Abstract
The short arm of human chromosome 21 (21p) contains many different types of repetitive sequences and is highly homologous to the short arms of other acrocentric chromosomes. Owing to its repetitive nature and the lack of chromosome 21p-specific molecular markers, most physical maps of chromosome 21 exclude this region. We constructed a physical map of chromosome 21p using sequence tagged site (STS) content mapping of yeast artificial chromosomes (YACs). To this end, 39 STSs located on the short arm or near the centromere of chromosome 21 were constructed, including four polymorphic simple tandem repeats (STRs) and two expressed sequence tags (ESTs). Thirty YACs were selected from the St. Louis YAC library, the chromosome 21-enriched ICRF YAC library, and the CEPH YAC and megaYAC libraries. These were assembled in a YAC contig map ranging from the centromere to the rDNA gene cluster at 21p12. The total size of the region covered by YACs is estimated between 2.9 and 5 Mb. The integrity of the YAC contig was confirmed by restriction enzyme fingerprinting and fluorescence in situ hybridization (FISH). One gap with an estimated size of 400 kb remained near the telomeric end of the contig. This YAC contig map of the short arm of human chromosome 21 constitutes a basic framework for further structural and functional studies of chromosome 21p.
Human chromosome 21 is the smallest of the five acrocentric chromosomes, the short arm of which (21p) is mainly composed of tandemly arranged or interspersed arrays of different types of repetitive satellite sequences (Choo et al. 1988; Greig and Willard 1992; Vissel and Choo 1992). They are located mainly, but not exclusively, on the short arms of the acrocentric chromosomes. Therefore, chromosome 21p shares extensive homology with the short arms of the other acrocentric chromosomes. The highest homology is shared with chromosome 13p, because, mapping of markers using somatic cell hybrids and macrorestriction mapping has not identified differences between chromosomes 21p and 13p (Jorgensen et al. 1987; Van Camp et al. 1992). The highly repetitive nature of chromosome 21p and its extensive homology with other acrocentric chromosomes have hampered the search for chromosome 21p-specific markers and the construction of genetic and physical maps of this region. Most maps of chromosome 21 span the entire long arm (21q) (Chumakov et al. 1992; Nizetic et al. 1994; Gardiner et al. 1995; Korenberg et al. 1995), whereas only a few of them extend a short distance into the p-arm (Nizetic et al. 1994; Doering et al. 1995).
More than 90 genes are known on chromosome 21q (Antonarakis 1998), whereas only the ribosomal gene cluster (rDNA) was localized to chromosome 21p (Henderson et al. 1972). It is believed that the short arm of chromosome 21 and that of the other acrocentric chromosomes is composed almost exclusively of satellite sequences and that they are devoid of genes. However, the absence of genes might also reflect lack of detailed investigation of these genomic regions.
Trisomy of chromosome 21 results in Down syndrome (DS), the most common mental retardation–dysmorphology disorder in humans. Ninety-five percent of the DS cases have three copies of chromosome 21 due to a nondisjunction event that occurred in meiosis I or meiosis II (Epstein 1989). The remaining 5% of DS patients have a trisomy of chromosome 21 as a result of a Robertsonian translocation, mostly involving one of the other acrocentric chromosomes (Antonarakis 1993). The parental origin of the extra copy can be determined accurately using highly informative chromosome 21-specific polymorphic markers (Lamb et al. 1997). However, because no chromosome 21p or centromere-specific polymorphic markers are available, the study of the meiotic nondisjunction is based on data obtained from chromosome 21q markers only (Antonarakis et al. 1992). The availability of chromosome 21p-specific polymorphic markers would highly increase the informativness of such studies.
We describe the development of PCR-based markers located on chromosome 21p including nonpolymorphic STSs, simple tandem repeat (STR) markers and expressed sequence tags (ESTs). Furthermore, we used these markers to construct a yeast artificial chromosome (YAC) contig map of chromosome 21p ranging from the centromere to the rDNA gene cluster at 21p12. The integrity of the YAC contig map was validated by fluorescence in situ hybridization (FISH) analyses and restriction enzyme fingerprinting. The size of the contig was estimated to span a region of 2.9–5 Mb. One gap with an estimated size of 400 kb remained near the telomeric end of the contig. This YAC contig map of chromosome 21p is a valuable resource for the identification of chromosome 21p-specific polymorphic markers and expressed sequences, a research area that has not yet been explored.
RESULTS
Development and Characterization of STSs
The inserts of 12 plasmid clones mapped previously to the pericentromere or the short arm of chromosome 21 were sequenced from both sides using vector-based sequencing primers (Table 1). Homology to known sequences in the public databases was analyzed using the BLASTN algorithm (Altschul et al. 1990). As expected, pCHB and pA2 were 98%–100% homologous to the spacer and 28S coding sequence of the rDNA gene cluster, respectively, whereas Xba21 was 98% homologous to an α-satellite sequence. D21S190 and D21S191 were highly homologous to chromosome 21 genomic sequences, whereas the other sequence homologies were unrelated to chromosome 21 (Table 1). The sequences of three clones did not identify homologous sequences in the public databases.
Table 1.
Sequence Analysis of chromosome 21p Clones
Six insert sequences were incomplete because the 5′ and 3′ sequences did not overlap. Homology searches were performed using the BLAST algorithm (Altschul et al. 1990). (Acc. No.) GenBank accession no.
The obtained sequences were used to develop PCR primer pairs. Also, primer pairs were developed from two repetitive α-satellite sequences, D21Z4 and D21Z3 (Vissel et al. 1992), one expressed sequence, hmc01a06 (Chen et al. 1996, 1997), and four (CA)n-repeat-containing clones ABM–C61, ABM–C62D, ABM–C78 and D21S411 (Bosch et al. 1996; A. Bosch and X. Estivill, unpubl.) (Table 2). First the markers were analyzed on a monochromosomal somatic-cell hybrid mapping panel, indicating that all markers were located on chromosome 21 as well as on chromosome 13. In addition, most of them also amplified sequences on other acrocentric chromosomes and one or more nonacrocentric chromosomes (Table 2). PCR amplification of an extended-mapping panel of partial chromosome 21 somatic cell hybrids localized the markers in four clusters separated by the 21p breakpoints of the cell hybrids (Table 3A). In addition to the novel markers, D21S286, D21S381, D21Z1, D21S275 (Chumakov et al. 1992), and D21S237, (Watkins et al. 1985) were regionally localized to the pericentromeric region of chromosome 21. Xba21 did not amplify the somatic cell hybrid DNA, and pA2 was positive on mouse and hamster DNA; therefore, it could not be mapped unambiguously.
Table 2.
PCR Primer Sequences and Chromosomal Assignments of Chromosome 21p Markers
| Locus | Clone | Type | Primer sequences | Size (bp) | Chromosomal assignment |
|---|---|---|---|---|---|
| D21S190 | pVC1.21c | STS | CTGACAAGGAATTAGTAACC | 133 | 13, 18, 21 |
| TACACCTATTTGCCATTTGC | |||||
| — | ABM-C61 | STR | CCTAGCTTTCTGTCAGCAGT | 450 | 13, 14, 15, 21, 22 |
| CAACATGATTCCACATAATGTA | |||||
| — | hmc01a06 | EST | TGGATGTCACTCTCATCCTTG | 122 | 13, 22, 21, Y |
| TCGAAGAAGAACATCCATGAG | |||||
| — | ABM-C62D | STR | CCACTGTTGTACAAATCAACTA | 143 | 13, 15, 21, Y |
| TCTAAGAATACCTGTAGGCATA | |||||
| — | Xba21 | STS | CAATCCCGTTTCCCACG | 133 | N.D. |
| CTTGTTTGTGATGTGTGCCC | |||||
| D21Z4 | pTRA-4 | STS | ATTTGGAGGCCTTTGTGGCTATGG | 287 | 13, 14, 21 |
| GGCCTCAAAACGCTCCAAGTATCC | |||||
| D21Z3 | pTRA-1 | STS | TGAGCCATTTGAGGCCTACTGTG | 138 | 13, 14, 21 |
| TGTCTCCAAACTGCTCAATCAGAA | |||||
| — | ABM-C78 | STR | CCTCTTCTATGCCAGTCTTAA | 109 | 9, 13, 14, 15, 20, 21, 22 |
| CAGCTCTAAAGTAAGCTTGGA | |||||
| D21S5 | pPW235D | STS | CCAAAGGTTACTGCAGTCTC | 123 | 13, 14, 21 |
| GGGGGTCATATAAAGGAAAC | |||||
| D21S187 | pVC10a | STS | TACATGCAAATGACCAAGAG | 400 | 4, 9, 13, 14, 15, 21, 22 |
| ATCAGCAATGCCTTCTAAGT | |||||
| D21S1276 | pLSB77 | STS | GGCCTGATGTCTGCCTTAGAT | 108 | 1, 3, 9, 13, 14, 15, 20, 21, 22, Y |
| GCCATAGGTGAGCAACAGGA | |||||
| D21S188 | pVC1.12c | STR | TTCTGTGTCATCTGAACTGG | 184 | 9, 13, 14, 15, 20, 21, 22, Y |
| ACGCACATTGAATACTGAGG | |||||
| D21S191 | pVC1.23c | STS | TGATTGGTATGACTTTGTTCCCAG | 179 | 3, 4, 13, 14, 15, 21, 22 |
| AGATGTCCACCCAGTGCTGC | |||||
| D21S411 | ABM-C74 | STR | GAACCATTATAAGTTGACCATC | 196–206 | 3, 4, 13, 14, 15, 21, 22 |
| ACATGTTAACATGCTATATCTGT | |||||
| D21S1277 | pAW14a | STS | AACTAAGGGCTCTAAAGCAT | 300 | 3, 4, 9, 13, 14, 15, 21, 22, Y |
| AATGGGAGCACAAGTTCTAA | |||||
| — | 3C6 | STS | CAAGAGGGCACTGAACAG | 59 | 1, 4, 13, 14, 15, 21, 22, Y |
| GCACTCTTAGCTTTGCCA | |||||
| D21S1278 | pAW32a | STS | TCATACAGAGTATAACACCAGGAC | 266 | 1, 10, 13, 14, 15, 20, 21, 22, Y |
| GTCTTATTGTGATAGGCTTGC | |||||
| rDNA | pCHB | STS | CGAGGTGACTCTCGGTTTGC | 370 | 13, 14, 15, 21, 22 |
| TTTCGCAAGCAGGCATTTG | |||||
| rDNA | pA2 | EST | TATGAACGCTTGGCCGC | 204 | N.D. |
| CGGTAACGCAGTGTCTAAGGC |
(N.D.) Not determined.
Table 3.
Somatic Cell Hybrid Mapping on Chromosome 21p
| Locus | Clone | WAV17 | AHVI-17 | JC-6A | ACEM2-10d | 153-E7b | R2-10W | 2FUr1 | Region |
|---|---|---|---|---|---|---|---|---|---|
| A D21S190 | PVC1.21c | + | + | + | + | + | + | + | 21q11.1 |
| D21S286 | G51E07 | + | + | + | + | + | + | + | 21q11.1 |
| D21S381 | E341 | + | + | + | + | + | + | − | 21cen |
| D21Z1 | L1.26 | + | + | + | + | + | + | − | 21cen |
| — | ABM-C61 | + | + | + | + | + | + | − | 21cen |
| D21S275 | G51G19 | + | + | + | + | + | + | − | 21cen |
| D21S237 | PPW265D | + | + | + | + | + | + | − | 21cen |
| — | Hmc01a06 | + | + | + | − | + | + | − | 21cen |
| — | ABM-C62D | + | + | + | − | + | + | − | 21cen |
| D21Z4 | PTRA-4 | + | + | + | + | + | + | − | 21cen |
| D21Z3 | PTRA-1 | + | + | + | + | + | + | − | 21cen |
| — | ABM-C78 | + | + | + | + | + | − | − | 21cen |
| D21S5 | PPW235D | + | + | + | + | + | − | − | 21p11.1 |
| D21S187 | PVC10a | + | + | + | + | + | − | − | 21p11.1 |
| D21S1276 | PLSB77 | + | + | + | + | + | − | − | 21p11.1 |
| D21S188 | PVC1.12c | + | + | + | + | − | − | − | 21p11.1 |
| D21S191 | PVC1.23c | + | + | + | + | − | − | − | 21p11.2–p12 |
| D21S411 | ABM-C74 | + | + | + | + | − | − | − | 21p11.2–p12 |
| D21S1277 | PAW14a | + | + | + | + | − | − | − | 21p11.2–p12 |
| — | 3C6 | + | + | + | + | − | − | − | 21p11.2–p12 |
| D21S1278 | PAW32a | + | + | + | + | − | − | − | 21p11.2–p12 |
| — | pCHB | + | + | + | + | − | − | − | 21p11.2–p12 |
| B | 1F8R | + | + | + | + | + | + | − | 21cen |
| 5A12R | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 891B10L | + | + | + | − | + | + | − | 21cen | |
| 831B6R | + | + | + | − | + | + | − | 21cen | |
| 2C9L | + | + | + | − | + | + | − | 21cen | |
| 829D2R | + | + | + | + | + | − | − | 21p11.1 | |
| 837F5L | + | + | + | + | + | − | − | 21p11.1 | |
| 2C9R | + | + | + | + | + | − | − | 21p11.1 | |
| 891B10R | + | + | + | + | + | − | − | 21p11.1 | |
| 4E9R | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 2E4L | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 4E9L | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 1B8L | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 2C2R | + | + | + | + | − | − | − | 21p11.2–p12 | |
| 5B7L | + | + | + | + | + | + | − | 21cen | |
| 2E4R | + | + | + | + | − | − | − | 21p11.2–p12 |
(A) STSs, STRs, and ESTs derived from chromosome 21p or centromeric clones. (B) STSs derived from 21p YAC terminal fragments. Names ending with L and R indicate, respectively, left and right YAC end STSs. (+ or −) Positive or negative PCR amplification.
Selection and Characterization of YACs
The St. Louis and CEPH human YAC libraries were screened by PCR or hybridization with Xba21, D21S187, D21S1276, D21S5, D21S188, D21S191, D21S1277, D21S1278, pCHB, and pA2, resulting in the identification of 14 YACs. Furthermore, we obtained 22 YACs from the chromosome 21-enriched ICRF YAC library and six CEPH YACs mapped previously to the centromere or short arm of chromosome 21 by PCR, hybridization, or FISH (Chumakov et al. 1992; Nizetic et al. 1994). PCR amplification of the selected YACs identified 42 YACs that were positive for the markers with which they were selected. The selected YACs were reexamined for their marker specificity by PCR amplification, and true positive YACs were used in the YAC content mapping. Sizes of the YACs were estimated by PFGE and ranged from 100 to 1600 kb (Table 4).
Table 4.
STS Content Mapping of Chromosome 21p YACs
Column heads 5–42 show the markers used in the STS content mapping. (Colume 1) Markers used in the YAC library screening. (Colume 3) (I) ICRF YAC library; (C) CEPH YAC library; (S) St. Louis YAC library. (Shaded areas) positive PCR amplifications; (−) negative PCR amplification. (n.d.) Not determined.
YAC Contig Construction
STS content mapping of the 42 selected YACs was performed with the 24 chromosome 21p markers (Table 4). All markers were positive on one or more YACs, except ABM–C62D, which was negative for all YACs. Again, Xba21 did not amplify YAC DNA and was excluded from further STS content mapping. Together, 26 YACs were identified that contained multiple markers and were assembled into a YAC contig map based on their marker content (Shimizu et al. 1995; Wang et al. 1995, 1997; Korenberg et al. 1997). The STS content of each YAC was verified using DNA of five different single YAC colonies. For four YACs—4C7, 781G5, 1B8, and 5B7—the STS content of the clones was not identical, possibly because of internal deletions of the YACs. For these YACs, we considered STSs that amplified four of the five clones. The YAC contig showed two gaps, one between hmc01a06/D21S237 and D21Z3 and one between D21S1278 and pCHB. Next, we isolated 34 end fragments of 15 YACs from the preliminary YAC contig map and 4 YACs containing one single STS. Their DNA sequence was determined and the homology to sequences in public databases was analyzed. The left ends of 831B6, 1F8, and 3A8 were 94%–98% homologous to the repetitive α-satellite sequence α21-I, whereas the left ends of 5A12 and 2C2 were 78%–84% homologous to a satellite III sequence. The left and right ends of A24F7 were identical to the 18S coding sequence and the intergenic spacer sequence of the rDNA gene cluster, respectively, confirming that this YAC is located within the rDNA gene cluster. Furthermore, a 74-bp region of the right end of 1F8 was 80% homologous to a cDNA sequence of the human protein kinase C-like gene (PRKC-L), related to the protein kinase C gene family (Bacher et al. 1991). No other end sequences showed any homology to known sequences.
Twenty-seven STSs were developed from the nonrepetitive YAC end sequences, and their chromosomal location was determined by PCR analysis of the monochromosomal somatic cell hybrid mapping panel (Table 5). Fourteen STSs were positive on chromosome 21, as well as on other acrocentric and nonacrocentric chromosomes, and were regionally localized to chromosome 21p using the extended partial chromosome 21 somatic cell hybrid panel (Table 3B). Their location was consistent with the position of the corresponding YACs in the contig map, except for 5B7L and 5A12R. YAC 5B7 is located in 21p11.2–21p12, whereas its left end mapped to the centromeric region. Conversely, 5A12R localized to 21p11.2–12, whereas the YAC mapped in the 21cen region. 3A8R did not PCR amplify the somatic cell hybrids and could not be localized. All chromosome 21p YAC-end STSs were included in the YAC content mapping. However, this did not result in the closure of the two gaps in the YAC contig. Therefore, STSs 2E4R, ABM–C78, 831B6R, and ABM–C62D flanking the gaps were used to screen the CEPH YAC and megaYAC libraries. Six additional YACs were selected, four positive with ABM–C78, one with 831B6R, and one with 2E4R (Table 4). STS content mapping of these YACs resulted in closing the gap between hmc01a06/D21S237 and D21Z3 by YAC 891B10, as it was positive for markers at both sides of the gap. Next, both ends of 891B10, the right end of 829D2, and the left end of 89D10 were isolated and sequenced. The left end of 89D10 was 88% homologous to the repetitive satellite sequence α21-II. The other three sequences did not identify homologous sequences in the public databases and were used to develop STSs. All three STSs were mapped to chromosome 21 and other chromosomes using the monochromosomal cell hybrid mapping panel (Table 5). The regional location was determined by PCR amplification of the extended partial chromosome 21 somatic cell hybrid panel and was consistent with the location of the YACs in the contig (Table 3B). Inclusion of the additional STSs confirmed the closure of the gap between hmc01a06/D21S237 and D21Z3. Unexpectedly, 891B10R was present only in 891B10, but not in overlapping YACs.
Table 5.
YAC End STSs and Their Chromosomal Locations
| Name | Primer sequences | Size (bp) | Chromosomal assignment |
|---|---|---|---|
| A 89B10L | TGAAACTGTTTTTGGCAGGTAGG | 306 | 4, 7, 13, 21, 22, X, Y |
| GTAGGATGGGCAGAAGTGGTCAG | |||
| 891B10R | TTATTTTGCCGNGCTTCAAGACCT | 96 | 4, 13, 21 |
| GAGAGCCAGTGTCCTTATACAAAG | |||
| 2C9L | ACGCCTGCTGTGTTCTCTTGGACT | 168 | 13, 21 |
| TGATGGATCGGTGAATTTGCTCTG | |||
| 2C9R | AATGAGGTCAAATGAACAATACAA | 150 | 4, 9, 13, 14, 21 |
| TCAGCAGTCAGAAGAAAAGGAA | |||
| 4E9L | TGATGAATGAGGTGACTGTG | 102 | 3, 4, 13, 15, 21, 22 |
| AACTTTACGTTTCCCTTTCC | |||
| 4E9R | AATCTCCATGGTGTTTGAAC | 183 | 3, 4, 13, 14, 15, 21, 22, Y |
| ATCCAGAATTTGTAGGGCTT | |||
| 2E4L | TTTGTGTTTTAATGCCTTCT | 156 | 13, 14, 15, 21, 22 |
| TGCTACAAACTTGAGACTTT | |||
| 2E4R | TGACACTTAAATGAGCTGCT | 100 | 13, 14, 21 |
| TTCAAATTTCTGAAATCCAA | |||
| B 1F8R | TCTTGGCCCTAGTCTGGTCCACTC | 181 | 13, 14, 15, 21, 22, Y |
| AGAGCCAGGGTGGGTCCTCTGT | |||
| 5A12R | CGTAGCAAGTATGTTTTCAAAGAA | 51 | 4, 13, 14, 15, 21, 22 |
| AGCCACCAAGACCCACCTTA | |||
| 3A8R | ACTTAATAATTTGCCCACCAT | 58 | 4, 7, 13, 14, 15, 21, 22, Y |
| AACCAAAGAAATGTAGATTTACC | |||
| 831B6R | CTTTGTGAGCGCGGCAGACT | 145 | 13, 21, Y |
| TAGAGAACGTGGACGGATTATTTT | |||
| 2C2R | ACGGTTTACGTTTCCCTTTCCTAA | 111 | 13, 14, 15, 21, 22 |
| TGGTGCATTATTGATGAATGAGGT | |||
| C 829D2R | AAAGCACGTTGAAAGGTCATAC | 243 | 9, 13, 15, 20, 21 |
| ACAGCCTTTACAGTGTTGAAGTA | |||
| 5B7L | CATGCGTGCCCAGCTACACTTGA | 60 | 4, 13, 14, 15, 21, 22, Y |
| ACATGGGCACGCATCTTT(C/T)TGC | |||
| D 4C7L | TGTCTGGGAGTGCTAGAAGT | 94 | 20 |
| ATGCCTCCAGGTTTCTCTAT | |||
| 4C7R | CCTCTATAGGACAGCAGGAA | 70 | 4 |
| CAGACAGCATTCTAGGTGCT | |||
| G32G11L | ATGTTTTGAGGGGTGGATTGTTTA | 247 | 5, 11, 22 |
| TCTGTTGCCTGGTGACTGGAC | |||
| G32G11R | CAGTTGAAATGGTTGTATGA | 100 | 20 |
| ATCCAGTTGAGTTTATCCA | |||
| B35E11L | CCAGGATATGAAATGTCAGCT | 110 | 5, 18 |
| ATCTCGATTTTTATAGCTCAACTC | |||
| 1B2L | AGGGACTGAAATAAAATGAAAC | 88 | 14 |
| CAATGTGCAAAAATAAACTGA | |||
| 1B2R | ACTCAATTACCTTTTTCCTCCATC | 112 | 9 |
| TTCAGAAAATTTGATTTGAGGAGA | |||
| 837F5L | CTTCAATGCCCAGATACAGATGAA | 93 | 9, 13, 14, 15, 21, 22, Y |
| CCCCGCCGACTTTATTTAGCT | |||
| 837F5R | TTGAACAAGGAAGAAATCCAAAAC | 100 | X |
| CAAGTCCTGGGCTTCCCTTTAC | |||
| 965D4L | TAAATTTGAAGTTGTTGCCTAATG | 163 | 14 |
| TCCTTAATGGATTTCTGATGGA | |||
| 965D4R | CTGCTTTCAGGGATGGGCTTAA | 100 | 4, 22, Y |
| AAAAGATGATTCCCAGTCCCACAT | |||
| F9B6R | GTGGGCCTTAATGCAATGATTTC | 129 | 1 |
| TTGGCATCCCCTGGCTTGTAT | |||
| 3G8L | CATCATAAATAAACAAAAACATTAGA | 128 | 11 |
| CATGCAAGCTCCATTTAGAG | |||
| 1B8L | AAGGCTTCTTGGAAAGGAACT | 93 | 1, 3, 4, 13, 14, 21, 22 |
| TTGTGATTGGTATGGGAGAATC | |||
| 1B8R | AGAAATAAAACTNGCANGGG | 100 | 3 |
| GTATGANTTCACCCACATCT |
YAC addresses followed by L and R indicate left and right YAC end STSs, respectively.
(A) STSs of YACs with both ends mapping to chromosome 21. (B) YACs with one end mapping to chromosome 21 and one end homologous to satellite repeat sequences. (C) YACs with one end isolated and mapped to chromosome 21. (D) YACs with one or two ends not mapping to chromosome 21.
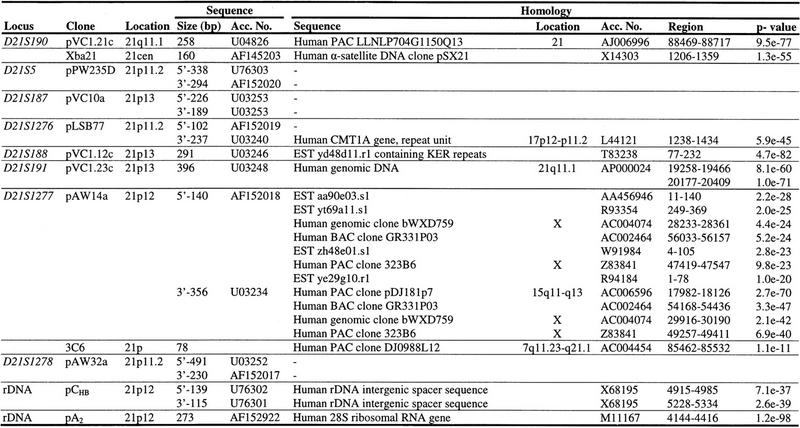
Together, the STS-content mapping resulted in a contiguous YAC map ranging from D21S190 to D21S1278 (Fig. 1). The contig comprises 30 YACs that contain multiple markers. Of four YACs, both ends were isolated and mapped to chromosome 21 and of five additional YACs, one end was localized to chromosome 21, while the other end represented a chromosome 21-repetitive sequence. Of nine YACs, one or both ends did not map to chromosome 21 (Table 5). The YAC-contig map contains 39 markers including 17 nonpolymorphic STSs, four polymorphic STRs, and two ESTs. STR marker ABM–C62D, which was localized by somatic cell hybrid mapping in a region covered by YACs, was absent from all YACs localized in that region.
Figure 1.
Chromosome 21p YAC contig map. The orientation of the YAC contig is based on the regional location of D21Z1 and pCHB. Horizontal lines represent YACs. Dotted lines indicate possible chimerical regions of the YACs. (Single arrowheads) Chromosome 21 YAC left (tails of arrows) and right (heads of arrows) ends; (triple tails of arrows) YAC end fragments homologous to α-satellite or satellite III repetitive sequences; (open circles) nonchromosome 21 YAC ends.
YAC Contig Validation
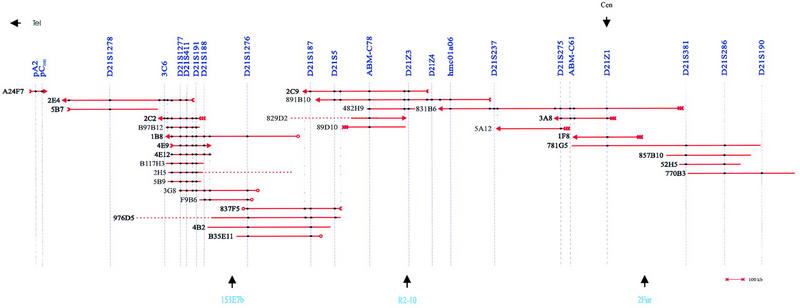
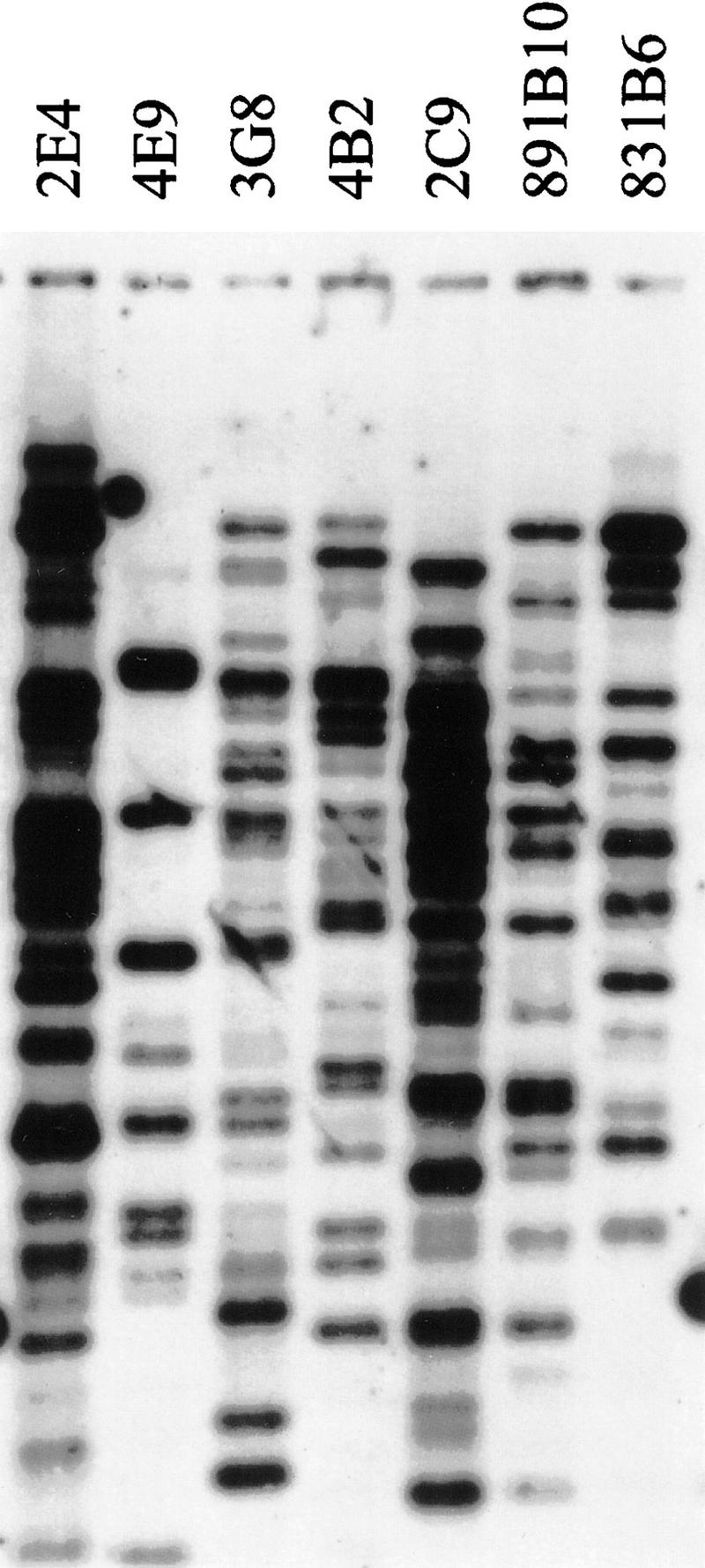
Seven overlapping YACs—831B6, 891B10, 2C9, 4B2, 3G8, 4E9, and 2E4—representing the complete region covered by the YAC contig map were analyzed using HindIII–BamHI fingerprinting using an Alu repeat as hybridization probe (Fig. 2). At least six bands of the same size were observed for each pair of YACs with overlapping marker contents. The total size of the commonly shared fragments ranged between 19- and 66 kb, that is, between 14.5% and 81.5% (average 54.5%) of the summed sizes of Alu-positive bands. Also, these seven YACs and the centromeric YAC 1F8 were used as hybridization probes in FISH analyses of metaphase chromosomes. Ten metaphase spreads of each YAC were examined. Hybridization with 4E9 resulted in chromosome 21p-specific signals using standard procedures (Fig. 3A). For the other YACs, extra signal amplification was needed. Of these, 831B6 and 1F8 specifically recognized the short arm of chromosome 21. For the remaining five YACs, the background signal was high despite the use of 50-fold excess of human Cot-1 DNA. However, in addition to signals on chromosome 21p, other signals with significantly stronger intensity than the background were detected on other acrocentric chromosomes (data not shown).
Figure 2.
HindIII–BamHI fingerprint analysis of YACs.
Figure 3.
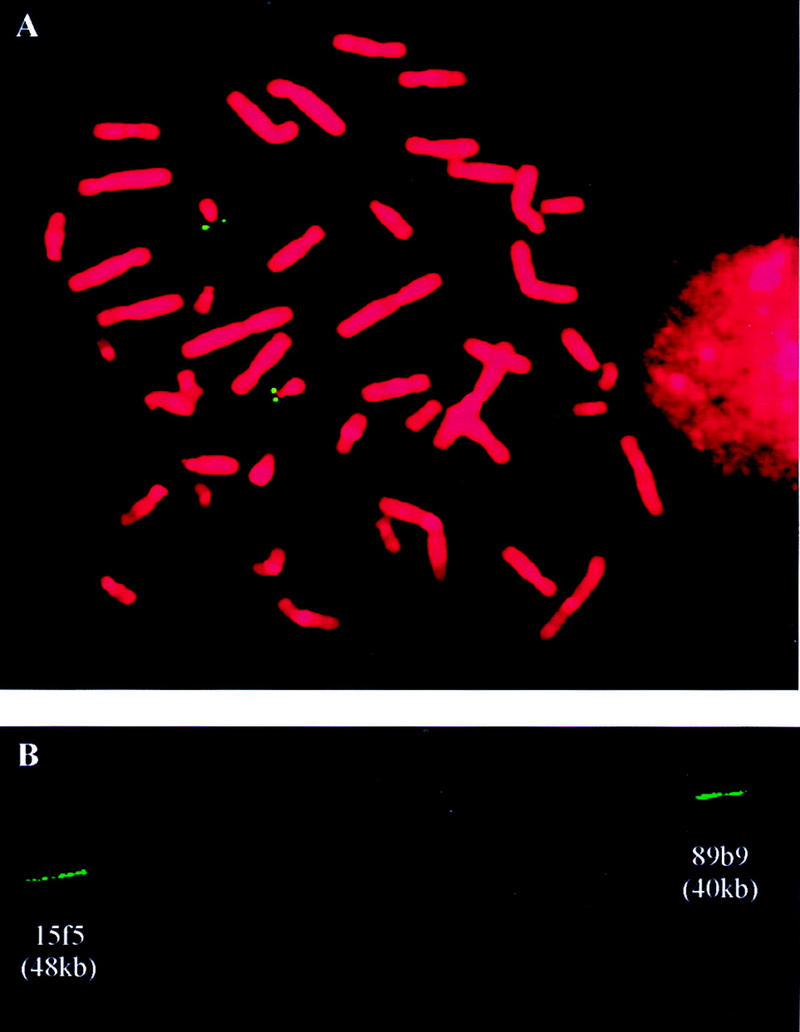
FISH analyses. (A) Metaphase FISH analysis of YAC 4E9. (B) Fiber FISH analysis of cosmid probes 15f5 and 89b9 to estimate the gap size between D21S1278 and pCHB.
Gap Size Estimation
The markers pCHB and 2E4R, flanking the remaining gap were used as hybridization probes to screen the chromosome 21 cosmid library LL21NC02 (HGMP Resource Center, UK). Eight cosmids were selected with pCHB and two with 2E4R. The cosmid insert sizes were estimated between 38 and 48 kb by PFGE analysis of SfiI digested cosmid DNA. Cosmids 89b9 (pCHB) and 15f5 (2E4R) were hybridized in situ on released DNA fibers (Fig. 3B). The gap size, calculated based on the length of cosmid signals and the distance between them averaged over 10 signal pairs, was estimated at 400 kb.
STR Analysis
Five STR markers containing (CA)n-repeat sequences (D21S188, D21S411, ABM–C78, ABM–C62D, and ABM–C61) were mapped to chromosome 21p, although none of them was chromosome 21-specific (Table 3). When the STRs were PCR amplified on genomic DNA of unrelated individuals and analyzed, a complex pattern of constant and variable bands emerged. D21S188 showed 16 different alleles with frequencies ranging from 1% to 97% and 1 constant allele in 39 unrelated individuals. Analysis of D21S188 in CEPH families 1333, 1334, and 1347 demonstrated Mendelian inheritance of each allele (Fig. 4). Comparison of different alleles with the patterns obtained using somatic cell hybrids WAV17, GB3, HDm-5, WegrothB3, HorlI, and WegrothD2, each containing a single acrocentric chromosome, did not allow for the identification of chromosome 21-specific alleles.
Figure 4.
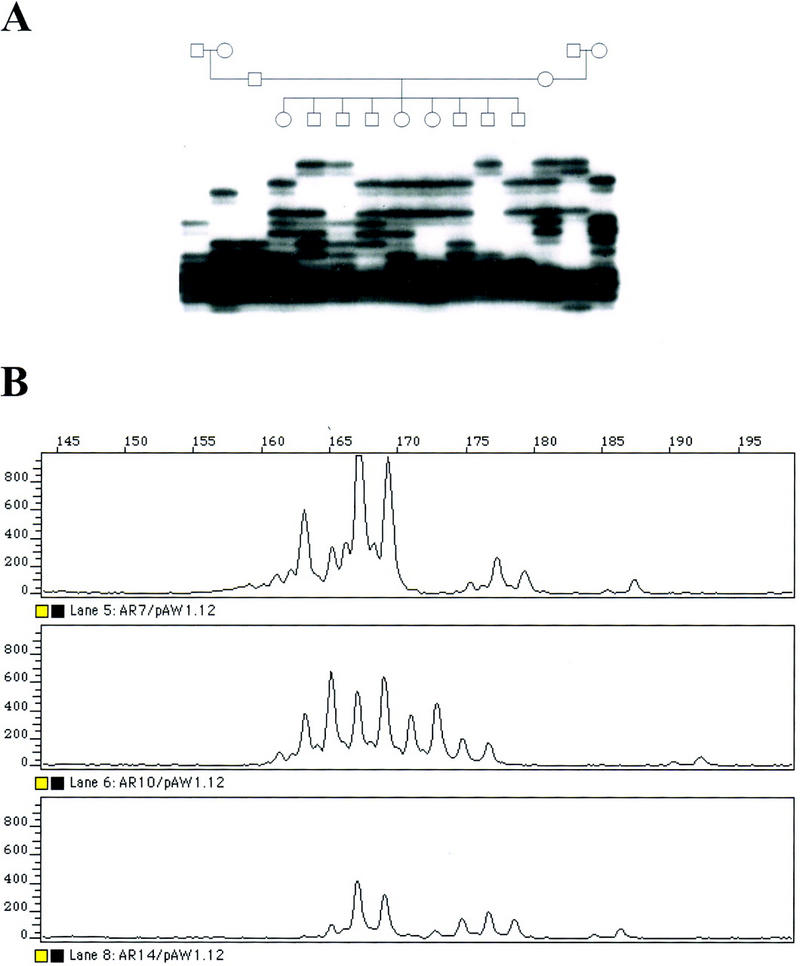
Analysis of (CA)n repeat D21S188. (A) Segregation analysis of D21S188 in CEPH pedigree 1347 using a radiolabeled PCR primer and autoradiography. (B) Fluorescent analysis of D21S188 in three at random individuals.
DISCUSSION
We describe the construction of an STS-based YAC-contig map of chromosome 21p ranging from the centromere (D21Z1) to the rDNA gene cluster at 21p12 (pCHB). Because chromosome 21p markers were lacking, we first developed 19 novel STSs from plasmid clones localized previously to chromosome 21p. Analysis of the markers on a monochromosomal somatic cell hybrid mapping panel indicated that in addition to chromosome 21, all markers amplified sequences of chromosome 13, substantiating previous observations that among the acrocentric chromosomes, 13p and 21p share the highest degree of homology. Previous homology studies related to repetitive satellite sequences (Jorgensen et al. 1987; Van Camp et al. 1992), whereas our data extend the observation of this homology to nonrepetitive sequences. In addition to chromosome 21 and 13, all markers amplified sequences of other acrocentric chromosomes. Furthermore, 59% of the markers amplified nonacrocentric chromosomes.
By using somatic cell hybrids containing different regions of chromosome 21, we were able to sublocalize the markers on chromosome 21p without the interference of the other human chromosomes. D21S187, which by hybridization on DNA of chromosome 21 somatic cell hybrids showed two bands corresponding to locations in 21p11.2 and 21p13, respectively (Van Camp et al. 1990), was assigned by PCR to the centromeric locus only. This might be explained if the PCR primer pair of D21S187 is locus specific and amplifies only the 21p11.2 sequence. Alternatively, if sequence homology is high between both loci, PCR amplification might result in fragments of similar or identical size that cannot be separated by agarose gel electrophoresis. D21S188 and D21S191, both localized to 21p13 by hybridization (Van Camp et al. 1990), were mapped to 21p11.2–p12 by PCR, as they were positive on the somatic cell hybrid ACEM2-10d (Table 3). It is possible that the plasmid clones map to the 21p13 region but have weak homology to a more centromeric region that is recognized only by PCR. However, the ambiguities between hybridization and PCR in both cases involve the same somatic cell hybrid ACEM2-10d containing a complex 21;21 translocation chromosome (Van Keuren et al. 1989), which might indicate that ACEM2-10d is highly unstable, as reported previously (Graw et al. 1995).
Using the newly developed markers, YACs were selected from the St. Louis and CEPH YAC libraries, and additional YACs from the chromosome 21-enriched ICRF YAC library were obtained. The YACs were assembled in a contig based on their STS content. The chimerism of the YACs was determined by sequence homology analysis and somatic cell hybrid mapping of YAC ends (Table 5). Ten YACs were localized to chromosome 21p, as both ends either mapped to chromosome 21p or were homologous to repetitive sequences present in chromosome 21p. Five YACs were considered chimerical, because one end fragment did not map to chromosome 21p. Four YACs with both ends not mapping to chromosome 21 were excluded from the YAC contig. Inclusion of the YAC ends in the contig mapping and selection of six additional CEPH megaYACs resulted in a map that was contiguous from the centromere to the rDNA genes at 21p12, with the exception of a region between pCHB and D21S1278, where a gap remained. The contig consists of 30 multiple-locus YACs and 39 markers including 32 nonpolymorphic STSs, 4 polymorphic STRs, and 2 ESTs.
The pericentromeric region spanned by the YAC contig map partly overlaps with the published 21q YAC contig maps (Chumakov et al. 1992; Gardiner et al. 1995; Korenberg et al. 1995). The locations of YACs 831B6, 781G5, and 52H5 in our contig map are compatible with these maps. In the hybridization-based physical map of chromosome 21 reported by Nizetic et al. (1994), D21S5, D21S187, D21S1276, D21S188, D21S191, D21S1277, and D21S1278 were localized in the 21p region. The order of these markers and YACs are the same as in our map; however, the orientation is reversed. In our YAC contig map (Fig. 2), D21S1278 is located at 21p11.2–p12, at the telomeric end of the YAC contig, whereas D21S5 is located in the centromeric region of the contig. The location of these STSs was determined by both PCR amplification of the chromosome 21 somatic cell hybrids and STS-content mapping of the YACs. 2E4 and 5B7 contain D21S1278 and were mapped in the 21p11.2–p12 region; however, 2E4 was also positive with the STSs D21S275 and ABM–C61 located in the pericentromeric region of chromosome 21. Furthermore, the left end of 5B7 was localized to the pericentromere. 5A12 was mapped to the pericentromeric region, whereas its right end localized to 21p11.2–p12. Together, these data suggest the presence of homologous sequences at two loci on chromosome 21p, which may explain why 2E4 overlapped with 5A12 by hybridization, resulting in the localization of D21S1278 toward the centromere on chromosome 21 in the hybridization-based YAC map (Nizetic et al. 1994). From centromere to telomere, the order of repetitive satellite sequences in this map is α21-I (D21Z1, 831B6L, 1F8L, and 3A8L), satellite III (5A12L), α21-II (D21Z4, D21Z3, 89DD10R), and satellite III (2C2L), which is consistent with the results reported by Shimizu et al. (1995), Trowell et al. (1993), and Ikeno et al. (1994).
The minimal physical size of the region covered by YACs was estimated at 2.9 Mb, based on the insert lengths of nonoverlapping and nonchimerical YACs 831B6, 2C9, 4E9, 5B7, and A24F7. The maximum size was estimated at 5 Mb, based on the sizes of overlapping YACs 831B6, 891B10, 2C9, 4B2, F9B6, 4E9, 2E4, and A24F7. The size of the gap was estimated at 400 kb by fiber FISH using cosmids selected with YAC ends flanking the gap. The size of the region between the centromere and the rDNA cluster at 21p12 estimated from our YAC contig is significantly smaller than that derived from the PFGE map of the satellite repeat sequences on chromosome 21p (Doering et al. 1995; Korenberg et al. 1997). Possibly, this is due to the large variability of the size of the short arm and the centromere of chromosome 21. The exact size could not yet be determined accurately (Antonarakis 1998). Also, it is possible that a reasonable part of chromosome 21p is located distal to rDNA, a region not covered by our YAC contig map. Another explanation for the discordance in size estimations is that instability of long repetitive sequences in YACs leads to internal deletions, an observation that has been reported (Doering et al. 1996) and that might result in a YAC contig map that is shorter than the genomic size of the region. Two STSs (ABM–C62D and 891B10R) that localized to the pericentromere and 21p11, respectively, by somatic cell hybrid mapping were absent from YACs covering that region. These results suggest that deletions or other kinds of rearrangements might have occurred in some YACs of the contig map. However, when the STS content of each YAC was performed on five individual clones, only four YACs showed different STS contents.
All YACs in the contig were initially selected by chromosome 21p markers that were also present on at least one other chromosome and therefore might not be derived from chromosome 21. We investigated the chromosomal origin of seven YACs (831B6, 891B10, 2C9, 4B2, 3G8, 4E9, and 2E4) covering the region mapped by the contig using metaphase FISH. YAC 1F8 was analyzed by FISH because 74 bp of its right end was 80% homologous to PRKC-L, a member of the protein kinase C gene family. Three YACs—831B6, 1F8, and 4E9—specifically recognized chromosome 21p, indicating that they are derived from chromosome 21. In addition, 4E9 gave clear hybridization results without excessive signal amplification, suggesting that a major fraction of this YAC contains single- or low-copy sequences. The other four YACs recognized loci on other acrocentric chromosomes, in addition to chormosome 21p. In each case, the chromosome 21 signal was the strongest, or of equal intensity to the other signals. No YAC could be excluded from chromosome 21p. Based on the results of the FISH analysis and the sequence analysis of YAC end fragments, the YAC contig can be considered representative of chromosome 21p. Also, five of the seven YACs were selected from the ICRF library, which is an enriched chromosome 21 YAC library. This further supports the belief that the YACs most probably are derived from chromosome 21p.
We identified five chromosome 21p STR markers of the (CA)n-repeat type and located four of them in the YAC contig map. We have analyzed these STR markers for their locus specificity by somatic cell hybrid mapping; however, none of them was chromosome 21p specific. All five STRs amplify multiple loci in the human genome and thus show a complex pattern of variable and constant bands. PCR amplification of somatic cell hybrids containing single human acrocentric chromosomes could not identify chromosome-specific alleles. D21S188 was the most polymorphic showing Mendelian inheritance and it is a useful marker for DNA fingerprint analyses. We used this marker successfully to confirm the identity of EBV-transformed cell lines versus genomic DNA of many samples in a fast and reliable manner. When used in multiplex analyses with other highly polymorphic STRs, D21S188 can be applied for paternity testing and forensic applications investigation of the genetic relation between different samples.
Finally, it is interesting to note that hmc01a06, which is derived from an exon-trapping experiment using chromosome 21 cosmids (Chen et al. 1996), mapped to the pericentromeric 21p region in our YAC contig map (Fig. 2). Also, pAW14a (D21S1277) and 1F8R are partly homologous to several cDNA clones and a CpG-rich region of a genomic clone. These results suggest genes and/or pseudogenes might be located on chromosome 21p.
In conclusion, we have constructed a high-resolution physical map spanning a region of 2.9–5 Mb of the centromere and the short arm of chromosome 21, one of the acrocentric chromosomes. The current YAC contig map can be used to identify chromosome 21p-specific polymorphic markers. Such markers will be useful for nondisjunction studies in DS but have not yet been identified. Also, the YACs of this contig can be used as starting material for structural and functional studies of chromosome 21p, and the investigation of the existence of chromosome 21p genes, a study that has not yet been performed.
METHODS
Chromosome 21 Plasmid Clones
Nine plasmid clones localized previously to chromosome 21p or the pericentromeric region of 21q by Southern blot hybridization were selected. D21S190 (pVC1.21c), D21S5 (pPW235D), D21S187 (pVC10a), D21S1276 (pLSB77), D21S188 (pVC1.12c), and D21S191 (pVC1.23c) are subclones from phages derived from chromosome 21-specific phage libraries (Watkins et al. 1985; Van Camp et al. 1990; Stuyver et al. 1991). Additionally, pA2 and pCHB are clones of the 28S coding and spacer region of the rDNA gene cluster, respectively (Sylvester et al. 1986). Finally, Xba21 contains an XbaI fragment of the chromosome 21 somatic cell hybrid 153E9A that was selected with an alphoid DNA probe (Thompson et al. 1989).
Two additional plasmid clones were identified by screening the chromosome 21 phage library LL21NL01 using total human DNA or Blur8 containing human Alu repetitive DNA (Rubin et al. 1980). Somatic cell hybrids WAV17 (Graw et al. 1995), Hdm-15 (Lugo et al. 1987), and AHVI-17 (Cox and Epstein 1985) were used to select for chromosome 21 phage clones. Two clones, F14 and F32, were characterized in detail, and low-copy fragments were subcloned in plasmid pUC18. pAW14a (D21S1277) contains a 2.1-kb EcoRI fragment and pAW32a (D21S1278) a 0.8-kb EcoRI–SstI fragment. Additionally, one plasmid clone, 3C6, was generated using microdissection of banded human metaphase chromosome 21p and microcloning (Bennett et al. 1995).
Finally, clones containing a (CA)n-repeat were isolated from the flow-sorted chromosome 21 phage library LA21NS01 by hybridization using a (GT)10 oligonucleotide probe. Four clones, D21S411, ABM–C78 (GenBank accession no. AF187011), ABM–C62D (GenBank accession no. AF187010), and ABM–C61, were localized to chromosome 21p by Southern blot hybridization of a somatic cell hybrid mapping panel as described elsewhere (Bosch et al. 1996; A. Bosch and X. Estivill, unpubl.). Clone pVC1.12c (Van Camp et al. 1990) was shown to contain a (CA)n repeat by hybridization with a (CA)n probe (Sigma-Aldrich, St. Louis, MO).
Development and Characterization of STSs
The DNA sequence of plasmid clones was determined using the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). The sequences were analyzed on an automated DNA sequencer model 373A (Applied Biosystems), and PCR primer pairs were designed. Of the sequences of D21S411, ABM–C78, ABM–C62D, and ABM–C61, PCR-primer pairs flanking the (CA)n repeat were selected. One EST, hmc01a06, was derived from the sequence of an exon-trapped product that mapped to the pericentromere of chromosome 21 (Chen et al. 1996, 1997). Additionally, the sequence of two α-satellite repeat sequences, D21Z3 and D21Z4 (Vissel et al. 1992), were used to develop PCR primer pairs. Finally, five additional STSs: [D21Z1 (L1.26), D21S275 (G51G10), D21S237 (pPW265D), D21S381 (E341), and D21S286 (G51E07)] were selected from the Genome Database (GDB) based on their location in the centromere of chromosome 21 (Warburton et al. 1991; Chumakov et al. 1992; Tanzi et al. 1992).
Human genomic DNA and DNA from somatic cell hybrids were PCR amplified in a total volume of 25 μl containing 50–100 ng of template DNA, 10 pmoles of each primer, 1.5–2.0 mm MgCl2, 200 μm dNTPs, and one unit of Taq DNA polymerase (GIBCO BRL, Gaitherburg, MD) using a DNA thermal cycler 480 (Perkin-Elmer-Cetus, Norwalk, CT). The first denaturing step was at 94°C for 1.5 min, followed by 30–40 amplification cycles consisting of a denaturing step at 94°C for 1 min, annealing at the empirically defined optimal temperature for 1.5 min and extension at 72°C for 1.5 min. A final extension step was performed at 72°C for 4 min. The PCR products were separated on a 1.5% agarose gel and visualized on a UV transilluminator after ethidium bromide staining. First, a monochromosomal somatic cell hybrid mapping panel (BIOS Laboratories, New Haven, CT) was used to establish the chromosomal assignment of the STSs. Next, the STSs were analyzed using an extended mapping panel of human chromosome 21 somatic cell hybrids, composed of WAV17, JC-6A, ACEM2-10d, 153-E7b, R2-10W, 2Fur1 (Graw et al. 1995), and AHVI-17 (Cox and Epstein 1985), each containing different regions of human chromosome 21.
Selection and Characterization of YACs
The St. Louis YAC library (Brownstein et al. 1989) was screened by PCR using STSs Xba21, D21S76, D21S187, D21S188, D21S191, D21S1277, D21S1278, pCHB, and pA2 in the context of the International Chromosome 21 Joint YAC-Screening Effort (JYSE) at the Eleanor Roosevelt Institute. The CEPH YAC and megaYAC libraries (Chumakov et al. 1992) were screened by both PCR amplification and hybridization using markers ABM–C78 and D21S5. In addition, YACs of the ICRF chromosome 21-enriched YAC library and CEPH library that were selected previously and mapped to the centromere or the p-arm of chromosome 21 (Chumakov et al. 1992; Nizetic et al. 1994) were included.
Single YAC colonies were grown at 30°C for 48 hr in 20 ml of selective AHC medium (6.7 grams/liter yeast nitrogen base without amino acids, 10 grams/liter casein hydrolysate, 10 mg/liter adenine and 2 grams/liter glucose) in the absence of tryptophan and uracil. Total YAC DNA and agarose plugs were prepared as described elsewhere (Cruts et al. 1995). To determine the size of the YACs, the YAC and yeast chromosomes were separated by PFGE using the CHEF Mapper XA apparatus (Bio-Rad) and the chromosomes were visualized by ethidium bromide (0.1 μg/ml) staining. Alternatively, the DNA was transferred to Hybond-N+ membranes (Amersham), and the YAC chromosomes were identified by hybridization with 32P-labeled human genomic DNA. The sizes of the YAC chromosomes were estimated using the chromosomes of Saccharomyces cerevisiae strain YP148 as size marker.
YAC Contig Mapping
The STS content of the YACs was determined by using standard PCR amplifications. YAC vectorette PCR (Riley et al. 1990) and a linker-mediated technique (Cruts et al. 1995) were performed to isolate YAC end fragments. Alternatively, we used inverse PCR (Ochman et al. 1988; Triglia et al. 1988) with the following modifications: An amount of 0.2 μg of YAC DNA was digested with 10 units of RsaI and SspI in One-Phor-All (OPA) buffer (Pharmacia, Upsalla, Sweden) and circulated by self-ligation using 10 units of T4 DNA ligase in a total volume of 200 μl by incubation overnight at room temperature. A second restriction-enzyme digestion was performed using FspI for the YAC left end (centromeric side) and SmaI for the YAC right end (URA3 side). The primer pairs 1207 (5′-AGCCAAGTTGGTTTAAGGCGCAAGGACT-3′) and 161 (5′-CGATGCTGTCGGAATGGACGATATC-3′), complementary to the YAC left vector arm, and 1208 (5′-TCGAACGCCCGATCTCAAGATTACG-3′) and 162 (5′-GCATGTCTCCATTCACTTCCCAGAC-3′), complementary to the YAC right vector arm, were used to amplify both ends of the human insert fragments. The PCR products were separated by agarose gel electrophoresis, excised from the gel, purified, and PCR amplified again using the same primers. The YAC end fragments were directly sequenced using the same primers as in the PCR amplification. Alternatively, the PCR products were subcloned into plasmid vector pUC18 or pGEM-T (Promega, Madison, WI) and sequenced using vector primers. PCR primer pairs were developed and PCR was performed on human genomic DNA and DNA from somatic cell hybrids and YACs.
Restriction Enzyme Fingerprinting of YACs
About 3 μg of YAC DNA embedded in agarose plugs was digested with 5 units of HindIII and BamHI at 37°C for 6 hr. The reactions were terminated by heat inactivation or the restriction enzyme at 65°C for 10 min. DNA fragments were separated on a 0.7% agarose gel at 2 V/cm for 20 hr. The DNA was transferred to Hybond-N+ membranes by Southern blotting, and hybridization was performed using Alu clone pPD39 (Batzer et al. 1994) as probe.
FISH Analysis
Metaphase chromosomes were prepared from human lymphoblast cell lines according to standard procedures. Free DNA fibers were achieved from nuclei fixed to silanized microscope slides. Briefly, slides were submerged in 55 ml of lysis buffer (0.5% SDS, 50 mm EDTA, 200 mm Tris at pH 7.4) for 10 min in an upright position. Fifty milliliters of absolute ethanol was added dropwise to the lysis solution, and the slides were kept in this solution for 10 min. Finally, the slides were fixed for 30 min in 70% ethanol at room temperature and air-dried. FISH was performed as described (Pinkel et al. 1986; Tissir et al. 1995) using hybridization probes that were biotinylated with the Biotin-Nick Translation kit (Boehringer Mannheim). Briefly, the hybridization mixture contained 50% formamide and 10% dextran sulfate in 2× SSC. Repetitive sequences were suppressed with 50-fold excess of human Cot-1 DNA. After overnight incubation at 37°C, the slides were washed three times at 45°C in 50% formamide and 2× SSC and three times in 0.1× SSC. Probes were detected with avidin–FITC (Vector Laboratories), amplified with biotinylated goat anti-avidin (Vector Laboratories) and a second layer of avidin-FITC. Fluorescent signals were analyzed on an Axioskop microscope (Zeiss, Germany). Images were captured using a CCD camera (Applied Imaging, UK). The length of probe signals and the gap were measured from digitized images with the CytoVision software (Applied Imaging).
STR Analysis
The polymorphic STRs D21S188, D21S411, ABM–C78, ABM–C62D, and ABM–C61 (Bosch et al. 1996; A. Bosch and X. Estivill, unpubl.) were detected using PCR on human genomic DNA of CEPH families 1333, 1334, 1347, or 7 unrelated individuals, and a set of somatic cell hybrids containing acrocentric chromosomes. GB3 contains chromosome 13 (Scheffer et al. 1986), HDm-5 (Lugo et al. 1987) and WegrothB3 (Geurts van Kessel et al. 1983) chromosome 14, HorlI chromosome 15 (Heisterkamp et al. 1982), and Wegroth D2 chromosome 22 (Geurts van Kessel et al. 1983). The PCR reaction was carried out in a total volume of 10 μl containing ∼100 ng template DNA, 0.1 unit of Taq DNA polymerase, 5 pmoles of each primer, one of which was γ-32P end-labeled, 100 μm dNTP, and 1.0 mm MgCl2. PCR amplification was carried out as described above. Aliquots of the PCR products were denatured at 94°C for 5 min and separated on a 6% denaturing polyacrylamide gel containing 8 m urea at 55 W for 4.5 hr. The polymorphic alleles were visualized by overnight autoradiography.
Alternatively, D21S188 was PCR amplified using one fluorescently labeled primer and separated using an automated DNA sequencer ABI 373 (Applied Biosystems). The fragments were analyzed using the GeneScan 672 software (Applied Biosystems).
Acknowledgments
This work was supported by European Union grants BIOMED GENO-CT91-0024, GENO-CT93-0015, and BMH4-CT96-0554 and the Fund for Scientific Research-Flanders Belgium (FWO-F) to C.V.B., National Institutes of Health grants HD17449 and HG00406 to D.P., and Swiss Fonds National de Recherche Scientifique 31.33965.92 and 31.40500.94 grants to S.E.A., as well as the Spanish Dirección General de Enseñanza Superior (DGES) (PM95-0106-C02-01) and the Fundaćo Catalana Síndrome de Down/La Marató de TV3 to X.E. We thank P.C. Watkins for kindly providing pPW235D, and R.D. Schmickel for providing pA2 and pCHB. Also, we thank P. Liang and A. Wehnert for their contribution to this work. M.C. is a postdoctoral fellow of the FWO-F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicated this fact.
Footnotes
E-MAIL cvbroeck@uia.ua.ac.be; FAX 323 8202541.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE. Human chromosome 21: Genome mapping and exploration, circa 1993. Trends Genet. 1993;9:142–148. doi: 10.1016/0168-9525(93)90210-9. [DOI] [PubMed] [Google Scholar]
- ————— 10 years of Genomics, chromosome 21, and Down syndrome. Genomics. 1998;51:1–16. doi: 10.1006/geno.1998.5335. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Petersen MB, McInnis MG, Adelsberger PA, Schinzel AA, Binkert F, Pangalos C, Raoul O, Slaugenhaupt SA, Hafez M. The meiotic stage of nondisjunction in trisomy 21: Determination by using DNA polymorphisms. Am J Hum Genet. 1992;50:544–550. [PMC free article] [PubMed] [Google Scholar]
- Bacher N, Zisman Y, Berent E, Livneh E. Isolation and characterization of PKC-L, a new member of the protein kinase C-related gene family specifically expressed in lung, skin, and heart. Mol Cell Biol. 1991;11:126–133. doi: 10.1128/mcb.11.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Alegria-Hartman M, Deininger PL. A consensus Alu repeat probe for physical mapping. Genet Anal Tech Appl. 1994;11:34–38. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Bennett LB, Claussen U, Farrer MJ, Kessling AM. A micro-dissected chromosome 21p mini-library. Cytogenet Cell Genet. 1995;70:168. [Google Scholar]
- Bosch A, Guimera J, Graw S, Gardiner K, Chumakov I, Patterson D, Estivill X. Integration of 30 CA-repeat markers into the cytogenetic, genetic and YAC maps of human chromosome 21. Eur J Hum Genet. 1996;4:135–142. doi: 10.1159/000472187. [DOI] [PubMed] [Google Scholar]
- Brownstein BH, Silverman GA, Little RD, Burke DT, Korsmeyer SJ, Schlessinger D, Olson MV. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989;244:1348–1351. doi: 10.1126/science.2544027. [DOI] [PubMed] [Google Scholar]
- Chen H, Chrast R, Rossier C, Morris MA, Lalioti MD, Antonarakis SE. Cloning of 559 potential exons of genes of human chromosome 21 by exon trapping. Genome Res. 1996;6:747–760. doi: 10.1101/gr.6.8.747. [DOI] [PubMed] [Google Scholar]
- Chen HM, Rossier C, Antonarakis SE. A testis-expressed gene encoding a transmembrane protein is located within a chromosome 21 centromeric YAC and has homologous sequences on chromosomes 13, 15, 22 and Y. Med Genet. 1997;9:137. [Google Scholar]
- Choo KH, Vissel B, Brown R, Filby RG, Earle E. Homologous alpha satellite sequences on human acrocentric chromosomes with selectivity for chromosomes 13, 14 and 21: Implications for recombination between nonhomologues and Robertsonian translocations. Nucleic Acids Res. 1988;16:1273–1284. doi: 10.1093/nar/16.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov I, Rigault P, Guillou S, Ougen P, Billaut A, Guasconi G, Gervy P, LeGall I, Soularue P, Grinas L. Continuum of overlapping clones spanning the entire human chromosome 21q. Nature. 1992;359:380–387. doi: 10.1038/359380a0. [DOI] [PubMed] [Google Scholar]
- Cox DR, Epstein CJ. Comparative gene mapping of human chromosome 21 and mouse chromosome 16. Ann NY Acad Sci. 1985;450:169–177. doi: 10.1111/j.1749-6632.1985.tb21491.x. [DOI] [PubMed] [Google Scholar]
- Cruts M, Backhovens H, Theuns J, Clark RF, Le Paslier D, Weissenbach J, Goate AM, Martin JJ, Van Broeckhoven C. Genetic and physical characterization of the early-onset Alzheimer's disease AD3 locus on chromosome 14q24.3. Hum Mol Genet. 1995;4:1355–1364. doi: 10.1093/hmg/4.8.1355. [DOI] [PubMed] [Google Scholar]
- Doering JL, Geronimo I, Przybysz M, Doll J, Chawla-Gupta ME, Cucci R, Cummings MR. Physical mapping of the short arm/centromere of chromosome 21. Cytogenet Cell Genet. 1995;70:171. [Google Scholar]
- Epstein CJ. Down syndrome, trisomy 21. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic basis of inherited disease. New York, NY: McGraw Hill; 1989. pp. 291–341. [Google Scholar]
- Gardiner K, Graw S, Ichikawa H, Ohki AM, Joetham A, Gervy P, Chumakov I, Patterson D. YAC analysis and minimal tiling path construction for chromosome 21q. Somat Cell Mol Genet. 1995;21:399–414. doi: 10.1007/BF02310207. [DOI] [PubMed] [Google Scholar]
- Geurts van Kessel A, Tetteroo PA, von dem, A. Hagemeijer B, Bootsma D. Expression of human myeloid-associated surface antigens in human-mouse myeloid cell hybrids. Proc Natl Acad Sci. 1983;80:3748–3752. doi: 10.1073/pnas.80.12.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw SL, Gardiner K, Hall-Johnson K, Hart I, Joetham A, Walton K, Donaldson D, Patterson D. Molecular analysis and breakpoint definition of a set of human chromosome 21 somatic cell hybrids. Somat Cell Mol Genet. 1995;21:415–428. doi: 10.1007/BF02310208. [DOI] [PubMed] [Google Scholar]
- Greig GM, Willard HF. Beta satellite DNA: Characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics. 1992;12:573–580. doi: 10.1016/0888-7543(92)90450-7. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Groffen J, Stephenson JR, Spurr NK, Goodfellow PN, Solomon E, Carritt B, Bodmer WF. Chromosomal localization of human cellular homologues of two viral oncogenes. Nature. 1982;299:747–749. doi: 10.1038/299747a0. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet. 1994;3:1245–1257. doi: 10.1093/hmg/3.8.1245. [DOI] [PubMed] [Google Scholar]
- Jorgensen AL, Bostock CJ, Bak AL. Homologous subfamilies of human alphoid repetitive DNA on different nucleolus organizing chromosomes. Proc Natl Acad Sci. 1987;84:1075–1079. doi: 10.1073/pnas.84.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Mitchell S, Fannin S, Gerwehr S, Cohen D, Chumakov I. A high-fidelity physical map of human chromosome 21q in yeast artificial chromosomes. Genome Res. 1995;5:427–443. doi: 10.1101/gr.5.5.427. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Aaltonen J, Brahe C, Cabin N, Creau N, Delabar JM, Doering J, Gardiner K, Hubert RS, Ives J, et al. Report of the Sixth International Workshop on Human Chromosome 21 Mapping 1996. Cytogenet Cell Genet. 1997;79:21–52. doi: 10.1159/000134681. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y, Hallberg A, Hersey J, Karadima G, Pettay D, et al. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet. 1997;6:1391–1399. doi: 10.1093/hmg/6.9.1391. [DOI] [PubMed] [Google Scholar]
- Lugo TG, Handelin B, Killary AM, Housman DE, Fournier RE. Isolation of microcell hybrid clones containing retroviral vector insertions into specific human chromosomes. Mol Cell Biol. 1987;7:2814–2820. doi: 10.1128/mcb.7.8.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizetic D, Gellen L, Hamvas RM, Mott R, Grigoriev A, Vatcheva R, Zehetner G, Yaspo ML, Dutriaux A, Lopes C, et al. An integrated YAC-overlap and “cosmid-pocket” map of the human chromosome 21. Hum Mol Genet. 1994;3:759–770. doi: 10.1093/hmg/3.5.759. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CM, Houck CM, Deininger PL, Friedmann T, Schmid CW. Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature. 1980;284:372–374. doi: 10.1038/284372a0. [DOI] [PubMed] [Google Scholar]
- Scheffer H, van der Lelie D, Aanstoot GH, Goor N, Nienhaus AJ, van der Hout AH, Pearson PL, Buys CH. A straightforward approach to isolate DNA sequences with potential linkage to the retinoblastoma locus. Hum Genet. 1986;74:249–255. doi: 10.1007/BF00282543. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Antonarakis SE, Van Broeckhoven C, Patterson D, Gardiner K, Nizetic D, Créau N, Delabar J-M, Korenberg JR, Reeves R, et al. Report of the fifth international workshop on human chromosome 21 mapping 1994. Cytogenet Cell Genet. 1995;70:147–182. [Google Scholar]
- Stuyver L, Van Camp G, Van De Voorde A, Van Broeckhoven C, Van Heuverswyn H. Isolation of chromosome 21 enconded expressed sequences. Cytogenet Cell Genet. 1991;58:2039–2040. [Google Scholar]
- Sylvester JE, Whiteman DA, Podolsky R, Pozsgay JM, Respess J, Schmickel RD. The human ribosomal RNA genes: Structure and organization of the complete repeating unit. Hum Genet. 1986;73:193–198. doi: 10.1007/BF00401226. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Romano DM, Berger R, Buraczynska MJ, Gaston SM, Kurnit DM, Patterson D, Gusella JF, Stewart GD. Sequence-tagged sites (STSs) for a set of mapped markers on chromosome 21. Genomics. 1992;14:498–502. doi: 10.1016/s0888-7543(05)80251-4. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Sylvester JE, Gonzalez IL, Costanzi CC, Gillespie D. Definition of a second dimeric subfamily of human alpha satellite DNA. Nucleic Acids Res. 1989;17:2769–2782. doi: 10.1093/nar/17.7.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Riviere M, Guo DF, Tsuzuki S, Inagami T, Levan G, Szpirer J, Szpirer C. Localization of the genes encoding the three rat angiotensin II receptors, Agtr1a, Agtr1b, Agtr2, and the human AGTR2 receptor respectively to rat chromosomes 17q12, 2q24 and Xq34, and the human Xq22. Cytogenet Cell Genet. 1995;71:77–80. doi: 10.1159/000134067. [DOI] [PubMed] [Google Scholar]
- Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowell HE, Nagy A, Vissel B, Choo KH. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: Identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet. 1993;2:1639–1649. doi: 10.1093/hmg/2.10.1639. [DOI] [PubMed] [Google Scholar]
- Van Camp G, Van Hul W, Backhovens H, Stinissen P, Wehnert A, Patterson D, Vandenberghe A, Van Broeckhoven C. Physical mapping of chromosome 21 DNA markers in Alzheimer's disease region using somatic cell hybrids. Somat Cell Mol Genet. 1990;16:241–249. doi: 10.1007/BF01233360. [DOI] [PubMed] [Google Scholar]
- Van Camp G, Cruts M, Backhovens H, Wehnert A, Van Broeckhoven C. Unique sequence homology in the pericentromeric regions of the long arms of chromosomes 13 and 21. Genomics. 1992;12:158–160. doi: 10.1016/0888-7543(92)90420-w. [DOI] [PubMed] [Google Scholar]
- Van Keuren ML, Stewart GD, Bradley CM, Kurnit DM, Neve RL, Watkins PC, Tanzi RE, Gusella JF, Patterson D. Characterization of an unusual and complex chromosome 21 rearrangement using somatic cell genetics and cloned DNA probes. Am J Med Genet. 1989;33:369–375. doi: 10.1002/ajmg.1320330316. [DOI] [PubMed] [Google Scholar]
- Vissel B, Choo KH. Evolutionary relationships of multiple alpha satellite subfamilies in the centromeres of human chromosomes 13, 14, and 21. J Mol Evol. 1992;35:137–146. doi: 10.1007/BF00183225. [DOI] [PubMed] [Google Scholar]
- Vissel B, Nagy A, Choo KH. A satellite III sequence shared by human chromosomes 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet Cell Genet. 1992;61:81–86. doi: 10.1159/000133374. [DOI] [PubMed] [Google Scholar]
- Wang S-Y, Liang P, Wehnert A, Stuyver L, Van Broeckhoven C. An STS-based YAC contig map of the chromosome 21p-arm. Cytogenet Cell Genet. 1995;70:171. [Google Scholar]
- Wang S-Y, Cruts M, Del-Favero J, Nizetic D, Potier M-C, Patterson D, Estivill X, Antonarakis S, Kessling A, Yaspo ML, et al. An STS-based YAC contig map of chromosome 21p from D21Z1 to the rDNA cluster. Cytogenet Cell Genet. 1997;79:39. [Google Scholar]
- Warburton PE, Greig GM, Haaf T, Willard HF. PCR amplification of chromosome-specific alpha satellite DNA: Definition of centromeric STS markers and polymorphic analysis. Genomics. 1991;11:324–333. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- Watkins PC, Tanzi RE, Gibbons KT, Tricoli JV, Landes G, Eddy R, Shows TB, Gusella JF. Isolation of polymorphic DNA segments from human chromosome 21. Nucleic Acids Res. 1985;13:6075–6088. doi: 10.1093/nar/13.17.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]