Our results also set the stage for comparing different types of RGCs that respond to motion in the same direction. Lines are now available that mark three distinct sets of DSGCs selective for ventral motion: OFF-DSGCs (J-RGCs) Kim et al., 2008, 2010), ON-DSGCs (SPIG1-RGCs) (Yonehara et al., 2008, 2009), and ooDSGCs (BD-RGCs, this article). Ongoing electrophysiological studies indicate that J-RGCs and BD-RGCs use radically different synaptic mechanisms to compute image motion (Y. Zhang, unpublished observation).
Correlated structural and functional asymmetry in an ooDSGC subset
Oyster et al. (1993) recorded from a large set of ooDSGCs in rabbit retina, then examined their dendritic morphologies. They noted that some cells had asymmetric dendritic arbors but found no “morphological feature that is correlated with the cells' preferred response directions.” It was therefore surprising to find that the asymmetry of BD-RGC arbors was strongly associated with their directional preference. Remarkably, this association was not evident in DRD4- and W9-RGCs, which otherwise seemed structurally indistinguishable from BD-RGCs.
The correspondence of dendritic asymmetry with preferred movement direction in BD-RGCs resembles that in J-RGCs, a far more strikingly asymmetric group of OFF-DSGCs that we described recently (Kim et al., 2008, 2010). We suspect, however, that the association differs in the two cases. Both J- and BD-RGCs include some cells whose arbors appear symmetric. The symmetric J-RGCs are not direction selective, supporting the idea that structure underlies function for these cells (Kim et al., 2008). In contrast, structurally symmetric BD-RGCs are as direction selective as asymmetric ones, suggesting that for these cells structural asymmetry does not determine directional preference. We do not know why the association seen for BD-RGCs is not evident for DRD4- and W9-RGCs.
Central targets of ooDSGCs
We identified several differences between projections of ooDSGCs and those of RGCs generally. First, ooDSGC axon arbors are absent from many retinal targets, including the superchiasmatic nucleus, the accessory optic nuclei LTN and DTN, and most pretectal nuclei. Second, within the major retinal target, the superior colliculus, ooDSGC arbors are confined to the upper portion of the retinorecipient zone. Third, ooDSGC axons are nearly if not entirely excluded from the ipsilateral projection, even though this projection does include axons of multiple RGC subtypes (Hong et al., 2011).
We also found two differences in targets among ooDSGC subsets. First, BD-RGC and DRD4-RGC arbors were partially segregated within the dLGN. Second, BD- and Cdh6-RGCs project to the MTN and NOT, whereas DRD4-RGCs do not. These nuclei are involved in generating optokinetic reflexes (OKRs) (Masseck and Hoffmann, 2009). The finding that ooDSGCs provide vertical but not nasal motion information to these nuclei may have implications for understanding OKR behavior. More generally, our results show that ooDSGC-derived directional information is segregated in distinct brain targets.
Footnotes
This work was supported by grants from the NIH to I.-J.K., M.M., and J.R.S., a grant from NIH to J.R.S., Collaborative Innovation Award 43667 from HHMI, and fellowships from the Life Sciences Research Foundation to J.N.K. and from the Charles A. King Trust to Y.Z. We thank Sara Haddad, Debbie Pelusi, and Laura Stoppel for assistance.
References
- Amthor FR, Oyster CW. Spatial organization of retinal information about the direction of image motion. Proc Natl Acad Sci U S A. 1995;92:4002–4005. doi: 10.1073/pnas.92.9.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Barlow HB, Levick WR. The mechanism of directionally selective units in rabbit's retina. J Physiol. 1965;178:477–504. doi: 10.1113/jphysiol.1965.sp007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow HB, Hill RM, Levick WR. Retinal ganglion cells responding selectively to direction and speed of image motion in the rabbit. J Physiol. 1964;173:377–407. doi: 10.1113/jphysiol.1964.sp007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, Sanes JR. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- Chan W, Costantino N, Li R, Lee SC, Su Q, Melvin D, Court DL, Liu P. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 2007;35:e64. doi: 10.1093/nar/gkm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140:123–136. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- Dann JF, Buhl EH. Retinal ganglion cells projecting to the accessory optic system in the rat. J Comp Neurol. 1987;262:141–158. doi: 10.1002/cne.902620111. [DOI] [PubMed] [Google Scholar]
- DeBoer DJ, Vaney DI. Gap-junction communication between subtypes of direction-selective ganglion cells in the developing retina. J Comp Neurol. 2005;482:85–93. doi: 10.1002/cne.20351. [DOI] [PubMed] [Google Scholar]
- Demb JB. Cellular mechanisms for direction selectivity in the retina. Neuron. 2007;55:179–186. doi: 10.1016/j.neuron.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Elstrott J, Anishchenko A, Greschner M, Sher A, Litke AM, Chichilnisky EJ, Feller MB. Direction selectivity in the retina is established independent of visual experience and cholinergic retinal waves. Neuron. 2008;58:499–506. doi: 10.1016/j.neuron.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Godement P, Salaün J, Imbert M. Prenatal and postnatal development of retinogeniculate and retinocollicular projections in the mouse. J Comp Neurol. 1984;230:552–575. doi: 10.1002/cne.902300406. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Wakabayashi T, Watanabe A, Kowa H, Hosoda R, Nakamura A, Kanazawa I, Arai T, Takio K, Mann DM, Iwatsubo T. CLAC: a novel Alzheimer amyloid plaque component derived from a transmembrane precursor, CLAC-P/collagen type XXV. EMBO J. 2002;21:1524–1534. doi: 10.1093/emboj/21.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Inta D, Monyer H, Wässle H. Expression analysis of green fluorescent protein in retinal neurons of four transgenic mouse lines. Neuroscience. 2009;160:126–139. doi: 10.1016/j.neuroscience.2009.01.081. [DOI] [PubMed] [Google Scholar]
- Hong YK, Kim IJ, Sanes JR. Stereotyped axonal arbors of retinal ganglion cell subsets in the mouse superior colliculus. J Comp Neurol. 2011;519:1691–1711. doi: 10.1002/cne.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo M, Tanihara H, Suzuki S, Tanaka T, Honda Y, Takeichi M. Differential expression of cadherin adhesion receptors in neural retina of the postnatal mouse. Invest Ophthalmol Vis Sci. 2000;41:546–551. [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, Barres BA. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Sanes JR. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science. 1997;276:1428–1431. doi: 10.1126/science.276.5317.1428. [DOI] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW, Sanes JR. Neurod6 expression defines novel retinal amacrine cell subtypes and regulates their fate. Nat Neurosci. 2011 doi: 10.1038/nn.2859. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452:478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, Dymecki SM. Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron. 2009;63:305–315. doi: 10.1016/j.neuron.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitten LL, Rath MF, Coon SL, Kim JS, Klein DC, Møller M. Localization and regulation of dopamine receptor D4 expression in the adult and developing rat retina. Exp Eye Res. 2008;87:471–477. doi: 10.1016/j.exer.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim K, Zhou ZJ. Role of ACh-GABA cotransmission in detecting image motion and motion direction. Neuron. 2010;68:1159–1172. doi: 10.1016/j.neuron.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Masseck OA, Hoffmann KP. Comparative neurobiology of the optokinetic reflex. Ann NY Acad Sci. 2009;1164:430–439. doi: 10.1111/j.1749-6632.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- Oyster CW, Barlow HB. Direction-selective units in rabbit retina: distribution of preferred directions. Science. 1967;155:841–842. doi: 10.1126/science.155.3764.841. [DOI] [PubMed] [Google Scholar]
- Oyster CW, Amthor FR, Takahashi ES. Dendritic architecture of ON-OFF direction-selective ganglion cells in the rabbit retina. Vision Res. 1993;33:579–608. doi: 10.1016/0042-6989(93)90181-u. [DOI] [PubMed] [Google Scholar]
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O'Leary DD, Goulding M, Turner EE. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci. 2005;25:11595–11604. doi: 10.1523/JNEUROSCI.2837-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikimaru A, Komori K, Sakamoto T, Ichise H, Yoshida N, Yana I, Seiki M. Establishment of an MT4-MMP-deficient mouse strain representing an efficient tracking system for MT4-MMP/MMP-17 expression in vivo using beta-galactosidase. Genes Cells. 2007;12:1091–1100. doi: 10.1111/j.1365-2443.2007.01110.x. [DOI] [PubMed] [Google Scholar]
- Rogge G, Jones D, Hubert GW, Lin Y, Kuhar MJ. CART peptides: regulators of body weight, reward and other functions. Nat Rev Neurosci. 2008;9:747–758. doi: 10.1038/nrn2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Sohail A, Sun Q, Zhao H, Bernardo MM, Cho JA, Fridman R. MT4-(MMP17) and MT6-MMP (MMP25), A unique set of membrane-anchored matrix metalloproteinases: properties and expression in cancer. Cancer Metastasis Rev. 2008;27:289–302. doi: 10.1007/s10555-008-9129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of mouse retinal ganglion cells. J Comp Neurol. 2002;451:115–126. doi: 10.1002/cne.10323. [DOI] [PubMed] [Google Scholar]
- Völgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol. 2009;512:664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H, Puller C, Müller F, Haverkamp S. Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J Neurosci. 2009;29:106–117. doi: 10.1523/JNEUROSCI.4442-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Dulac C, Roth KA, Sanes JR. Labeled lines in the retinotectal system: markers for retinorecipient sublaminae and the retinal ganglion cell subsets that innervate them. Mol Cell Neurosci. 2006;33:296–310. doi: 10.1016/j.mcn.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Shintani T, Suzuki R, Sakuta H, Takeuchi Y, Nakamura-Yonehara K, Noda M. Expression of SPIG1 reveals development of a retinal ganglion cell subtype projecting to the medial terminal nucleus in the mouse. PLoS One. 2008;3:e1533. doi: 10.1371/journal.pone.0001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, Noda M. Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLoS One. 2009;4:e4320. doi: 10.1371/journal.pone.0004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZJ, Lee S. Synaptic physiology of direction selectivity in the retina. J Physiol. 2008;586:4371–4376. doi: 10.1113/jphysiol.2008.159020. [DOI] [PMC free article] [PubMed] [Google Scholar]