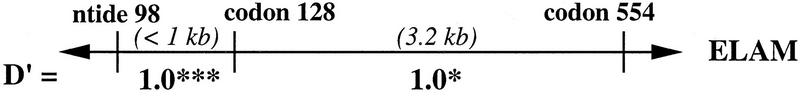Abstract
A number of chronic diseases, including cardiovascular disease, appear to have a multifactorial genetic risk component. Consequently, techniques are needed to facilitate evaluation of complex genetic risk factors in large cohorts. We have designed a prototype assay for genotyping a panel of 35 biallelic sites that represent variation within 15 genes from biochemical pathways implicated in the development and progression of cardiovascular disease. Each DNA sample is amplified using two multiplex polymerase chain reactions, and the alleles are genotyped simultaneously using an array of immobilized, sequence-specific oligonucleotide probes. This multilocus assay was applied to two types of cohorts. Population frequencies for the markers were estimated using 496 unrelated individuals from a family-based cohort, and the observed values were consistent with previous reports. Linkage disequilibrium between consecutive pairs of markers within the apoCIII, LPL, and ELAM genes was also estimated. A preliminary analysis of single and pairwise locus associations with severity of atherosclerosis was performed using a composite cohort of 142 individuals for whom quantitative angiography data were available; evaluation of the potentially interesting associations observed will require analysis of an independent and larger cohort. This assay format provides a research tool for studies of multilocus genetic risk factors in large cardiovascular disease cohorts, and for the subsequent development of diagnostic tests.
Multiple genetic and environmental risk factors appear to contribute to common diseases such as cardiovascular disease and cancer. Although specific genetic causes have been identified among certain families with a history of disease, the association of these genes with disease in the general population is not fully understood. One reason is that genetic predisposition to such diseases can result from the cumulative effect of common allelic variants, variants that individually confer only a modest increased risk. Thus, one challenge is to identify the common multilocus profiles that confer a high risk for disease; a second challenge is to understand how environmental factors modulate expression of a genetic predisposition to disease.
A growing number of genetic variants have been implicated in the development of complex diseases. As these candidate genes are identified, there is an increasing need for assays capable of simultaneously genotyping multiple loci. Studies focused on single markers can be used to assign relative risk values, but this approach provides only a limited context for evaluating genetic risk factors. Studies encompassing multiple markers provide a broader context that is critical to assess information on candidate markers for multifactorial diseases, and multilocus assays can greatly facilitate the necessary genotyping process. Multilocus results can provide insight into mechanisms of disease susceptibility and identify key subsets of predictive markers that are clinically informative. These informative genetic markers can then be used to supplement routine biochemical assays for patient care, for example, in lieu of protein activity or concentration measurements that are difficult to make or show significant intra- and interindividual variability independent of disease state.
In developing a prototype multilocus genotyping assay, we focused on cardiovascular disease (CVD), a leading cause of death worldwide. Monogenic disorders, such as familial hypercholesterolemia and hypertrophic cardiomyopathy (for reviews, see Hobbs et al. 1992; Day et al. 1997; Bonne et al. 1998), have been identified among some families. Established risk factors for disease in the general population include age, gender, diabetes mellitus, obesity, high serum cholesterol levels, and hypertension, as well as cigarette smoking and physical inactivity (Pasternak et al. 1996). These factors, however, do not explain all premature CVD cases (Hoeg 1997). A number of these established factors have genetic components, and as yet unknown risk factors may be primarily genetic. In addition, recent evidence indicates that genetic factors influence patient responsiveness to therapeutic intervention, both dietary (Humphries et al. 1996) and pharmaceutical (Kuivenhoven et al. 1998).
The “CVD35” assay described here is comprised of 35 biallelic sites within 15 genes representing pathways implicated in the development and progression of atherosclerotic plaques: lipid metabolism, homocysteine metabolism, blood pressure regulation, thrombosis, and leukocyte adhesion (Table 1). The panel includes well-known polymorphisms in the apolipoprotein E (apoE; for review, see Mahley 1988) and angiotensinogen (AGT; Jeunemaitre et al. 1992) genes, mutations such as apoB Gln-3500 (Soria et al. 1989) and factor V Leiden (Bertina et al. 1994), and more recently identified sequence variations in the methylene tetrahydrofolate reductase (MTHFR; Frosst et al. 1995; Goyette et al. 1995) and E-selectin (ELAM; Wenzel et al. 1996) genes. The CVD35 assay uses pooled polymerase chain reaction (PCR; Mullis and Faloona 1987; Saiki et al. 1988) primer pairs to coamplify 27 targets from genomic DNA in two reactions. Amplified fragments within each PCR product pool are then detected colorimetrically with sequence-specific oligonucleotide probes immobilized in a linear array on nylon membranes (Saiki et al. 1989). Probe sequences have been optimized carefully to permit genotyping of all sites under a single assay condition. Therefore, large cohorts can be typed rapidly at all 35 sites, providing an extended database for evaluating the disease association of these markers, and a multilocus context for evaluating new candidate markers. We have applied this multilocus assay to a population-based cohort to estimate allele frequencies and intragenic haplotypes, and to a lipid clinic-based cohort to model a case-control study.
Table 1.
Targets Genotyped by the CVD35 Assay
| Gene polymorphism: Reported implication, GenBank accession no., and references |
| Lipid metabolism |
| apolipoprotein B (apoB) on chr. 2p: component of plasma lipoproteins, particularly LDL; mediates binding to LDL receptor |
| Thr 71–Ile: possibly associated with increased plasma LDL cholesterol and apoB levels; Arg-3531-cys LDL receptor binding defect appears to segregate with Thr allele HUMAPB03 (M19810); (Young and Hubl 1989; Pullinger et al. 1995) |
| Arg-3500–Gln mutation: disorder of hypercholesterolemia known as familial defective apoB-100, due to reduced binding to LDL receptor |
| HSLDL100 (X04506); (Soria et al. 1989; Humphries and Talmud 1995; Pullinger et al. 1995) |
| apolipoprotein CIII (apoCIII) on chr. 11q: component of plasma lipoproteins |
| T(−625)del, C(−482)T, T(−455)C: increased plasma triglyceride levels |
| HUMAPOCP (M60674), HSAPC3A (X01392); (Dammerman et al. 1993) |
| C3175G (Sst)I: increased plasma triglyceride levels |
| HSAPC3A (X01392); (Rees et al. 1985; Ordovas et al. 1991) |
| C1100T, T3206G: increased plasma triglyceride levels |
| HSAPC3A (X01392); (Xu et al. 1994) |
| apolipoprotein E (apoE) on chr. 19q: component of plasma lipoproteins; mediates binding to the LDL and remnant (apoE) receptors |
| e3/e2, e4: interindividual variation in plasma total and LDL cholesterol levels, atherosclerotic progression |
| HUMAPOE4 (M10065); (Mahley 1988; Hixson and Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group 1991; de Knijff et al. 1994) |
| cholesteryl ester transfer protein (CETP) on chr. 16q: reverse cholesterol transport pathway; possible proatherogenic role in presence of dyslipidemia (Tall 1995; Lassel et al. 1998) |
| Ile-405–Val: increased plasma HDL cholesterol and apoA-I levels |
| HUMCETP6 (M32997); Agellon et al. 1990; Funke et al. 1994; Gudnason et al. 1997) |
| Asp-442–Gly: increased HDL cholesterol levels, yet also disease risk |
| HUMCETP7 (M32998); (Takahashi et al. 1993; Zhong et al. 1996) |
| lipoprotein lipase (LPL) on chr. 8p: hydrolysis of plasma triglycerides |
| T(−93)G: increased LPL promoter activity, reduced plasma triglycerides |
| HUMLPLA (M29549); (Yang et al. 1995a,b; Ehrenborg et al. 1997; Hall et al. 1997) |
| T(−39)C: reduced LPL promoter activity |
| HUMLPLA (M29549); (Yang et al. 1995a,b) |
| Asp9–Asn: increased plasma triglycerides, increased atherosclerotic progression |
| HUMLPL (M15856); (Oka et al. 1990; Mailly et al. 1995; Jukema et al. 1996) |
| Asn-291–Ser: reduced plasma HDL cholesterol, increased triglyceride levels |
| HUMLPL (M15856): (Oka et al. 1990; Reymer et al. 1995; Zhang et al. 1995) |
| Ser-447–Ter: increased plasma HDL cholesterol, reduced plasma triglyceride levels; possible impact on responsiveness to β-blockers |
| HUMLPLF1 (M76722); (Hata et al. 1990; Groenemeijer et al. 1997; Kuivenhoven et al. 1997) |
| paraoxonase (PON) on chr. 7q: HDL-associated enzyme known to hydrolyze organophosphate poisons; possible contribution to HDL's protective capacity against LDL oxidation |
| Gln-192–Arg: increased enzymatic activity; in vitro, reduced protection against lipid peroxidation |
| HUMPONA (M63012); (Humbert et al. 1993; Hegele et al. 1995; Serrato and Marian 1995; Mackness et al. 1997; Sanghera et al. 1997) |
| Renin–angiotensin system |
| angiotensin-converting enzyme (ACE) on chr. 17q: proteolyzes angiotensin I to produce angiotensin II |
| Alu element insertion/deletion in intron 16: increased plasma ACE levels; mixed evidence of association with myocardial infarction |
| HSATICE (X62855), HUMAICEB (J04144); (Cambien et al. 1992; Rigat et al. 1992; Evans et al. 1994; Lindpaintner et al. 1995; Ludwig et al. 1995) |
| angiotensin II receptor type 1 (ATIIR1) on chr. 3q: one of two receptors for angiotensin II, particularly in vascular smooth muscle cells |
| A1166C: hypertension; possible synergism with ACE conferring risk of myocardial infarction HSANTENII (Z11162); (Bonnardeaux et al. 1994; Tiret et al. 1994) |
| angiotensinogen (AGT) on chr. 1q: substrate for renin, yielding angiotensin I |
| Met-235–Thr: increased plasma AGT levels; hypertension |
| HUMAGT02 (M24686); (Jeunemaitre et al. 1992; Morgan et al. 1996) |
| Homocysteine metabolism |
| cystathionine β-synthase (CBS) on chr. 21q: transulfuration pathway, converting homocysteine to cystathioine, with pyridoxine as cofactor |
| Ala-114–Val: pyridoxine-responsive homocystinuria |
| HUMCYSTATH (L19501); (Kozich et al. 1993) |
| Arg-125–Gln, glu131asp: pyridoxine-unresponsive homocystinuria |
| HUMCYSTATH (L19501); (Marble et al. 1994) |
| Ile-278–Thr: pyridoxine-responsive homocystinuria |
| HUMCYSTATH (L19501); (Kozich and Kraus 1992; Hu et al. 1993) |
| 68-bp insertion: linkage disequilibrium with 278thr; (Sebastio et al. 1995; Sperandeo et al. 1996; Tsai et al. 1996) |
| Gly-307–Ser: pyridoxine-unresponsive homocystinuria |
| HUMCYSTATH (L19501); (Hu et al. 1993) |
| methylene tetrahydrofolate reductase (MTHFR) on chr. 1p: remethylation pathway, generating the 5-methyltetrahydrofolate that serves as the methyl group donor |
| C677T (Ala/Val) increased thermolability; associated with hyperhomocysteinemia given low dietary folate; increased risk for deep-vein thrombosis in carriers of factor V Leiden HSU09806 (U09806); (Frosst et al. 1995; Goyette et al. 1995; Kluijtmans et al. 1996; Cattaneo et al. 1997; Christensen et al. 1997) |
| C692T: absence of enzyme activity |
| HSU09806 (U09806); (Goyette et al. 1995) |
| Thrombosis |
| glycoprotein IIIa (GPIIIa) on chr. 17q: component of GPIIb/IIIa platelet adhesion receptor, binding fibrinogen, fibronectin, and von Willebrand factor; (Nurden 1996) |
| Leu-33–Pro: interindividual variation in platelet adhesion and/or adhesion; mixed evidence of association with risk of coronary thrombosis |
| HUMGPP3A07 (M32672); (Newman et al. 1989; Jin et al. 1993; Weiss et al. 1996; Ridker et al. 1997) |
| fibrinogen on chr. 4q: determinant of plasma viscosity, cofactor for platelet aggregation, precursor of fibrin (component of plaques); (Cook and Ubben 1990; Mannucci 1995) |
| β chain G(−455)A: increased plasma fibrinogen levels and progression of atherosclerosis |
| HSFIBBR1 (X05018); (Green et al. 1993; Humphries et al. 1995; Humphries et al. 1997) |
| factor V on chr. 1q: activated form is procoagulant cofactor in prothrombin activation, inactivated through cleavage by activated protein C |
| Arg-506–Gln (Leiden mutation): resistance to activated protein C; hypercoagulability |
| HUMF510 (L32764); (Bertina et al. 1994; Ridker et al. 1995; Kalafatis and Mann 1997) |
| Leukocyte adhesion |
| endothelial leukocyte adhesion molecule-1 (ELAM) on chr. 1q: adhesion of leukocytes to activated arterial endothelium (Carlos and Harlan 1990); also known as E-selectin |
| G98T, Ser-128–Arg, Leu-554–Phe: increased risk for severe atherosclerosis |
| HUMELAM1 (M61895), HUMELAM3 (M61888), HUMELAM7 (M61892); (Wenzel et al. 1994; 1996) |
Markers are grouped by gene within broad categories based upon biological function. Chromosomal locations of each gene are given; among these genes, no two are known to be in strong linkage disequilibrium. For each target, with wild-type or most frequent allele is listed to the left, and the variant to the right, of the numerical nucleotide or codon position. Single letters indicate nucleotide variations; three-letter codes designate amino acid variations. For each target, a possible role for the variant allele, the GenBank sequence used for primer and probe selection, and references for the sequence variations are listed. Review articles are cited in the interest of space.
RESULTS
CVD35 Assay
A three-primer, two-probe system for apoE was used as the basis for the CVD35 assay; the specificity of this system has been described previously (Cheng et al. 1998). No Arg-112/Cys-158 alleles were detected among any of the 1400 control or cohort samples genotyped, consistent with previous studies of the apoE gene (Houlston et al. 1989). In future versions of this assay, the allele-specific primers for codon 112 will be replaced by probes to simplify genotyping of the apoE marker (data not shown).
The PCR products range from 95 to 535 bp in size. As shown in Figure 1, nearly all of the PCR products in each of the final multiplexes (14 in multiplex A, 13 in multiplex B) could be clearly distinguished by gel electrophoresis. Although the largest product bands appeared relatively weak in fluorescence intensity, these yields were sufficient for detection by the immobilized probes.
Figure 1.

Agarose gel image illustrating the PCR product pool resulting from coamplification of multiple loci in the CVD35 research assay. The molecular weight standard is HaeIII-digested ΦX174. The PCR products in lane A (multiplex A) range from 95 to 395 bp; those in lane B (multiplex B) range from 105 to 535 bp. Not all of the PCR products are clearly visible, and one low molecular weight nontarget band is visible in lane A.
Detection of the amplified alleles is illustrated in Figure 2, as well as the specificity of the probe panel; for example, the results distinguish each of the three possible genotypes at LPL(−93) (Fig. 2A: strips three to five), PON192 (Fig. 2A: strips one to three), AGT235 (Fig. 2B: strips one to three), and factor V 506 (Fig. 2B: strips one, two, and four). Figure 2C shows the third strip (strip B2) for two different individuals; of the ∼720 unrelated individuals genotyped from all sources, only one variant allele was detected among these four candidate markers (data not shown).
Figure 2.



Representative probe strips demonstrating the identification of different alleles. Genomic DNA samples were amplified with either the multiplex A (A) or the multiplex B (B,C) primer pool, then hybridized to the corresponding immobilized probe strips, as described under Methods. Five examples each of probe strips A and B are shown; only two examples of probe strip B2 are shown. For the purpose of illustrating a range of different genotypes, the individuals typed here in A do not correspond to those in B or C. On each vertical strip, alleles amplified from the original sample are indicated by horizontal lines. The template guides at the left and right identify the allele detected at each probe position. With the exception of apoE3, ACE-I, and ACE-D, the less prevalent genetic variant for each marker is listed on the right. Differences in the relative efficiencies of amplification and probe hybridization contribute to the variation in the actual intensity among the loci. (In actual size, each strip is ∼8 cm long.)
Samples representing a subset of 303 families from the Stanislas cohort (Siest et al. 1998) were used to assess the performance of the CVD35 assay in a large-scale genotyping effort. Eight families were excluded from the haplotype analysis because of inconsistencies of genotypes between parents and offspring, although the unrelated parents were included in the analysis of allele frequencies. In addition, four samples were omitted as a result of weak second allele signals for several markers. Because the assay was designed to yield comparable signals for both alleles in heterozygotes, these weak signals may have resulted from sample contamination. When available, all questionable samples will be retyped using new DNA preparations. A total of 1190 samples were used for subsequent analyses of the allele frequencies for all markers and haplotypes within the apoCIII, ELAM, and LPL genes.
Genotyping data for apoE, ACE, and LPL447, which had been obtained previously through independent methods, were used to evaluate the accuracy of the CVD35 assay for these three markers. No discordant results were noted for LPL447; those few samples yielding discordant genotype results for apoE and ACE were investigated further. For apoE, detection of the e4 allele by the CVD35 assay was problematic if insufficient template DNA was used for amplification; this difficulty should be corrected in future versions of the assay that no longer rely on codon 112-specific primers (data not shown). Three initially discordant results were traced to the specific aliquots tested.
For the ACE I/D marker, one D allele and two I alleles that had been identified by capillary electrophoresis were not detected by the CVD35 assay. This particular ACE-D allele was detectable if an alternative primer pair was used, suggesting the presence of a novel sequence variation within the default priming sites. One of the undetected ACE-I alleles was also amplifiable with an alternative upstream primer, although with poor efficiency, and gel electrophoresis revealed a truncated insertion of ∼50 bp in size (data not shown); this ACE-I allele had been detected previously based on the 3′ insertion junction sequence. The second undetected ACE-I allele was identified correctly after single-target amplification with the original primer pair, or using a multiplex amplification with an alternative ACE-I-specific primer spanning the 3′ insertion junction, as in Evans et al. (1994; data not shown). Failure of the CVD35 assay to amplify the full-length Alu element from a wild-type sequence in a multiplex reaction appeared to be unique to this sample. With the 3′ junction-specific primer, the ACE-I target was reduced from 533 to 155 bp; this primer was used to confirm all D/D genotypes within the UCSF cohort (data not shown). Use of the ACE-I-specific primer spanning the 3′ insertion junction in future assays should address these few difficulties.
Allele Frequencies
No variant alleles were observed within either cohort for 6 of the 35 sites: LPL nucleotide (−39); CETP codon 442; CBS codons 125, 131, and 307; and MTHFR nucleotide 692. Within the Stanislas cohort, a single CBS Val-114 allele and no apoB codon Gln-3500 alleles were detected. The genotypes of 496 unrelated parents from this group were used to estimate population frequencies for each of the remaining 28 markers; these data (Table 2) extend the previous report based on 455 individuals (Cheng et al. 1998). The observed frequencies are consistent with previous reports for caucasian populations (as cited in Table 2). Four markers (CBS Ile/Thr-278, CBS del/ins, ELAM 98, and ELAM 128) appeared to differ (χ2 > 3.84) from expectations based on Hardy–Weinberg equilibrium, and complete concordance in genotypes between the two CBS markers and between the two ELAM markers was also noted. This may be type I error, given the number of loci tested; in general, the observed allele frequencies did predict the observed genotype frequencies.
Table 2.
Allele Frequencies (pA, pa) Observed among Unrelated Individuals within the Population-Based Stanislas Cohort
| Gene | Allele (A/a) | Stanislas (2n = 992) | UCSF (2n = 284) | Literature comparison | |
|---|---|---|---|---|---|
| p2 | p2 | p2 | |||
| apoE | e3/e2,4 | 0.101 | 0.060 | 0.08 | de Knijff et al. (1994) |
| p4 | p4 | p4 | |||
| 0.130 | 0.183 | 0.15 | |||
| pa | pa | pa | |||
| apoB | Thr-71/Ile | 0.307 | 0.324 | 0.36 | Young and Hubl (1989) |
| Arg-3500/Gln | 0 | 0.014 | 0.0015 | Humphries and Talmud (1995) | |
| apoCIII | (−625) T/del | 0.383 | 0.384 | 0.42 | Dammerman et al. (1993) |
| (−482) C/T | 0.257 | 0.261 | 0.24 | Dammerman et al. (1993) | |
| (−455) T/C | 0.370 | 0.380 | 0.42 | Dammerman et al. (1993) | |
| 1100 C/T | 0.247 | 0.317 | 0.21 | Xu et al. (1994) | |
| 3175 C/G | 0.080 | 0.116 | 0.07 | Ordovas et al. (1991) | |
| 3206 T/G | 0.352 | 0.444 | 0.35 | Xu et al. (1994) | |
| CETP | Ile-405/Val | 0.312 | 0.338 | 0.31 | Gudnason et al. (1997) |
| LPL | (−93) T/G | 0.020 | 0.035 | 0.02 | Ehrenborg et al. (1997) |
| Asp-9/Asn | 0.019 | 0.035 | 0.02 | Mailly et al. (1995) | |
| Asn-291/Ser | 0.015 | 0.011 | 0.02 | Reymer et al. (1995) | |
| Ser-447/term | 0.121 | 0.102 | 0.17 | Hata et al. (1990) | |
| PON | Gln-192/Arg | 0.274 | 0.324 | 0.31 | Serrato and Marian (1995) |
| ACE | intron 16 I/D | 0.534 | 0.511 | 0.54 | Ludwig et al. (1995) |
| ATIIR1 | 1166 A/C | 0.300 | 0.303 | 0.29 | Tiret et al. (1994) |
| AGT | Met-235/Thr | 0.403 | 0.412 | 0.36 | Jeunemaitre al. (1992) |
| CBS | Ile-278/Thr | 0.084 | 0.113 | 0.06 | Tsai et al. (1996) |
| 68-bp I | 0.084 | 0.109 | 0.06 | Tsai et al. (1996) | |
| Ala-114/Val | 0.001 | 0.000 | <0.1 | Sebastio et al. (1995) | |
| MTHFR | 677 C/T | 0.384 | 0.352 | 0.38 | Frosst et al. (1995) |
| GPIIIa | Leu-33/Pro | 0.157 | 0.141 | 0.10 | Weiss et al. (1996) |
| figrinogen | (−455) G/A | 0.199 | 0.229 | 0.22 | Humphries et al. (1995) |
| factor V | Arg-506/Gln | 0.026 | 0.014 | 0.03 | Ridker et al. (1995) |
| ELAM | 98 G/T | 0.087 | 0.099 | 0.11 | Wenzel et al. (1996) |
| Ser-128/Arg | 0.087 | 0.099 | 0.09 | Wenzel et al. (1996) | |
| Leu-554/Phe | 0.048 | 0.035 | 0.02 | Wenzel et al. (1996) | |
For apoE, p2 is the observed frequency of the e2 allele and p4 is the observed frequency of the e4 allele; the e3 allele frequency is not shown explicitly. For each biallelic marker, except for ACE, the alleles are written in the order of most frequent (allele A)/least frequent (allele a). For ACE, the risk-associated D allele was designated the variant allele (a). Markers for which no variant alleles were detected are not listed [CETP442, LPL(−39), CBS125, CBS131, CBS307, and MTHFR692]. Representative allele frequencies reported previously are cited for comparison. Allele frequencies are also given for the smaller UCSF Cohort. Although the significance of comparing these values with those of the Stanislas Cohort is limited by the sample sizes, the lipid clinic-based UCSF Cohort showed an overall trend of increased frequencies for lipid metabolism-related alleles that have been implicated in disease.
Within the UCSF cohort of 142 individuals, no variant CBS Val-114 alleles were observed. Four carriers of the apoB Gln-3500 mutation were noted within this clinic-based cohort, in contrast to the population-based Stanislas cohort. One carrier of the CBS Thr-278 allele without the 68-bp insertion was also detected; this genotype was confirmed by sequencing (data not shown). One sample initially yielded a null result for the ACE target, but subsequently was assigned the I/I genotype with use of the insertion-junction-specific primer. All ACE D/D genotypes were confirmed by reanalysis with the junction-specific primer (data not shown). Allele frequencies are given in Table 2. One marker (ELAM554) appeared to differ (χ2 = 4.14) from expectations based on Hardy–Weinberg equilibrium, but given the small sample size and number of loci, this may reflect type I error; in general, the observed allele frequencies predicted the observed genotype frequencies.
Linkage Disequilibrium
Using 1100 chromosomes from 275 families, three loci were examined for haplotypes defined by multiple sites typed within each gene: ELAM (three sites), LPL (four sites), and apoCIII (six sites). Linkage disequilibrium estimates between proximal marker sites are presented in Figure 3; the haplotype data will be explored in greater detail elsewhere (W. Klitz et al., in prep.). Allele frequencies among this subset of the Stanislas cohort were comparable to those listed in Table 2.
Figure 3.
Linkage disequilibrium values (D′) estimated from the intragenic haplotypes for the ELAM, LPL, and apoCIII genes that were observed among 275 families from the Stanislas cohort. Above each gene represented by a horizontal line, the genotyped sites are labeled by nucleotide (ntide) or codon position. Distances between adjacent sites are given in kilobases; the spacing is not to scale. (†) P > 0.1; (*) P < 0.05; (***) P < 0.001.
Maximum disequilibrium (D′ = 1.0) was observed among the three sites genotyped within the ELAM gene. The lower statistical significance associated with codon 554 was due to the low frequency of the Phe-554 allele. Codon Phe-554 was observed only on chromosomes carrying the G98 and Ser-128 alleles. Although complete concordance in genotypes between the ELAM 98 and 128 sites was noted in this study, chromosomes bearing only one of the two variants have been reported (Wenzel et al. 1996).
Maximum linkage disequilibrium was also observed between the LPL promoter site (−93) and codon 9. Three chromosomes were observed with the (−93)G (less frequent allele) and Asp-9 (more frequent allele) haplotype; all others were concordant in genotype between the two sites. Codons 291 and 447 appeared to be in modest linkage disequilibrium, although statistical significance was not achieved due to the low frequency of the Ser-291 allele (0.016). The strong disequilibrium between promoter site (−93) and codon 9 among caucasians has been reported previously (Hall et al. 1997). The suggestion of greater recombination rates upstream of codon 291 and between codons 291 and 447 (exons 6 and 9), as inferred from the linkage disequilibrium results, is consistent with recently reported data (Clark et al. 1998; Nickerson et al. 1998).
For the apoCIII gene, linkage disequilibrium was greatest within the promoter and exon 4 (markers 3175, 3206), regions with marker sites separated by fewer than 150 bp. Within the promoter, interestingly, linkage disequilibrium was not maximal between the (−482) and (−455) sites, separated by only 27 bp. Disequilibrium was low between the promoter region and site 1100, which are separated by 1.5 kb, yet strong disequilibrium was observed between sites 1100 and 3175 (exons 3 and 4), a 2-kb separation. The relatively infrequent 3175-G variant did appear to be in strong disequilibrium with the promoter variants (data not shown), as had been reported previously (Dammerman et al. 1993). Strong linkage disequilibrium between sites 1100 and 3206 has also been previously noted (Xu et al. 1994).
Although the likelihood of detecting association with disease is expected to be greatest with allelic variants of demonstrated functional significance, this information may not be readily available. In the absence of clear evidence as to the most functionally significant variations within a single gene, linkage disequilibrium data assist in determining the most informative sites to genotype for disease association studies. Redundant sites may then be replaced by new candidate markers. Linkage disequilibrium data may also lead to hypotheses tracing the evolution of haplotypes that may be associated with disease.
Disease Association
The variant allele frequencies observed within the lipid clinic-based UCSF cohort are listed alongside those of the population-based Stanislas cohort in Table 2. Although the Stanislas and UCSF cohorts were not specifically matched for population substructure, higher frequencies of apoE e2 and e4, apoB Gln-3500, and apoCIII 1100T and 3206G alleles (P < 0.05) were observed within the clinic-based UCSF cohort, consistent with previous reports associating these variants with elevated lipid levels; none of the nonlipid-related markers showed a nominally significant difference in frequencies between the two cohorts. The overall trend of increased frequencies for reportedly disease-associated alleles that was observed in the UCSF cohort might be expected among these individuals who are at higher risk for coronary events than the general population.
The UCSF cohort was comprised of 142 unrelated caucasians for whom angiograms had been quantitated and scored by the Gensini method (Gensini 1975). These scores were used to subdivide the cohort into quintiles that represented differing severities of coronary arterial occlusion. No significant deviations from Hardy–Weinberg equilibrium were noted within these quintiles (data not shown). The Gensini-based quintiles did not show significant correlation with total, low-density lipoprotein (LDL), or high-density lipoprotein (HDL) cholesterol levels, although there was an unexpected, suggestive trend toward lower average very low-density lipoprotein (VLDL)-triglyceride (TG) and VLDL-cholesterol levels with increasing Gensini score (data not shown).
Disease association with the allelic variants of 15 markers was explored among female-only (FQ1 vs. FQ5) and the combined gender (Q1 vs. Q5) quintiles, as described under Methods. Although the small size of this UCSF cohort limited the statistical power to detect one- and two-locus effects on risk for CVD, the intent of this analysis was to demonstrate how the CVD35 assay could be used to evaluate disease association and genotype interactions with a case-control study design. Given the exploratory nature of these preliminary analyses, no formal statistical correction for multiple testing was applied.
The markers were first considered individually for association with disease. The test for apoB codon 71 among women yielded a nominally significant difference in frequency between the extreme Gensini quintiles (12 carriers of the Ile-71 allele among 20 individuals in FQ1, 4 carriers among 18 in FQ5; uncorrected, two-tailed P < 0.03). These results are potentially interesting, but no conclusions can be drawn in light of the small sample size available. Previous evidence for association of the apoB Ile-71 site with plasma lipoprotein levels has been mixed (Young et al. 1987; Tikkanen et al. 1988).
Multilocus data are of particular value in enabling evaluation of combinations of markers for their association to complex disease. Although only large effects would be expected to yield statistically significant results with this limited sample size, we sought to explore this opportunity by considering two-locus effects. As shown in Table 3, analysis for two-locus effects within the UCSF cohort yielded 14 pairs of variant alleles that showed nominally significant associations (uncorrected P < 0.05) with angiographic scores in the combined gender or female-only quintile comparisons. One marker pair, GPIIIa Pro-33 with ATIIR 1166C, yielded a possibly predisposing association in both the combined gender and female-only quintile comparisons; the small cohort size did not permit direct evaluation of the role of gender. This preliminary study did suggest a number of potentially interesting two-locus effects, such as increased risk for disease associated with having two hypertension gene variants, ATIIR1 1166C and AGT Thr-235. A particularly high relative risk was estimated if CBS Thr-278 (associated with hyperhomocysteinemia; Hu et al. 1993) was paired with either apoE e4 or apoCIII promoter variants (associated with hypertriglyceridemia; Dammerman et al. 1993). In contrast, when this CBS variant was paired with ATIIR1 1166C (hypertension pathway; Bonnardeaux et al. 1994), the effect appeared to be protective. Overall, the number of nominally significant (uncorrected P < 0.05) marker pairs just exceeded expectations given a type I error rate of 5%. Analysis of larger cohorts would provide the necessary power to detect true effects of clinical relevance, leading to hypotheses that could be tested subsequently in independent cohorts.
Table 3.
Pairs of Variant Alleles Observed to Have Suggestively Significant Association with the Severity of Atherosclerosis as Represented by Gensini Scores
| Marker I (background) | Marker 2 (additional variant) | Q1 vs Q5 | |
|---|---|---|---|
| combined gender | females only | ||
| apoE (3/4, 4/4) | CBS278thr/68-bp I | ▴* | — |
| apoCIII (−625)del or apoCIII (−455)C | apoB Ile-71 | — | ▾* |
| apoCIII 3206G | apoB Ile-71 | — | ▾* |
| PON 192arg | apoB Ile-71 | — | ▾* |
| PON 192arg | ACE (I/D, D/D) | ▴* | — |
| ACE (I/D, D/D) | apoB Ile-71 | — | ▾** |
| ATIIR 1166C | AGT Thr-235 | — | ▴* |
| ATIIR 1166C | CBS Thr278/68-bp I | — | ▾* |
| AGT 235thr | apoB Ile-71 | — | ▾** |
| AGT 235thr | ATIIR 1166C | — | ▴* |
| CBS 278thr/68-bp I | apoCIII (−625)del or apoCIII (−455)C | — | ▴* |
| CBS 278thr/68-bp I | apoCIII (−428T)C | — | ▴* |
| GPIIIa 33pro | ATIIR 1166C | ▴* | ▴* |
| fibrinogen (−455)A | apoB Ile-71 | — | ▾* |
| ▴ Predisposing: | 3 | 5 | |
| ▾ Protective: | 0 | 7 | |
For each pair listed, the odds ratio corresponds to the risk associated with carrying a second variant allele (relative to the more frequent allele of Marker 2) within the set of individuals who carried a first variant allele (the background marker). A total of 15 markers were analyzed in this pairwise manner. Both combined-gender and female-only quintiles were evaluated, as described in Methods. (**) P < 0.01; (*) 0.001 ≤ P < 0.05; (—) P > 0.05.
DISCUSSION
With this immobilized probe assay format, large cohorts can be assayed rapidly for multiple biallelic sites, providing the necessary epidemiological data for evaluation of these markers in association with disease or therapeutic response. An additional advantage of this technology is the relative ease with which the panel of targets can be modified to include new markers of interest. The assay described here is currently being expanded to type >60 sites in 36 genes, and could be expanded even further. One limitation of this approach is that sequence-specific probes will not identify new mutations or polymorphisms; only those new sequence variations resulting in unusually weak signal intensities would be detected. Furthermore, this format does not detect variable number tandem repeat polymorphisms, and higher density probe arrays are more appropriate for detection of specific mutations in genes such as the LDL receptor gene, for which >600 mutations, including large deletions, have been reported (University College of London 1999). Our multilocus assay can be adopted more readily by individual laboratories, however, particularly for candidate gene evaluations similar to those described here, as compared with genome-wide scanning efforts using high-density arrays. The use of minisequencing on primer arrays with 33P incorporation to simultaneously genotype 12 variable sites was reported recently (Pastinen et al. 1998); this approach is also promising for rapid analysis of large cohorts.
With larger cohorts, multiple regression or logistic regression methods have greater power to identify those combinations of genotypes that are most clinically informative with regard to disease phenotypes and endpoints. Alternative analytical approaches for multilocus genotype data sets may also reveal interesting associations. Extensive (n > 10,000) epidemiological and intervention studies such as the Framingham Heart Study (Dawber et al. 1951), Women's Health Initiative (The Women's Health Initiative Study Group 1998), and Multiple Risk Factor Intervention Trial (The MRFIT Research Group 1982) may offer the greatest power to detect multilocus risk factors, but smaller cohorts of carefully characterized individuals should also be informative for factors having significant impact on disease. Even with relatively large cohorts, direct evaluation of disease risk associated with combinations of four or more genotypes may be difficult, and inferences may need to be drawn from analyses of smaller subsets of markers. As increasing numbers of markers are analyzed, the issue of multiple testing must also be addressed to provide appropriate statistical interpretion of the results. This issue arises whether samples are genotyped using a multilocus assay, as described here, or through a series of single-locus studies.
Understanding the molecular basis of genetic predisposition to common multifactorial diseases such as cardiovascular disease will depend on the joint efforts of those performing genome-wide scans to identify candidate loci in regions detected through linkage studies and those studying specific mutations and polymorphisms through association studies. Functional studies will also be critical to identify the genetic variations contributing to disease development. The assay described here was designed to provide multilocus genotype information for CVD, but this format can be applied to other diseases such as asthma, bipolar disorder, and osteoporosis. Given the complexity of these diseases, well-defined cases and phenotypes will be essential components of studies seeking to provide insight into disease development from the complex genetic data. Genotype data can then guide the development of algorithms incorporating genetic contributions to calculate aggregate scores of risk, expanding on the approach developed by the Framingham Heart Study investigators for coronary heart disease (Wilson et al.), for example. Clinically informative subsets of these research markers may then form the basis of panels for diagnostic or prognostic use in patient care.
METHODS
Primers
The sites targeted for PCR amplification are listed in Table 1. Primers were synthesized with 5′ biotinylation using the cyanoethoxyphosphoramidite method (1-μmole scale) on an Applied Biosystems 394 DNA Synthesizer (Perkin-Elmer, Foster City, CA). The use of allele-specific primers at codon 112 combined with probes for codon 158 to genotype apoE alleles has been described (Cheng et al. 1998). Primers for the CBS exon 8, factor V Leiden, and MTHFR targets were published previously (Hu et al. 1993; Goyette et al. 1995; Ridker et al. 1995); the forward primer for CBS exon 8 was later relocated further upstream, to eliminate duplication of the 68-bp insertion sequence (Tsai et al. 1996). The remaining primer sequences were selected with the assistance of two software packages, Oligo (v. 5.0, National Biosciences, Plymouth, MN) and Amplify (v. 1.2, W. Engels, University of Wisconsin, Madison).
Two PCR pools were developed: Multiplex A consisted of 14 biotinylated primer pairs designed to amplify the e2 and e3 alleles of apoE, and targets within the apoB, apoCIII, CETP, LPL, and PON genes. Multiplex B consisted of 13 biotinylated primer pairs designed to amplify the e4 allele of apoE, and targets within the ACE, ATIIR1, AGT, CBS, MTHFR, GPIIIa, fibrinogen, factor V, and ELAM genes. To the extent possible, PCR targets were chosen to be within the 100- to 400-bp size range and to permit resolution of all products by agarose gel electrophoresis. Gel analysis was then used to guide the optimization of PCR conditions. Primer concentrations were adjusted for generally comparable yields of all targets, and ranged from 0.04 to 0.75 μm.
As others have reported (Houlston et al. 1989; Hixson and Vernier 1990), amplification of the apoE region, which is relatively high in GC-bp content, was most efficient in the presence of DMSO. Even in the presence of DMSO, however, the apoCIII promoter target was amplified most effectively if divided into two separate amplicons of 163 and 165 bp.
Unexpectedly weak probe signal intensities necessitated primer redesign for the AGT and GPIIIa targets. Reducing the size of each amplicon resulted in much stronger probe intensities, suggesting the possibility that the longer amplicons were able to form stable secondary structures that inhibited probe binding (data not shown). The AGT target was reduced from 360 to 171 bp; the GPIIIa target was reduced from 312 to 131 bp.
Oligonucleotide Probes
Two probes were designed for each biallelic site, to detect and distinguish between the variant sequences. Most of the markers required discrimination of single base differences. To confirm successful amplification for the two largest PCR targets, probes were also designed for invariant regions of apoE and ACE. Candidate probe sequences were selected initially using published guidelines (Thein and Wallace 1986), with the assistance of the MELT program by J. Wetmur (Mt. Sinai School of Medicine, New York, NY; see also Wetmur 1991) for calculation of dissociation temperatures. Sequences were then modified to meet sensitivity and specificity requirements under the assay temperature and buffer conditions. Concentrations of the final 70 probes were chosen to achieve signal balance between alleles at each variable site, and for generally comparable intensities among all of the loci. Probes were conjugated at their 5′ ends to bovine serum albumin (BSA) by methods similar to Tung et al. (1991), then applied in a linear array to sheets of backed nylon membrane using a Linear Striper and Multispense2000 controller (IVEK, N. Springfield, VT). Each sheet was cut into strips between 0.35 and 0.5 cm in width. The probes on “Probe Strip A” corresponded to the targets amplified by the multiplex A primer pool; “Probe Strips B and B2” corresponded to the targets amplified by the multiplex B primer pool.
Control DNA Templates
Total genomic DNA from three cell lines was used for preliminary experiments: Molt-4 (GM02219C from the Human Genetic Mutant Cell Repository, Coriell Institute, Camden, NJ), KASO11 (no. 9009 from the 10th International Histocompatibility Workshop; Dupont 1987), and CRK (kindly provided by the Clinical Immunogenetics Laboratory, Fred Hutchinson Cancer Center, Seattle, WA). Genomic DNA samples previously characterized at individual sites by other methods were generously provided by G. Assmann and H. Funke (Westfälisches Wilhelms-Universität, Münster, Germany) for ACE, apoB3500, apoE, CETP405, LPL9, LPL291, LPL447, and MTHFR677; P.F. Bray (Johns Hopkins University, Baltimore, MD) for GPIIIa; F. Chehab (University of California, San Francisco, CA) for factor V Leiden; R.M. Krauss and P. Blanche (Lawrence Berkeley Laboratory, Berkeley, CA) for apoB3500, apoCIII3206, and apoE; and B. Shane (University of California, Berkeley, CA) for MTHFR677. Single-stranded templates containing the point mutations in CBS exons 3 and 8 were prepared on an Applied Biosystems 394 DNA Synthesizer, then converted to double-stranded templates by PCR, using the appropriate primer pairs from the multiplex primer pools. For the remaining markers, variant alleles that were identified during development of the assay itself were confirmed by sequencing using Dye Terminator and dRhodamine Terminator Cycle Sequencing Kits with an ABI Prism DNA Sequencer (Perkin-Elmer). All of these samples were used as controls to guide the optimization of probe sequences and concentrations for specificity and sensitivity.
Additional Reagents
MicroAmp tubes for PCR, dNTPs (N = A, G, C, U), and AmpliTaq Gold DNA polymerase were obtained from Perkin-Elmer. Deaza-dGTP was obtained from Boehringer Mannheim Biochemicals (Indianapolis, IN; now Roche Molecular Biochemicals). For higher volume assays, PCRs were performed in 96-well Thermowell Polypropylene Plates with Sealing Mats (Corning Costar, Cambridge, MA). Typing Trays (20-well capacity, amber lid), denaturation solution (1.6% NaOH), SSPE concentrate (20× sodium phosphate solution with NaCl, EDTA), SDS concentrate (20%), streptavidin–horseradish peroxidase conjugate (SA–HRP), substrates A (0.01% H2O2 in citrate solution) and B (0.1% 3,3′,5,5′-tetramethylbenzidine in 40% dimethylformamide) for color development, and citrate concentrate (40×) were obtained from Roche Diagnostic Systems (Branchburg, NJ). For manual assays, color development reagent was prepared by mixing five volumes of substrate A per volume of substrate B; for automated assays, substrate A was reduced to four volumes per volume of substrate B.
PCR Amplifications
Approximately 50 ng of total genomic DNA was used for each assay, 25 ng for each multiplex A and multiplex B reaction. In addition to the primer pools, each 50-μl reaction contained 20 mm Tris-HCl [0.2 m stock (pH 8.3) at 25°C], 50 mm KCl, 8.5% DMSO (vol/vol), 0.1 mm dATP, 0.1 mm dCTP, 0.07 mm dGTP, 0.03 mm 7–deaza-dGTP, 0.2 mm dUTP, 1.7 mm MgCl2 or MgOAc2, and 7 units of AmpliTaq Gold. The final concentration of 8.5% DMSO was chosen to enable reliable amplification of the apoE alleles with minimal adverse impact on the yields of other products such as the β-fibrinogen promoter region, which is relatively high in AT-bp content. Deaza–dGTP was also incorporated to facilitate amplification of regions high in GC content. Deoxy–UTP was included for compatibility with the use of uracil N-glycosylase to eliminate PCR product contamination (Longo et al. 1990). Samples were amplified in a Perkin-Elmer GeneAmp PCR System 9600 using a 2.4-hr thermal cycling profile: an initial hold of 94°C for 12.5 min; then 33 cycles of 96°C for 15 sec, 60°C for 1 min, and 72°C for 1.25 min; and a final extension step of 68°C for 5 min.
During assay development, 3- to 5-μl aliquots were run on horizontal agarose gels using 3% NuSieve, 1% SeaKem GTG agarose (FMC BioProducts, Rockland, ME) in TBE (89 mm Tris-borate, 1 mm EDTA) with ethidium bromide. ΦX174 RF DNA/HaeIII fragments and 123-bp DNA ladder (GIBCO BRL, Gaithersburg, MD) were used as molecular weight standards.
Allele-Specific Detection
The assay was initially developed at 50°C, and the final 52°C assay temperature reflects a compromise made to improve specificity at the apoCIII (−625) site. This marker is the presence (more frequent allele) or absence (less frequent allele) of an A:T bp between a G:C bp doublet and quartet within a generally GC-rich region. Sufficient discrimination between these alleles was achieved only by introducing G:T mismatches into the deletion-specific probe sequence; these relatively stable mismatches (Thein and Wallace 1986) destabilized the region sufficiently to reduce cross-hybridization with wild-type PCR product while maintaining sensitivity for the variant allele. Improved discrimination between apoCIII (−625) alleles was also observed at assay temperatures >52°C, but the signal intensities from probes for other markers were adversely affected by these higher temperatures (data not shown).
Therefore, detection of amplified alleles was performed at 52°C using a water bath rotating at 50–60 rpm (Hot Shaker Plus; Bellco, Vineland, NJ). Probe strips were first washed to remove unbound probe in 2× SSPE (0.36 m NaCl, 0.02 m Na2HPO4, 2 mm EDTA, adjusted to pH 7.4 with NaOH), 0.5% SDS. Twenty-microliter aliquots of the biotinylated PCR product pools from multiplex A and B reactions were denatured with equal volumes of denaturation solution, then added to Typing Tray wells containing 3 ml of hybridization buffer (4× SSPE, 0.5% SDS) and a correspondingly labeled probe strip A or probe strip B. Probe strip B2 was included with strip B. After 20 min at 52°C, the hybridization solution was replaced with fresh buffer containing 10 μl of SA-HRP and the strips were returned to the water bath for 5 min. This enzyme conjugate solution was then replaced with the stringent wash buffer (2× SSPE, 0.5% SDS), and the strips were returned to the water bath for 12 min. The washed strips were equilibrated in 50 mm Na-citrate at room temperature on a rotating (50–60 rpm) platform (Gyrotory Shaker Model G2; New Brunswick Scientific, Edison, NJ), then agitated in color development reagent for 8–10 min at room temperature. Developed strips were rinsed with distilled water, aligned on a flat surface next to a guide identifying the allele detected by each probe line, and photographed using type 559 or 55 film from Polaroid (Cambridge, MA). Genotype interpretations were made manually and independently by two individuals. Given this protocol, at least 40 DNA samples per day can be genotyped by one individual. An SLT ProfiBlot IIT (Tecan US, Research Triangle Park, NC) can also be used to automate the hybridization, stringent wash, and color development steps for 12 samples (24 wells) at a time. This level of automation can be used to increase the throughput of one individual to at least 75 samples per day.
Test Cohorts
To estimate population frequencies, the assay was used to genotype a subset of 1190 samples from 286 families of the Stanislas cohort recruited from families within eastern France (Siest et al. 1998). DNA was prepared from whole blood by the method of salting-out (Miller et al. 1988). These samples had been genotyped for apoE, LPL447, and ACE by methods described previously (Hixson and Vernier 1990; Evans et al. 1994; Salah et al. 1997).
The UCSF cohort was a composite cohort of 142 unrelated caucasian individuals recruited from clinics within the San Francisco Bay area (California, USA). These individuals had been recruited on the basis of a family history of disease, hyperlipidemia, or a treadmill test indication for angiography. Total cholesterol levels ranged from 162 to 548 mg/dl, with an average of 323 ± 69 mg/dl. DNA was prepared from whole blood either by the method of Bell (Bell et al. 1981) or using the Puregene DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, MN). Each DNA sample was associated with a Gensini score, which assigns greater weight to proximal lesions identified through quantitative angiography (Gensini 1975). Fifty of the samples were from men with an average age of 45.5 ± 8.5 years at the time of angiography, and Gensini scores ranging from 5 to 135. Ninety-two samples were from women with an average age of 54.0 ± 11.4 years, and Gensini scores ranging from 0 to 120. Some of these individuals had already been genotyped for apoE and apoB3500 by methods described previously (Hixson and Vernier 1990; Pullinger et al. 1995).
Allele Frequencies and Linkage Disequilibrium Analysis
Population frequencies were estimated from allele counts among 496 unrelated parents from the Stanislas cohort for whom all 35 sites had been genotyped. Allele frequencies were also calculated for the UCSF cohort. Deviation from Hardy–Weinberg equilibrium was assessed using the χ2 statistic.
Intragenic haplotypes for multiple markers within the apoCIII, LPL, and ELAM loci were estimated using the Family Analysis Program (v. PL1; M. Neugebauer and M.P. Baur; Neugebauer et al. 1984) from 275 families (1100 chromosomes) within the Stanislas cohort. This data set included families for whom markers on probe strip B2 had not been genotyped because of the rarity of variation at these four markers; therefore, this data set included samples that were not counted in the estimation of population frequencies. The intragenic haplotypes were used to estimate pairwise linkage disequilibrium values (D′; Lewontin 1964; Klitz et al. 1995) between consecutive sites within each locus.
Evaluation of Disease Association
The UCSF cohort was divided into quintiles based on the Gensini scores. The first combined gender quintile (Q1) contained the 28 lowest scores (0–8), and the fifth quintile (Q5) contained the 28 highest scores (35–135). The male subset was deemed insufficient in size for separate analysis, but the female subset was considered separately: female-only quintile 1 (FQ1) contained the 20 lowest scores (0–7); FQ5 contained the 18 highest scores (36–120).
A preliminary analysis of disease association was undertaken for a subset of 15 markers: apoE, apoB71, all apoCIII sites except 3175, CETP405, PON192, ACE, ATIIR, AGT, CBS278, MTHFR677, GPIIIa, and fibrinogen. For the remaining markers, the observed variant allele frequencies were deemed too rare for such an analysis. In addition, nearly complete linkage disequilibrium was observed between apoCIII sites (−625) and (−455); therefore, these two were subsequently treated as one marker. Disease association was examined by comparing the extreme quintiles, interpreting Q1 and FQ1 as individuals having little or no disease, and Q5 and FQ5 as individuals having the most severe disease. Heterozygous and homozygous carriers of the variant alleles were counted together, with the exception of ACE. The ACE alleles were grouped in two ways, consistent with combining either carriers of the reported risk allele (D/D, I/D) or carriers of the less frequent allele (I/I, I/D). For apoE, e3/e4 and e4/e4 were counted together; no e2/e4 genotypes were observed in any of the quintiles used for this analysis. For single sites (more frequent allele A, less frequent allele a), the odds ratios corresponded to the risk associated with carriers of the less frequent allele (Aa or aa genotypes) relative to the AA genotype. For pairwise combinations of sites, odds ratios were calculated for the risk associated with having variant alleles at two sites compared with just one site. Odds ratios were calculated with Haldane's correction when necessary (Haldane 1955). P values were calculated using Fisher's two-tailed exact test (Sokal and Rohlf 1995). These analyses were intended to be exploratory; therefore, no formal correction for multiple testing was applied.
Acknowledgments
We are indebted to our collaborators for their expert advice and generous gifts of characterized DNA samples: G. Assmann and H. Funke (Westfälische Wilhelms-Universität), S. Humphries and I. Day (University College of London Medical School), R. Krauss and P. Blanche (Lawrence Berkeley National Laboratory), P. Bray (Johns Hopkins Medical School), F. Chehab (University of California, San Francisco), and B. Shane (University of California, Berkeley). This work would not have been possible without the support of the Oligo Synthesis and Sequencing Groups at Roche Molecular Systems, and we thank A. Turck and J. Novotny for their technical advice. We also thank R. Higuchi, J. Sninsky, and T. White for their enthusiastic support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL suzanne.cheng@roche.com; FAX (510) 522-1285.
REFERENCES
- Agellon LB, Quinet EM, Gillette TG, Drayna DT, Brown ML, Tall AR. Organization of the human cholesteryl ester transfer protein gene. Biochemistry. 1990;29:1372–1376. doi: 10.1021/bi00458a004. [DOI] [PubMed] [Google Scholar]
- Bell GI, Karam JH, Rutter WJ. Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci. 1981;78:5759–5763. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertina RM, Koeleman BPC, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, van der Velden PA, Reitsma PH. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- Bonnardeaux A, Davies E, Jeunemaitre X, Féry I, Charru A, Clauser E, Tiret L, Cambien R, Corvol P, Soubrier F. Angiotensin II type I receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24:63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- Bonne G, Carrier L, Richard P, Hainque B, Schwartz K. Familial hypertrophic cardiomyopathy: From mutations to functional defects. Circ Res. 1998;83:580–593. doi: 10.1161/01.res.83.6.580. [DOI] [PubMed] [Google Scholar]
- Cambien F, Poirier O, Lecerf L, Evans A, Cambou J-P, Arveiler D, Luc G, Bard J-M, Bara L, Ricard S, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359:641–644. doi: 10.1038/359641a0. [DOI] [PubMed] [Google Scholar]
- Cattaneo M, Tsai MY, Bucciarelli P, Taioli E, Zighetti ML, Bignell M, Mannucci PM. A common mutation in the methylenetetrahydrofolate reductase gene (C677T) increases the risk for deep-vein thrombosis with mutant factor V (factor V:Q506) Arterioscler Thromb Vasc Biol. 1997;17:1662–1666. doi: 10.1161/01.atv.17.9.1662. [DOI] [PubMed] [Google Scholar]
- Cheng S, Pallaud C, Grow MA, Scharf SJ, Erlich HA, Klitz W, Pullinger CR, Malloy MJ, Kane JP, Siest G, Visvikis S. A multilocus genotyping assay for cardiovascular disease. Clin Chem Lab Med. 1998;36:561–566. doi: 10.1515/CCLM.1998.096. [DOI] [PubMed] [Google Scholar]
- Christensen B, Frosst P, Lussier-Cacan S, Selhub J, Goyette P, Rosenblatt DS, Genest J, Jr, Rozen R. Correlation of a common mutation in the methylenetetrahydrofolate reductase gene with plasma homocysteine in patients with premature coronary artery disease. Arterioscler Thromb Vasc Biol. 1997;17:569–573. doi: 10.1161/01.atv.17.3.569. [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengård J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook NS, Ubben D. Fibrinogen as a major risk factor in cardiovascular disease. Trends Pharmacol Sci. 1990;11:444–451. doi: 10.1016/0165-6147(90)90125-r. [DOI] [PubMed] [Google Scholar]
- Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3′ untranslated region polymorphisms. Proc Natl Acad Sci. 1993;90:4562–4566. doi: 10.1073/pnas.90.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FF. Epidemiological approaches to heart disease: The Framingham study. Am J Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day INM, Whittall RA, O'Dell SD, Haddad L, Bolla MK, Gudnason V, Humphries SE. Spectrum of LDL receptor gene mutations in heterozygous familial hypercholesterolemia. Hum Mutat. 1997;10:116–127. doi: 10.1002/(SICI)1098-1004(1997)10:2<116::AID-HUMU4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- de Knijff P, van den Maagdenberg AMJM, Frants RR, Havekes LM. Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels. Hum Mutat. 1994;4:178–194. doi: 10.1002/humu.1380040303. [DOI] [PubMed] [Google Scholar]
- Dupont B. Immunobiology of HLA, Vol. 1: Histocompatibility testing. New York, NY: Springer-Verlag; 1987. [Google Scholar]
- Ehrenborg E, Clee SM, Pimstone SN, Reymer PWA, Benlian P, Hoogendijk CF, Davis HJ, Bissada N, Miao L, Gagné SE, et al. Ethnic variation and in vivo effects of the −93t → g promoter variant in the lipoprotein lipase gene. Arterioscler Thromb Vasc Biol. 1997;17:2672–2678. doi: 10.1161/01.atv.17.11.2672. [DOI] [PubMed] [Google Scholar]
- Evans AE, Poirier O, Kee F, Lecerf L, McCrum E, Falconer T, Crane J, O'Rourke DF, Cambien F. Polymorphisms of the angiotensin-converting-enzyme gene in subjects who die from coronary heart disease. Q J Med. 1994;87:211–214. [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJH, den Heijer M, Kluijtmans LAJ, van den Heuvel LP, Rozen R. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Funke H, Wiebusch H, Fuer L, Muntoni S, Schulte H, Assmann G. Identification of mutations in the cholesterol ester transfer protein in Europeans with elevated high-density-lipoprotein cholesterol. Circulation. 1994;90:241. [Google Scholar]
- Gensini GG. Coronary arteriography. New York, NY: Futura Publishing Co.; 1975. [Google Scholar]
- Goyette P, Frosst P, Rosenblatt DS, Rozen R. Seven novel mutations in the methylenetetrahydrofolate reductase gene and genotype/phenotype correlations in severe methylenetetrahydrofolate reductase deficiency. Am J Hum Genet. 1995;56:1052–1059. [PMC free article] [PubMed] [Google Scholar]
- Green F, Hamsten A, Blombäck M, Humphries S. The role of β-fibrinogen genotype in determining plasma fibrinogen levels in young survivors of myocardial infarction and healthy controls from Sweden. Thromb Haemost. 1993;70:915–920. [PubMed] [Google Scholar]
- Groenemeijer BE, Hallman MD, Reymer PWA, Gagné E, Kuivenhoven JA, Bruin T, Jansen H, Lie KI, Bruschke AVG, Boerwinkle E, Hayden MR, Kastelein JJP. Genetic variant showing a positive interaction with β-blocking agents with a beneficial influence on lipoprotein lipase activity, HDL cholesterol, and triglyceride levels in coronary artery disease patients. Circulation. 1997;95:2628–2635. doi: 10.1161/01.cir.95.12.2628. [DOI] [PubMed] [Google Scholar]
- Gudnason V, Thormar K, Humphries SE. Interaction of the cholesteryl ester transfer protein I405V polymorphism with alcohol consumption in smoking and non-smoking healthy men, and the effect on plasma HDL cholesterol and apoAI concentration. Clin Genet. 1997;51:15–21. doi: 10.1111/j.1399-0004.1997.tb02407.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The estimation and significance of the logarithm of a ratio of frequencies. Ann Human Genet. 1955;20:309–311. doi: 10.1111/j.1469-1809.1955.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Hall S, Chu G, Miller G, Cruickshank K, Cooper JA, Humphries SE, Talmud PJ. A common mutation in the lipoprotein lipase gene promoter, −93T/G, is associated with lower plasma triglyceride levels and increased promoter activity in vitro. Arterioscler Thromb Vasc Biol. 1997;17:1969–1976. doi: 10.1161/01.atv.17.10.1969. [DOI] [PubMed] [Google Scholar]
- Hata A, Robertson M, Emi M, Lalouel J-M. Direct detection and automated sequencing of individual alleles after electrophoretic strand separation: Identification of a common nonsense mutation in exon 9 of the human lipoprotein lipase gene. Nucleic Acids Res. 1990;18:5407–5411. doi: 10.1093/nar/18.18.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Brunt JH, Connelly PW. A polymorphism of the paraoxonase gene associated with variation in plasma lipoproteins in a genetic isolate. Arterioscler Thromb Vasc Biol. 1995;15:89–95. doi: 10.1161/01.atv.15.1.89. [DOI] [PubMed] [Google Scholar]
- Hixson JE Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group. Apolipoprotein E polymorphisms affect atherosclerosis in young males. Arterioscler Thromb Vasc Biol. 1991;11:1237–1244. doi: 10.1161/01.atv.11.5.1237. [DOI] [PubMed] [Google Scholar]
- Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- Hoeg JM. Evaluating coronary heart disease risk. J Am Med Assoc. 1997;277:1387–1390. [PubMed] [Google Scholar]
- Houlston RS, Snowden C, Green F, Alberti KGMM, Humphries SE. Apolipoprotein (apo) E genotypes by polymerase chain reaction and allele-specific oligonucleotide probes: No detectable linkage disequilibrium between apo E and apo CII. Hum Genet. 1989;83:364–368. doi: 10.1007/BF00291382. [DOI] [PubMed] [Google Scholar]
- Hu FL, Gu Z, Kozich V, Kraus JP, Ramesh V, Shih VE. Molecular basis of cystathionine β-synthase gene deficiency in pyridoxine responsive and nonresponsive homocystinuria. Hum Mol Genet. 1993;2:1857–1860. doi: 10.1093/hmg/2.11.1857. [DOI] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- Humphries SE, Talmud PJ. Hyperlipidaemia associated with genetic variation in the apolipoprotein B gene. Curr Opin Lipidol. 1995;6:215–222. doi: 10.1097/00041433-199508000-00005. [DOI] [PubMed] [Google Scholar]
- Humphries SE, Ye S, Talmud P, Bara L, Wilhelmsen L, Tiret L European Atherosclerosis Research Study (EARS) EARS: Genotype at the fibrinogen locus (G−455-A β-gene) is associated with differences in plasma fibrinogen levels in young men and women from different regions in Europe. Arterioscler Thromb Vasc Biol. 1995;15:96–104. doi: 10.1161/01.atv.15.1.96. [DOI] [PubMed] [Google Scholar]
- Humphries SE, Talmud PJ, Cox C, Sutherland W, Mann J. Genetic factors affecting the consistency and magnitude of changes in plasma cholesterol in response to dietary challenge. Quart J Med. 1996;89:671–680. doi: 10.1093/qjmed/89.9.671. [DOI] [PubMed] [Google Scholar]
- Humphries SE, Panahloo A, Montgomery HE, Green F, Yudkin J. Gene-environment interaction in the determination of levels of haemostatic variables involved in thrombosis and fibrinolysis. Thromb Haemost. 1997;78:457–461. [PubMed] [Google Scholar]
- Jeunemaitre X, Soubrier F, Kotelevtsev YV, Lifton RP, Williams CS, Charru A, Hunt SC, Hopkins PN, Williams RR, Lalouel J-M, Corvol P. Molecular basis of human hypertension: Role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dietz HC, Nurden A, Bray PF. Single-strand conformation polymorphism analysis is a rapid and effective method for the identification of mutations and polymorphisms in the gene for glycoprotein IIIa. Blood. 1993;82:2281–2288. [PubMed] [Google Scholar]
- Jukema JW, van Boven AJ, Groenemeijer B, Zwinderman AH, Reiber JHC, Bruschke AVG, Henneman JA, Molhoek GP, Bruin T, Jansen H, Gagné E, Hayden MR, Kastelein JJP and on behalf of the REGRESS Study Group. The asp9asn mutation in the lipoprotein lipase gene is associated with increased progression of coronary atherosclerosis. Circulation. 1996;94:1913–1918. doi: 10.1161/01.cir.94.8.1913. [DOI] [PubMed] [Google Scholar]
- Kalafatis M, Mann KG. Factor VLeiden and thrombophilia. Arterioscler Thromb Vasc Biol. 1997;17:620–627. doi: 10.1161/01.atv.17.4.620. [DOI] [PubMed] [Google Scholar]
- Klitz W, Stephens JC, Grote M, Carrington M. Discordant patterns of linkage disequilibrium of the peptide-transporter loci within the HLA class II region. Am J Hum Genet. 1995;57:1436–1444. [PMC free article] [PubMed] [Google Scholar]
- Kluijtmans LAJ, van den Heuvel LPWJ, Boers GHJ, Frosst P, Stevens EMB, van Oost BA, den Heijer M, Trijbels FJM, Rozen R, Blom HJ. Molecular genetic analysis in mild hyperhomocysteinemia: A common mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am J Hum Genet. 1996;58:35–41. [PMC free article] [PubMed] [Google Scholar]
- Kozich V, Kraus JP. Screening for mutations by expressing patient cDNA segments in E. coli: Homocystinuria due to cystathionine β-synthase deficiency. Hum Mutat. 1992;1:113–123. doi: 10.1002/humu.1380010206. [DOI] [PubMed] [Google Scholar]
- Kozich V, de Franchis R, Kraus JP. Molecular defect in a patient with pyridoxine-responsive homocystinuria. Hum Mol Genet. 1993;2:815–816. doi: 10.1093/hmg/2.6.815. [DOI] [PubMed] [Google Scholar]
- Kuivenhoven JA, Groenemeyer BE, Boer JMA, Reymer PWA, Berghuis R, Bruin T, Jansen H, Seidell JC, Kastelein JJP. Ser447stop mutation in lipoprotein lipase is associated with elevated HDL cholesterol levels in normolipidemic males. Arterioscler Thromb Vasc Biol. 1997;17:595–599. doi: 10.1161/01.atv.17.3.595. [DOI] [PubMed] [Google Scholar]
- Kuivenhoven JA, Jukema JW, Zwinderman AH, de Knijff P, McPherson R, Bruschke AVG, Lie KI, Kastelein JJP. The role of a common variant of the cholesteryl ester transfer protein gene in the progression of coronary atherosclerosis. N Engl J Med. 1998;338:86–93. doi: 10.1056/NEJM199801083380203. [DOI] [PubMed] [Google Scholar]
- Lassel TS, Guérin M, Auboiron S, Chapman MJ, Guy-Grand B. Preferential cholesteryl ester acceptors among triglyceride-rich lipoproteins during alimentary lipemia in normolipidemic subjects. Arterioscler Thromb Vasc Biol. 1998;18:65–74. doi: 10.1161/01.atv.18.1.65. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations: Heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindpaintner K, Pfeffer MA, Kreutz R, Stampfer MJ, Grodstein F, LaMotte F, Buring J, Hennekens CH. A prospective evaluation of an angiotensin-converting-enzyme gene polymorphism and the risk of ischemic heart disease. N Engl J Med. 1995;332:706–711. doi: 10.1056/NEJM199503163321103. [DOI] [PubMed] [Google Scholar]
- Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- Ludwig E, Corneli PS, Anderson JL, Marshall HW, Lalouel J-M, Ward RH. Angiotensin-converting enzyme gene polymorphism is associated with myocardial infarction but not with development of coronary stenosis. Circulation. 1995;91:2120–2124. doi: 10.1161/01.cir.91.8.2120. [DOI] [PubMed] [Google Scholar]
- Mackness MI, Arrol S, Mackness B, Durrington PN. Alloenzymes of paraoxonase and effectiveness of high-density lipoproteins in protecting low-density lipoprotein against lipid peroxidation. Lancet. 1997;349:851–852. doi: 10.1016/S0140-6736(05)61755-2. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mailly F, Tugrul Y, Reymer PWA, Bruin T, Seed M, Groenemeyer BF, Asplund-Carlson A, Vallance D, Winder AF, Miller GJ, et al. A common variant in the gene for lipoprotein lipase (Asp9 → Asn) Arterioscler Thromb Vasc Biol. 1995;15:468–478. doi: 10.1161/01.atv.15.4.468. [DOI] [PubMed] [Google Scholar]
- Mannucci PM. Recent progress in the pathophysiology of fibrinogen. Eur Heart J (Supp1.A) 1995;16:25–30. doi: 10.1093/eurheartj/16.suppl_a.25. [DOI] [PubMed] [Google Scholar]
- Marble M, Geraghty MT, de Franchis R, Kraus JP, Valle D. Characterization of a cystathionine β-synthase allele with three mutations in cis in a patient with B6 nonresponsive homocystinuria. Hum Mol Genet. 1994;3:1883–1886. doi: 10.1093/hmg/3.10.1883. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L, Pipkin FB, Kalsheker N. Angiotensinogen: Molecular biology, biochemistry and physiology. Int J Biochem Cell Biol. 1996;28:1211–1222. doi: 10.1016/s1357-2725(96)00086-6. [DOI] [PubMed] [Google Scholar]
- Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Neugebauer M, Willems J, Baur MP. Analysis of multilocus pedigree data by computer. In: Albert ED, editor. Histocompatibility testing. Berlin, Germany: Springer-Verlag; 1984. pp. 52–58. [Google Scholar]
- Newman PJ, Derbes RS, Aster RH. The human platelet alloantigens, PlA1 and PlA2, are associated with a leucine33/proline33 amino acid polymorphism in membrane glycoprotein IIIa, and are distinguishable by DNA typing. J Clin Invest. 1989;83:1778–1781. doi: 10.1172/JCI114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson RG, Stengård J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- Nurden AT. Polymorphisms of human platelet membrane glycoproteins: Structure and clinical significance. Thromb Haemost. 1996;74:345–351. [PubMed] [Google Scholar]
- Oka K, Tkalcevic GT, Nakano T, Tucker H, Ishimura-Oka K, Brown WV. Structure and polymorphic map of human lipoprotein lipase gene. Biochim Biophys Acta. 1990;1049:21–26. doi: 10.1016/0167-4781(90)90079-h. [DOI] [PubMed] [Google Scholar]
- Ordovas JM, Civeira F, Genest J, Jr, Craig S, Robbins AH, Meade T, Pocovi M, Frossard PM, Masharani U, Wilson PWF, et al. Restriction fragment length polymorphisms of the apolipoprotein A-I, C-III, A-IV gene locus. Atherosclerosis. 1991;87:75–86. doi: 10.1016/0021-9150(91)90234-t. [DOI] [PubMed] [Google Scholar]
- Pasternak RC, Grundy SM, Levy D, Thompson PD. 27th Bethesda Conference: Matching the intensity of risk factor management with the hazard for coronary disease events. Task Force 3. Spectrum of risk factors for coronary heart disease. J Am Coll Cardiol. 1996;27:978–990. doi: 10.1016/0735-1097(96)87731-x. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Perola M, Niini P, Terwilliger J, Salomaa V, Vartiainen E, Peltonen L, Syvänen A-C. Array-based multiplex analysis of candidate genes reveals two independent and additive genetic risk factors for myocardial infarction in the Finnish population. Hum Mol Genet. 1998;7:1453–1462. doi: 10.1093/hmg/7.9.1453. [DOI] [PubMed] [Google Scholar]
- Pullinger CR, Hennessy LK, Chatterton JE, Liu W, Love JA, Mendel CM, Frost PH, Malloy MJ, Schumaker VN, Kane JP. Familial ligand-defective apolipoprotein B. J Clin Invest. 1995;95:1225–1234. doi: 10.1172/JCI117772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees A, Stocks J, Sharpe CR, Vella MA, Shoulders CC, Katz J, Jowett NI, Baralle FE, Galton DJ. Deoxyribonucleic acid polymorphism in the apolipoprotein A-I-C-III gene cluster. J Clin Invest. 1985;76:1090–1095. doi: 10.1172/JCI112062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymer PWA, Groenemeyer BE, Gagné E, Miao L, Appelman EEG, Seidell JC, Kromhout D, Bijvoet SM, van de Oever K, Bruin T, Hayden MR, Kastelein JJP. A frequently occurring mutation in the lipoprotein lipase gene (Asn291Ser) contributes to the expression of familial combined hyperlipidemia. Hum Mol Genet. 1995;4:1543–1549. doi: 10.1093/hmg/4.9.1543. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Hennekens CH, Lindpaintner K, Stampfer MJ, Eisenberg PR, Miletich JP. Mutation in the gene coding for coagulation factor V and the risk of myocardial infarction, stroke, and venous thrombosis in apparently healthy men. N Engl J Med. 1995;332:912–917. doi: 10.1056/NEJM199504063321403. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Hennekens CH, Schmitz C, Stampfer MJ, Lindpaintner K. PlA1/A2 polymorphism of platelet glycoprotein IIIa and risks of myocardial infarction, stroke, and venous thrombosis. Lancet. 1997;349:385–388. doi: 10.1016/S0140-6736(97)80010-4. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Corvol P, Soubrier F. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1) Nucleic Acids Res. 1992;20:1433. doi: 10.1093/nar/20.6.1433-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Walsh PS, Levenson CH, Erlich HA. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah D, Bohnet K, Gueguen R, Siest G, Visvikis S. Combined effects of lipoprotein lipase and apolipoprotein levels in the Stanislas Cohort. J Lipid Res. 1997;38:132–141. [PubMed] [Google Scholar]
- Sanghera DK, Saha N, Aston CE, Kamboh MI. Genetic polymorphism of paraoxonase and the risk of coronary heart disease. Arterioscler Thromb Vasc Biol. 1997;17:1067–1073. doi: 10.1161/01.atv.17.6.1067. [DOI] [PubMed] [Google Scholar]
- Sebastio G, Sperandeo MP, Panico M, de Franchis R, Kraus JP, Andria G. The molecular basis of homocystinuria due to cystathionine β-synthase deficiency in Italian families, and report of four novel mutations. Am J Hum Genet. 1995;56:1324–1333. [PMC free article] [PubMed] [Google Scholar]
- Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest. 1995;96:3005–3008. doi: 10.1172/JCI118373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siest G, Visvikis S, Herbeth B, Gueguen R, Vincent-Viry M, Sass C, Beaud B, Lecomte E, Steinmetz J, Locuty J, Chevrier P. Objectives, design and recruitment of a familial and longitudinal cohort for studying gene-environment interactions in the field of cardiovascular risk: The Stanislas Cohort. Clin Chem Lab Med. 1998;36:35–42. doi: 10.1515/CCLM.1998.007. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 3rd ed. San Francisco, CA: W.H. Freeman; 1995. [Google Scholar]
- Soria LF, Ludwig EH, Clarke HRG, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo MP, de Franchis R, Andria G, Sebastio G. A 68-bp insertion found in a homocystinuric patient is a common variant and is skipped by alternative splicing of the cystathionine β-synthase mRNA. Am J Hum Genet. 1996;59:1391–1393. [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Jiang X-C, Sakai N, Yamashita S, Hirano K, Bujo H, Yamazaki H, Kusunoki J, Miura T, Kussie P, et al. A missense mutation in the cholesteryl ester transfer protein gene with possible dominant effects on plasma high density lipoproteins. J Clin Invest. 1993;92:2060–2064. doi: 10.1172/JCI116802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall A. Plasma lipid transfer proteins. Annu Rev Biochem. 1995;64:235–257. doi: 10.1146/annurev.bi.64.070195.001315. [DOI] [PubMed] [Google Scholar]
- The MRFIT Research Group. Multiple Risk Factor Intervention Trial. Risk factor changes and mortality results. J Am Med Assoc. 1982;248:1465–1477. [PubMed] [Google Scholar]
- The Women's Health Initiative Study Group. Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- Thein SL, Wallace RB. In: The use of synthetic oligonucleotides as specific hybridization probes in the diagnosis of genetic disorders. Davies KE, editor. Oxford, UK: IRL Press; 1986. pp. 33–50. [Google Scholar]
- Tikkanen MJ, Viikari J, Åkerblom HK, Pesonen E. Apolipoprotein B polymorphism and altered apolipoprotein B and low density lipoprotein cholesterol concentrations in Finnish children. Br Med J. 1988;296:169–170. doi: 10.1136/bmj.296.6616.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiret L, Bonnardeaux A, Poirier O, Ricard S, Marques-Vidal P, Evans A, Arveiler D, Luc G, Kee F, Ducimetie↓re P, Soubrier F, Cambien F. Synergistic effects of angiotensin-converting enzyme and angiotensin-II type I receptor gene polymorphisms on risk of myocardial infarction. Lancet. 1994;344:910–913. doi: 10.1016/s0140-6736(94)92268-3. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Bignell M, Schwichtenberg K, Hanson NQ. High prevalence of a mutation in the cystathionine β-synthase gene. Am J Hum Genet. 1996;59:1262–1267. [PMC free article] [PubMed] [Google Scholar]
- Tung CH, Rudolph MJ, Stein S. Preparation of oligonucleotide–peptide conjugates. Bioconjugate Chem. 1991;2:464–465. doi: 10.1021/bc00012a015. [DOI] [PubMed] [Google Scholar]
- University College of London, accessed 1999. LDL-receptor mutation catalogue. http://www.ucl.ac.uk/fh.
- Weiss EJ, Bray PF, Tayback M, Schulman SP, Kickler TS, Becker LC, Weiss JL, Gerstenblith G, Goldschmidt-Clermont PJ. A polymorphism of a platelet glycoprotein receptor as an inherited risk factor for coronary thrombosis. N Engl J Med. 1996;334:1090–1094. doi: 10.1056/NEJM199604253341703. [DOI] [PubMed] [Google Scholar]
- Wenzel K, Ernst M, Rohde K, Baumann G, Speer A. DNA polymorphisms in adhesion molecule genes—A new risk factor for early atherosclerosis. Hum Genet. 1996;97:15–20. doi: 10.1007/BF00218826. [DOI] [PubMed] [Google Scholar]
- Wenzel K, Felix S, Kleber FX, Brachold R, Menke T, Schattke S, Schulte KL, Gläser C, Rohde K, Baumann G, Speer A. E-selectin polymorphism and atherosclerosis: An association study. Hum Mol Genet. 1994;3:1935–1937. doi: 10.1093/hmg/3.11.1935. [DOI] [PubMed] [Google Scholar]
- Wetmur JG. DNA probes: Applications of the principles of nucleic acid hybridization. Crit Rev Biochem Mol Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- Wilson PWF, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- Xu C-F, Talmud P, Schuster H, Houlston R, Miller G, Humphries S. Association between genetic variation at the Apo AI-CIII-AIV gene cluster and familial combined hyperlipidaemia. Clin Genet. 1994;46:385–397. doi: 10.1111/j.1399-0004.1994.tb04404.x. [DOI] [PubMed] [Google Scholar]
- Yang W-S, Nevin DN, Peng R, Brunzell JD, Deeb SS. A mutation in the promoter of the lipoprotein lipase (LPL) gene in a patient with familial combined hyperlipidemia and low LPL activity. Proc Natl Acad Sci. 1995a;92:4462–4466. doi: 10.1073/pnas.92.10.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A mutation in the promoter of the lipoprotein lipase (LPL) gene in a patient with familial combined hyperlipidemia and low LPL activity. Proc Natl Acad Sci. 1995b;93:524. doi: 10.1073/pnas.92.10.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Bertics SJ, Scott TM, Dubois BW, Beltz WF, Curtiss LK, Witztum JL. Apolipoprotein B allotypes MB191 and MB192 in subjects with coronary artery disease and hypercholesterolemia. Arteriosclerosis. 1987;7:61–65. doi: 10.1161/01.atv.7.1.61. [DOI] [PubMed] [Google Scholar]
- Young SG, Hubl ST. An ApaLI restriction site polymorphism is associated with the MB19 polymorphism in apolipoprotein B. J Lipid Res. 1989;30:443–449. [PubMed] [Google Scholar]
- Zhang H, Reymer PWA, Liu M-S, Forsythe IJ, Groenemeyer BE, Frohlich J, Brunzell JD, Kastelein JJP, Hayden MR, Ma Y. Patients with apoE3 deficiency (E2/2, E3/2, and E4/2) who manifest with hyperlipidemia have increased frequency of an asn 291 → ser mutation in the human LPL gene. Arterioscler Thromb Vasc Biol. 1995;15:1695–1703. doi: 10.1161/01.atv.15.10.1695. [DOI] [PubMed] [Google Scholar]
- Zhong S, Sharp DS, Grove JS, Bruce C, Yano K, Curb JD, Tall AR. Increased coronary heart disease in Japanese–American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levels. J Clin Invest. 1996;97:2917–2923. doi: 10.1172/JCI118751. [DOI] [PMC free article] [PubMed] [Google Scholar]