Abstract
Genome-wide physical and genetic mapping efforts have not yet fully addressed the problem of closure at the telomeric ends of human chromosomes. Targeted efforts at cloning human and mouse telomeres have succeeded in identifying unique sequences at most telomeres, but gap sizes between these telomere clones and the distal markers on integrated genetic/physical maps remain largely unknown. As telomeric regions are known to be the most gene-rich regions of the human genome, filling these gaps should have a high priority in completion of the Human Genome Project. We reported previously a first generation set of unique sequence probes for human telomeric regions. Of 41 human telomere regions, 33 were represented by unique clones with a known distance (≤300 kb) from the end of the chromosome; clones for the remaining eight telomeric regions had not yet been identified and were represented by the most distal markers on the integrated genetic/physical map. We have identified unique telomere clones for four of the remaining telomeres, 9p, 12p, 15q, and 16p. To determine the telomeric gap size for these chromosomes and five other human telomeres, interphase FISH analysis was performed to measure the distance between each telomere clone and the corresponding most distal marker. These studies provide distance estimates ranging from <100 kb to >1 Mb, thus defining the physical mapping task for filling telomeric gaps.
Physical maps for many regions of the human genome have been elucidated, however certain areas lack the coverage necessary for complete sequencing. Therefore, specific regions of human chromosomes will need to be targeted to complete the physical map of the entire genome. The telomeric regions of human chromosomes contain the highest gene concentrations in the genome (Saccone et al. 1992; Flint et al. 1997a). Yet, the majority of genome-wide physical and genetic mapping efforts have not yet addressed the problem of closure at the telomeric ends of human chromosomes.
The most distal human telomeric DNA consists of between 3 and 20 kb of tandemly repeated (TTAGGG)n sequences (Moyzis et al. 1988). Immediately proximal to this simple sequence are larger and more heterogeneous subtelomeric repeats, or telomere-associated repeats, that may have a polymorphic chromosomal distribution among individuals (Brown et al. 1990). Chromosome-specific (unique sequence) DNA for each telomere is located proximal to the subtelomeric repeats, about 100–300 kb from the end of the chromosome (National Institutes of Health et al. 1996; Flint et al. 1997b). Extensive half-YAC cloning has been carried out from the TTAGGG telomeric repeat to the unique telomere sequence and targeted efforts at cloning human telomeres have succeeded in identifying unique sequences at most telomeres (National Institutes of Health et al. 1996; Riethman 1997; Rosenberg et al. 1997). However, the distance between these telomere clones and the most distal marker on previously published physical and genetic maps is largely unknown.
We have reported a first generation set of unique sequence probes for human telomeric regions (National Institutes of Health et al. 1996). A complete telomere set was defined as 41 rather than 48 clones, because the X and Y chromosomes share homologous sequences at both telomeric pseudoautosomal regions, and no efforts were made to obtain the telomeres of the short arms of the five acrocentric chromosomes. Half-YAC cloning and subtelomeric sequence-mediated walking were employed to isolate the 33 chromosome-specific telomere probes. However, at that time, these methods were not successful for the eight remaining chromosome arms. Therefore, the most distal marker on the integrated chromosome map from the Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu/cgi-bin/contig/phys_map) was used for these eight chromosome arms (1p, 5p, 6p, 9p, 12p, 15q, 16p, and 20q).
Here, we report the development of unique telomere clones for four of the remaining chromosome arms: 9p, 12p, 15q, and 16p. The relationship between these four new telomere clones and the most distal marker identified by radiation hybrid mapping and/or genetic mapping was determined using interphase FISH to estimate the genomic distance. In addition, gap sizes at five additional telomeres were measured to survey other telomeric regions. These data provide integration of the genetic, physical, and cytogenetic maps for these nine telomeric regions and provide estimates of the physical gap sizes for targeted efforts at high-resolution physical mapping and sequencing.
RESULTS
STSs developed from half-YAC vector-insert junction sequences were used to isolate clones for chromosome arms 9p and 12p. An STS from the vector-insert junction sequence of a newly identified half-YAC, yRM2039, was used to isolate a unique clone for chromosome 9p. Clone yRM2039 contains the subtelomeric repeat HC1103 and shows multiple signals by FISH on chromosome 9, including the 9p telomere and the two pericentromeric regions of chromosome 9 (H. Riethman, unpubl.). An initial clone, RG305J7 (see Methods for nomenclature used for clones), which was isolated using the yRM2039 STS, was tested by FISH and showed a strong signal at the 9p telomere; however, it also showed significant hybridization to the two regions flanking the pericentromeric region of chromosome 9 and an interstitial site at 2q13 (Fig. 1A). The band 2q13 represents the ancestral telomere fusion site of two great ape chromosomes that gave rise to human chromosome 2 (Ijdo et al. 1991). Because RG305J7 was not chromosome 9p specific, it was end sequenced to walk proximally out of the subtelomeric repetitive DNA sequences. Primers were designed from the sequence from both ends of RG305J7 and tested against a monochromosomal hybrid panel to determine which end sequence to use for further BAC screening. One primer set amplified DNA from both chromosomes 2 and 9, whereas the other amplified only chromosome 9 DNA. The latter was chosen for screening and identified a unique 9p telomere clone, BAC RG41L13 (Fig. 1B).
Figure 1.
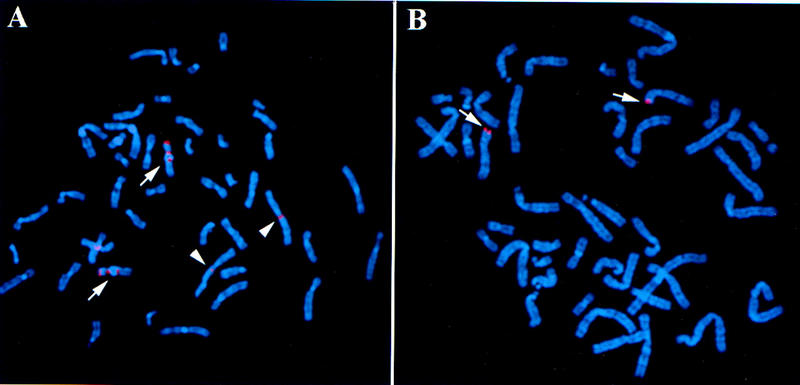
Hybridization of the 9p telomere clones, RG305J7 and RG41L13, labeled with digoxigenin and detected with anti-digoxigenin rhodamine (red), to normal metaphase chromosome preparations. (A) Clone RG305J7 shows a strong hybridization signal at the 9p telomere; additional signals are present on both sides of the pericentromeric region of chromosome 9 and at 2q13, the site of an ancestral telomere fusion. The chromosome 9 homologs are designated with arrows; the 2q13 signals are marked with arrowheads. (B) Hybridization with clone RG41L13 shows strong hybridization to the 9p telomere, indicated by arrows, with no cross-hybridization to any other chromosomes.
Another half-YAC, TYAC 14, was reported for chromosome arm 12p (Vocero-Akbani et al. 1996). An STS from the vector–insert junction sequence was used to identify BAC GS8M16. When used as a FISH probe, this clone exhibited strong hybridization to the 12p telomere with weak cross-hybridization to the 6p and 20q telomeres.
Interphase FISH was used to obtain genomic distance measurements between the 9p and 12p telomere clones and the corresponding distal marker clones. Representative images of interphase FISH analysis for the 9p and 12p telomeres are shown in Figure 2, A and B, respectively. Prior to the identification of the 9p telomere clone, we had reported a distal marker clone (PAC GS34H2, WI-5646) that was used to represent the telomeric region of 9p (National Institutes of Health et al. 1996). Interphase distance measurements between this distal marker clone and the 9p telomere clone revealed a genomic distance of ∼1.2–1.5 Mb. Since the report in 1996, additional markers have been placed on the chromosome 9 genetic/physical maps distal to WI-5646. Table 1 lists the telomere probe used, the distal marker and corresponding clone identified by radiation hybrid (RH) mapping and/or genetic mapping, and the estimated interphase distance measurements. As shown in Table 1, the estimated gap size at the 9p telomere was estimated to be <200 kb between the telomere clone (RG41L13) and the distal marker clone (GS100H5, WI-17492). Similarly, a gap size of ∼250 kb was identified between the 9p telomere clone (RG41L13) and a clone for the most distal genetic marker (GS56N11, AFM274XE1).
Figure 2.
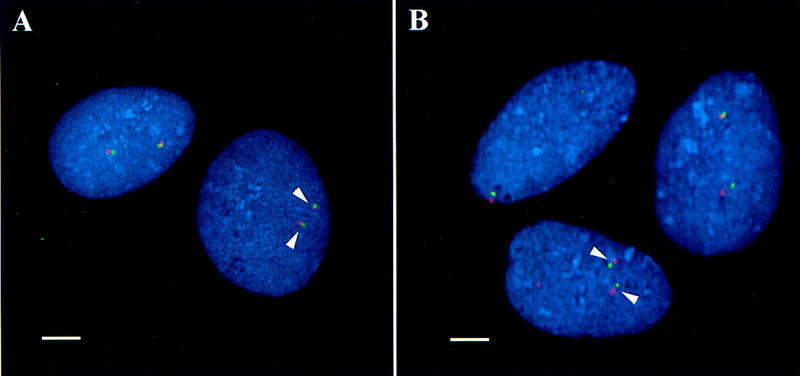
Interphase FISH distance measurements on interphase nuclei from G0 fibroblast cells to determine the distance between either the 9p or the 12p telomere probe and the corresponding probe for the most distal marker. Telomere probes are shown labeled in digoxigenin and detected with anti-digoxigenin rhodamine (red); probes for the most distal makers are shown labeled in biotin and detected with avidin–FITC (green). The arrowheads show hybridization signals for each probe set from a single nucleus. Bar, 5 μm. (A) Hybridization of clone RG41L13 (9p telomere, red) and clone GS56N11 (AFM274XE1, green); the genomic distance is estimated to be 250 kb between these two probes. Notice the close proximity of the green and red signals for the 9p clones. (B) Hybridization of clone GS8M16 (12p telomere, red) and clone GS93E1 (AFM303XD9, green); the genomic distance is estimated to be 600–800 kb between these two probes. Notice the greater signal separation between the 12p clones as compared to the 9p clones.
Table 1.
Comparison of the Estimated Distance to the Telomere Based on RH Data, Genetic Mapping, and Interphase FISH Analysis
| Telomere/clone | Distal marker/clonea | RH distance | Interphase FISH |
|---|---|---|---|
| 2p/68J13 | AFMA070WC9/357O1 | 16.02 cR (4.8 Mb) | 1 Mb |
| AFMB283ZH5/138E4 | <100 kb | ||
| 4p/118B13 | AFMA060ZE5/136B17 | 500–600 kb | |
| 4q/cT55 | AFMA224XH1bc/31J3 | 2 cR (600 kb) | <200 kb |
| 8p/63M14 | AFM197XG5/77L23 | <200 kb | |
| 9p/41L13 | WI-17492/100H5 | 0.0 cR | <200 kb |
| AFM274XE1/56N11 | 250 kb | ||
| 12p/8M16 | WI-5272/90I5 | 0.00 cR | 800 kb–1 Mb |
| AFM303XD9/93E1 | 600–800 kb | ||
| 15q/196D24d | WI-5214/154P1 | 10 cR (3 Mb) | <100 kb |
| AFMA140YE5c/121F5 | 600 kb | ||
| 16p/191K2 | D16S521b/268L4 | 6.29 cR (1.9 Mb) | colocalizes |
| 16q/191P24 | SGC32044/230M24 | 0 cR (0 kb) | 300 kb |
| AFM342XD9c/121A14 | 700–800 kb |
(AFM) Genethon; (WI) Whitehead Institute; (SGC) Stanford Genome Center.
Same distal marker on RH and genetic map.
New distal marker from comprehensive genetic map (Broman et al. 1998) may be distal to this marker.
Subtelomeric repeat probe that cross-hybridizes to multiple telomeres, including 15q.
For the 12p telomere, no additional markers have been placed distal to the previous marker (WI-5272) we used to identify a clone (PAC GS90I5) to represent the telomeric region of 12p (National Institutes of Health et al. 1996). A substantial gap, estimated at 800 kb–1 Mb, was identified for the 12p telomeric region between the newly identified 12p telomere clone (GS8M16) and this distal marker clone. The relationship between another 12p clone corresponding to a distal genetic marker (GS93E1, AFM303XD9) and the 12p telomere clone was also examined; a gap size of 600–800 kb was observed.
In our previous report, a clone corresponding to the most distal marker for chromosome 15q (PAC GS154P1, WI-5214) was used to represent the telomeric region of chromosome 15q (National Institutes of Health et al. 1996). At that time, no resources, such as a 15q-specific half-YAC clone, existed that could provide evidence for a link to the 15q telomere. In this report we took advantage of a chromosome 19-derived half-YAC clone, yRM2001, which contains subtelomeric repeats from chromosomes 3q, 15q, and 19p, as a reference point for studies of 15q. Using the vector-insert junction sequence for yRM2001, we identified a genomic clone (GS196D24) that shows strong cross-hybridization to the subtelomeric regions of chromosomes 3q, 6q, 15q, and 19p and weak cross-hybridization to 1p, 11p, 12p, and 16q. This pattern is consistent with the presence of subtelomeric repeats shared among these telomeric regions. Interphase distance measurements were used to estimate the genomic distance between this telomeric clone GS196D24 and the distal 15 marker GS154P1. Twenty-one signals out of a total of 62 signals (34%) colocalized in the interphase nucleus indicating a genomic distance of <100 kb. Using another distal marker clone for 15q (GS121F5, AFMA140YE5), the interphase distance to GS196D24 was shown to be ∼600 kb.
The telomeric region of human chromosome 16p is well characterized. In fact, the most distal 285 kb of chromosome 16p has been completely sequenced (Flint et al. 1997a). To develop a telomeric clone that met our criteria for a unique telomere clone, we used primers for a chromosome 16p STS, 16PTEL05 (D16S3400) (Rosenberg et al. 1997), which lies within 300 kb of the end of the chromosome to identify RG191K2. The appropriate telomeric location of BAC RG191K2 was confirmed by FISH; the probe shows strong signals on 16p with no cross-hybridization to other sites. Prior to the identification of GS191K2, a clone for the distal marker WI-12447 (PAC GS119L16) was used to represent the telomeric portion of 16p (National Institutes of Health et al. 1996). Distance measurements between GS191K2 and GS119L16 revealed that the two clones colocalized and are therefore <100 kb apart. GS268L4, corresponding to the distal marker D16S521, was isolated and was shown by FISH to hybridize to the telomere of 16p with cross-hybridization to many other telomeres. In this case, our unique telomere clone lies proximal to the most distal marker clone. Therefore, for 16p, telomeric closure has been obtained as there are markers that extend across the transitional zone from unique DNA into subtelomeric repeat DNA.
Telomeric gap sizes at five additional telomeres were also examined and are shown in Table 1. The unique telomere clones used for 2p, 4q, and 8p were reported previously (National Institutes of Health et al. 1996). For 4p and 16q, we have converted our previous telomere clones into larger format BAC clones to provide more robust FISH signals for analysis. For 4p, we used end sequence from a subtelomeric repeat clone (GS10K2) isolated with the telomeric STS, 4PTEL02 (D4S3359) (Rosenberg et al. 1997), to identify GS118B13, a unique 4p telomere clone. The unique 16q telomere clone (PAC GS191P24) was identified using primers developed from the telomeric sequence, 16QTEL013 (GenBank accession no. Z96319).
For the 2p telomere, the distance between the telomere clone (PAC GS68J13) and the distal marker clone (RG357O1, AFMA070WC9) was estimated to be 1 Mb. However, using a clone for a distal genetic marker (GS138E4, AFMB283ZH5), the distance was estimated to be <100 kb.
Clone GS136B17, isolated with a distal marker from chromosome arm 4p, AFMA060ZE5, shows cross-hybridization to the telomere of 1p in addition to hybridization to the 4p telomere. Interphase distance estimates between this clone and the 4p telomere clone revealed a gap size of 500–600 kb. The 4p subtelomeric repeat clone GS 10K2, from which we used end-sequence to isolate a unique 4p telomere clone, shows hybridization signal on the 4p telomere and cross-hybridization to the short arms of chromosomes 13, 15, 21, and 22. The subtelomeric region of chromosome 4p has been completely sequenced and shows this subtelomeric repeat pattern to be present within 60 kb of the 4p telomere (Flint et al. 1997b). Therefore, the distal marker 4p clone (GS 136B17) must be 500–600 kb proximal of the 4p telomere clone. The observation of cross-hybridization to the 1p telomeric region by the 4p distal marker clone most likely represents an interstitial repeat sequence present on 4p.
For the 4q telomere, distance measurements were made between the unique telomere clone, cosmid cT55 and two distal marker clones, GS56F20, isolated with AFMA239XA5, and GS31J3, isolated with AFMA224XH1. The distance from cT55 to GS56F20 was estimated to be 300–400 kb, whereas the distance from cT55 to GS31J3 was <200 kb.
The most distal genetic marker on 8p, AFM197XG5, was used to isolate a clone (GS77L23) for interphase FISH analysis. PAC 63M14 is derived from the 8p half-YAC, yRM2205, and lies in the subtelomeric repeat region of chromosome arm 8p, as demonstrated by FISH showing hybridization to the 8p and 1p telomeres (National Institutes of Health et al. 1996). The distance between the genetic marker and 63M14 was shown to be <200 kb.
Telomeric gap size estimations were completed for the 16q telomere using the unique telomere clone GS191P24. Distance measurements were made between this telomere clone and a clone for the most distal RH marker (GS230M24, SGC32044) and revealed a gap size of 300 kb. A 700- to 800-kb gap was identified between the telomere clone and GS121A14, corresponding to the distal genetic marker AFM342XD9.
Since the inception and completion of this study, a new genetic map has been published (http://www.marshmed.org/genetics/). We compared the distal markers used in this study with those of the new map. For six of nine telomeres (2p, 4p, 8p, 9p, 12p, and 16p) the distal marker on the new Marshfield map is the same as that used in this study. New distal markers were available for three telomeres (4q, D4S1523; 15q, D15S642; 16q, D16S671). We tested these STSs by PCR to determine whether they were contained within the distal marker or telomere clones used in this study. None of the markers tested amplified DNA from either the distal marker clones or the telomere clones. These new distal markers likely lie between the distal markers used in the present study and the telomere clones and will therefore contribute to closing the telomeric gaps.
DISCUSSION
We have developed new BAC clones within 300 kb of the end of each chromosome for four human telomeres (9p, 12p, 15q, and 16p). The telomeric location and chromosome specificity of each telomere clone was verified by FISH. The identification of these four new human telomeric clones will contribute to telomeric closure of these human physical maps.
RH mapping has been used previously to define the physical map for the telomeric regions of many human chromosomes (Rosenberg et al. 1997). RH mapping is useful, as either polymorphic or nonpolymorphic STSs can be mapped and placed on a physical map by estimating the distance between markers (Cox et al. 1990; Stewart et al. 1997). However, because of the statistical nature of RH mapping data, the resulting map is only a most likely order. Although not formally tested, this inaccuracy may be greater at telomeric regions due to the complex structure of telomeric DNA.
Another unique feature of human telomeres is an increased rate of genetic recombination. It has been demonstrated previously that recombination rates are increased in the telomeric regions of both sexes in human, but dramatically so in males (Donis-Keller et al. 1987; Rouyer et al. 1990; Blouin et al. 1995). Thus, the relationship between genetic distances and physical distances at human telomeres is greatly altered from other chromosomal regions. In fact, it has been shown previously that neither RH nor genetic mapping provided the correct order for telomeric markers on chromosome arms 4p and 16p, as compared to complete sequence data (Rosenberg et al. 1997).
Therefore, we used interphase FISH as an independent estimate of the physical distance between each of the four new telomere clones reported here and corresponding clones for the most distal markers mapped by RH and/or genetic mapping. In addition, we examined five other telomeres to determine the telomeric gap sizes present on other chromosome arms. Chromosome condensation may be different at telomeric regions, as compared to other regions of the genome, thus affecting interphase FISH distance estimates. However, because we used a calibration curve derived from telomeric clones (see Methods) for the conversion from interphase distance to genomic distance, any discrepancies should be minimized. The results presented here for nine chromosome ends illustrate the usefulness of interphase FISH mapping in the integration of the genetic, physical, and cytogenetic maps.
We have compared the distance measurements from using interphase FISH with the distances provided from RH mapping and genetic mapping data. These distance comparisons are outlined in Table 1. Because the integrated map was created using the GeneBridge 4 whole-genome RH panel, we used a conversion factor of 300 kb/cR for distance comparisons between RH and interphase mapping (Hudson et al. 1995).
For this study of telomeric gap size estimations, we examined nine human telomeric regions. Two of these telomeres, 4p and 16p, were predicted a priori to have small, if any, gaps as they have been well characterized and much of these regions have been sequenced. FISH confirmed telomeric closure observed as cross-hybridization of the clone to other telomeres (indicating the presence of subtelomeric repeat sequences). No predictions were made for the other seven telomeric regions with regards to the presence or size of gaps in the physical maps at the telomere. Our results demonstrate that gaps ranging from 200 kb to 1 Mb are present on existing genetic/physical maps, underscoring the need for targeted efforts at high-resolution physical mapping and sequencing. In addition, by using distal markers from both the RH and genetic maps, we have integrated the genetic, physical, and cytogenetic maps for these nine telomeric regions.
Our results demonstrate that interphase FISH mapping can be a valuable tool for completing the human physical map. We have established the validity of using interphase FISH in defining gap sizes on genetic/physical maps for the telomeric regions of human chromosomes. However, this methodology can be applied to other genomic regions; once a gap is identified, and its size estimated, efficient strategies can be designed for filling the gaps and completing that portion of the human genetic map.
The identification and mapping of telomeric clones will allow the completion of detailed characterization of human telomeric regions. The construction of complete telomeric physical maps will not only expedite positional cloning and large-scale sequencing efforts but will also aid in the identification of single nucleotide polymorphisms (SNPs) for telomeric regions. The identification of SNPs throughout the entire genome has become a high priority of the Human Genome Project (Collins et al. 1997; Wang et al. 1998). Because physical maps have already proven useful in the localization and characterization of many genes associated with human diseases, closure of telomeric physical maps will provide researchers with the landmarks necessary to pursue telomeric research related to various fields.
METHODS
Public Databases
The integrated physical map (http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map) and the Généthon map (http://www.genethon.fr/genethon_en.html) were used to identify distal markers mapped by RH mapping and genetic mapping, respectively (Hudson et al. 1995; Dib et al. 1996). The more recently published comprehensive genetic map (http://www.marshmed.org/genetics/) was also used to identify any new distal markers that were mapped after the completion of our study (Broman et al. 1998).
PAC and BAC Library Screening/Primer Development
A total human genomic BAC or PAC library was screened with telomeric STSs using a PCR-based screening strategy (Genome Systems, St. Louis, MO; Research Genetics, Huntsville, AL). All genomic clones discussed in this report are BAC clones unless otherwise stated. The letters RG or GS preceding a clone address identify that clone as being isolated from the Research Genetics library or Genome Systems library, respectively. PCR screening was carried out using STSs developed from either distal markers, telomeric sequence information, half-YAC vector–insert junction sequences, or end sequences from telomeric clones. The methods for identifying half-YAC clones, isolating vector-insert junction fragments, and obtaining sequence information have been described previously (Cross et al. 1989; Riethman et al. 1989; Negorev et al. 1994). Because the subtelomeric repeats are known to be shared among nonhomologous chromosomes, each primer set used for PCR screening was first tested and optimized on a monochromosomal hybrid panel to identify conditions that would produce chromosome-specific PCR amplification for screening purposes.
The software package Primer 3 (Rozen and Skaletsky 1996, 1997; code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html) was used to create primers from sequence information. Primers designed from the SP6 and T7 end sequence of BAC RG305J7 are 305J7/SP6-F, 5′-TCCCCAAATTTAGTACCACC-3′ and 305J7/SP6-R, 5′-CTTTGGAAAAGATCTCATGG-3′ (1.5 mm MgCl2, 55°C annealing temperature, product size 130 bp); 305J7/T7-F, 5′-TTACATTCCCTTTCATCACC-3′ and 305J7/T7-R, 5′-CACTGCACTCCAGTTTGG-3′ (1.5 mm MgCl2, 55°C annealing temperature, product size 128 bp). Vector–insert junction sequence from half-YAC yRM2001 is available in public databases (GenBank accession no. U11826, Genome Database no. 371271); the PCR primers used to amplify this sequence are V-I 2001F, 5′-TCAGAGCAGTGAGAAAAGAGTG-3′ and V-I 2001R, 5′-ACTCTCCCTTAGGAGGATTCC-3′ (1.5 mm MgCl2, product size 126 bp). For this primer set, the PCR reaction was cycled 35 times at 92°C for 1 min, 55°C for 2 min, and 72°C for 2 min. PCR primers for the vector–insert junction sequence of the chromosome 9p half-YAC yRM2039 are 2039Bg/F, 5′-GCAAAATGAATTGGTAGTTTGG-3′ and 2039Bg/R, 5′-TGCCTAAAGGAAGTGAGACATC-3′ (1.5 mm MgCl2, 55°C annealing temperature, product size 122 bp). The primer sequences from the vector–insert junction of TYAC14 have been published previously (Vocero-Akbani et al. 1996).
Distal markers identified by RH and genetic mapping were chosen based on the odds ratio at which they were placed on the maps. Markers mapped by RH mapping had to either be framework markers or be placed with odds greater than or equal to that of a framework marker (defined as >300:1) (Hudson et al. 1995). Markers from the genetic map had to be placed at an odds greater than or equal to 1000:1. Distance from the telomere is measured from the p-arm telomere on the genetic and physical maps. Therefore, for those q-arm telomeres, we equated the last marker on the map as being 0 cM (genetic) or 0 cR (physical) from the telomere. In some cases, there were markers distal to the one that we used that did not meet our minimum odds-ratio requirements. For these telomeres, we used the difference between the most distal map marker and the marker studied as the estimated distance from the telomere.
The distal marker primer sets used, which are available in public databases, were AFMA070WC9 (2p, RH), AFB283ZH5 (2p, genetic); AFMA060ZE5 (4p, genetic); AFMA239XA5 (4q, RH), AFMA224XH1 (4q, genetic); AFM197XG5 (8p, genetic); WI-17492 (9p, RH), AFM274XE1 (9p, genetic); WI-5272 (12p, RH), AFM303XD9 (12p, genetic); WI-5214 (15q, RH), AFMA140YE5 (15q, genetic); D16S521 (16p, RH and genetic); SGC32044 (16q, RH), AFM342XD9 (16q, genetic).
BAC End Sequencing
BAC DNA was isolated using an automated nucleic acid isolation system (AutoGen 740, Integrated Separation Systems, Natick, MA) and purified with Microcon 100 columns (Amicon, Inc., Beverly, MA). One microgram of BAC DNA and 40 pmole of T7 and SP6 primers were used for sequencing with the ABI Prism Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Inc., Foster City, CA). The sequences of the T7 and SP6 primers have been reported previously (Matsumoto et al. 1997). The sequencing reaction was cycled 50 times at 96°C for 10 sec, 50°C for 5 sec, and 60°C for 4 min. Sequence analysis was completed on an ABI 377 automated DNA sequencer.
FISH Characterization/Testing of New Telomere Probes
DNA was isolated for FISH analysis using an Autogen 740. FISH analysis was carried out on normal peripheral blood specimens to test each new probe identified. Probe and slide preparation, DNA hybridization, and analysis were carried out using previously described methods (Chong et al. 1997). At least 10 metaphase cells were analyzed using microscope and image analysis to verify probe location and chromosome specificity. Inverted DAPI staining was used to obtain a G-banded pattern for chromosome identification. A probe was characterized as telomere specific only if the signal was chromosome specific (i.e., no cross-hybridization to any other chromosomes) both down the microscope and on the individual fluorochrome raw image. Probes were tested for polymorphisms by analyzing the size and intensity of the FISH signals on at least five unrelated individuals.
Interphase Distance Measurements
Dual-color FISH was used for measuring genomic distances in interphase nuclei from either G0 fibroblast or peripheral blood lymphocyte cells. We have carried out measurements on both of these cell types and found the results to be comparable (C. Lese, unpubl.). Interphase distance measurements were made between each new telomere probe and the probe for the corresponding distal marker. Genomic distances were measured using the measure length command contained in the IP Lab Spectrum software package (v. 3, Signal Analytics, Vienna, VA). The measure length command was calibrated using images from a Zeiss stage micrometer. Between 50 and 100 interphase distances were measured for each telomere. Genomic distance was estimated from a calibration curve that was produced in our laboratory using the same cosmids used to create the published relationship between the mean interphase distance squared and the genomic distance for the chromosomal region 4p16.3 (Trask et al. 1993).
Acknowledgments
This work was supported in part by a National Research Service Award grant 1 F32 HG00174-01 to C.M.L. through the National Human Genome Research Institute and by National Institutes of Health grant NIHHG00567 to H.C.R. We acknowledge Julie Kuc, Anthea Kelsick, and Kathrin Precht for excellent technical assistance. We are grateful to Drs. B. Trask and M. MacDonald for providing the 4p16.3 clones.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dhl@genetics.uchicago.edu; FAX (773) 834-0505.
REFERENCES
- Blouin J-L, Christie DH, Gos A, Lynn A, Morris MA, Ledbetter DH, Chakravarti A, Antonarakis SE. A new dinucleotide repeat polymorphism at the telomere of chromosome 21q reveals a significant difference between male and female rates of recombination. Am J Hum Genet. 1995;57:388–394. [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- Chong SS, Pack SD, Roschke AV, Tanigami A, Carrozzo R, Smith ACM, Dobyns WB, Ledbetter DH. A revision of the lissencephaly and Miller-Dieker syndrome critical region in chromosome 17p13.3. Hum Mol Genet. 1997;6:147–155. doi: 10.1093/hmg/6.2.147. [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Chakravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Cross SH, Allshire RC, McKay SJ, McGill NI, Cooke HJ. Cloning of human telomeres by complementation in yeast. Nature. 1989;338:771–774. doi: 10.1038/338771a0. [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H, Green P, Helms C, Cartinhour S, Weiffenbach B, Stephens K, Keith TP, Bowden DW, Smith DR, Lander ES, et al. A genetic linkage map of the human genome. Cell. 1987;51:319–337. doi: 10.1016/0092-8674(87)90158-9. [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR. The relationship between chromosome structure and function at a human telomeric region. Nat Genet. 1997a;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum Mol Genet. 1997b;6:1305–1313. doi: 10.1093/hmg/6.8.1305. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu S-H, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Soeda E, Ohashi H, Fujimoto M, Kato R, Tsujita T, Tomita H, Kondo S, Fukushima Y, Niikawa N. A 1.2-megabase BAC/PAC contig spanning the 14q13 breakpoint of t(2; 14) in a mirror-image polydactyly patient. Genomics. 1997;45:11–16. doi: 10.1006/geno.1997.4897. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Roschke A, Smith ACM, Macha M, Precht K, Riethman H, Ledbetter D, Flint J, Horsley S, et al. National Institutes of Health, Institute of Molecular Medicine Collaboration. A complete set of human telomeric probes and their clinical application. Nat Genet. 1996;14:86–89. doi: 10.1038/ng0996-86. [DOI] [PubMed] [Google Scholar]
- Negorev DG, Macina RA, Spais C, Ruthig LA, Hu XL, Riethman HC. Physical analysis of the terminal 270 kb of DNA from human chromosome 1q. Genomics. 1994;22:569–578. doi: 10.1006/geno.1994.1430. [DOI] [PubMed] [Google Scholar]
- Riethman H. Closing in on telomeric closure. Genome Res. 1997;7:853–855. doi: 10.1101/gr.7.9.853. [DOI] [PubMed] [Google Scholar]
- Riethman HC, Moyzis RK, Meyne J, Burke DT, Olson MV. Cloning human telomeric DNA fragments into Saccharomyces cerevesiae using a yeast artificial chromosome vector. Proc Natl Acad Sci. 1989;86:6240–6244. doi: 10.1073/pnas.86.16.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Hui L, Ma J, Nusbaum HC, Clark K, Robinson L, Dziadzio L, Swain PM, Keith T, Hudson TJ, et al. Characterization of short tandem repeats form thirty-one human telomeres. Genome Res. 1997;7:917–923. doi: 10.1101/gr.7.9.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F, de la Chapelle A, Andersson M, Weissenbach J. An interspersed repeated sequence specific for human subtelomeric regions. EMBO J. 1990;9:505–514. doi: 10.1002/j.1460-2075.1990.tb08137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S, De Sario A, Della Valle G, Bernardi G. The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes. Proc Natl Acad Sci. 1992;89:4913–4917. doi: 10.1073/pnas.89.11.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Allen S, Massa H, Fertitta A, Sachs R, van den Engh G, Wu M. Studies of metaphase and interphase chromosomes using fluorescence in situ hybridization. Cold Spring Harb Symp Quant Biol. 1993;58:767–775. doi: 10.1101/sqb.1993.058.01.084. [DOI] [PubMed] [Google Scholar]
- Vocero-Akbani A, Helms C, Wang JC, Sanjurjo FJ, Korte-Sarfaty J, Veile RA, Liu L, Jauch A, Burgess AK, Hing AV, et al. Mapping human telomere regions with YAC and P1 clones: Chromosome- specific markers for 27 telomeres including 149 STSs and 24 polymorphisms for 14 proterminal regions. Genomics. 1996;36:492–506. doi: 10.1006/geno.1996.0495. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]


