Abstract
Background
Information about the effects of unsaturated fatty acids (UFA) supplementation on the health and integrity of the mammary gland in lactating dairy cows is lacking. Therefore, the aim of this study was to determine the effects of unprotected dietary UFA on the global expression pattern of genes in the mammary gland tissue of grazing dairy cows, and to translate this information into relevant biological knowledge.
Methods
Twenty-eight Holstein-Friesian dairy cows were randomly assigned to 4 different concentrated UFA-sources for 23 days after which all cows were switched to a non-UFA-supplemented concentrate for an additional 28 days. On the last day of both periods, mammary gland biopsies were taken to study genome-wide differences in gene expression on Bovine Genome Arrays.
Results
Supplementation with UFA reduced the concentration of short chain fatty acids (FA), C16 FA and saturated FA in the milk, whereas that of trans-FA increased. One major finding was that canonical pathways associated with remodelling and immune functions of the mammary gland were predominantly down-regulated during UFA supplementation and negatively correlated with the concentration of milk trans-FA.
Conclusions
Supplementing grazing dairy cows with unprotected dietary UFA can affect the remodelling and immune functions of the mammary gland with potential consequences for its integrity and health, as well as milk quality.
Background
Because of the potential health benefits of unsaturated fatty acids (UFA), there is considerable interest in increasing their concentration levels in milk. The effect of supplementing dietary UFA on the resulting milk fatty acid (FA) composition and expression of several genes involved in the mammary gland lipid metabolism has recently been studied in dairy cows [1,2]. However, information about the effects of UFA supplementation on genome-wide expression of genes in the mammary gland tissue of dairy cows, as well as their relationship with the resultant milk FA composition, is lacking. Identifying these effects is expected to contribute to our understanding of milk fatty acid metabolism in the mammary gland tissue, and to enhance opportunities to improve milk fat composition through nutrition. Therefore, the aim of this study was to determine the effects of unprotected dietary UFA on the global expression pattern of genes in the mammary gland tissue of grazing dairy cows, to translate this into more biological knowledge, and to correlate gene expression patterns to the resulting milk fatty acid composition.
Methods
Twenty-eight Holstein-Friesian dairy cows were randomly assigned to 4 concentrated UFA-sources based on either unprotected rapeseed oil, soybean oil, linseed oil, or a proportional mix of these plants oils for 23 days (Period I), after which all cows were switched to a non-UFA-supplemented concentrate for an additional 28 days (Period II). On the last day of both periods, mammary gland biopsies were taken to study genome-wide differences in gene expression using Affymetrix GeneChip® Bovine Genome Arrays. In addition, milk samples were taken and stored at -20°C until analysis for FA composition by gas chromatography. Milk FA composition was analyzed using a mixed-effects ANOVA (SAS Inst. Inc. Cary, NC, release 9.1). The model included UFA-sources, experimental periods, and the interaction between UFA-sources and the experimental period, as fixed effects, and cow within pen as a random effect. The same model was performed to analyse gene expressions (MAANOVA package of R, release 1.16). The list of differentially expressed genes was generated using a False Discovery Rate (FDR) < 5% together with an absolute fold-change (FC) threshold of 1.3. Additionally, the Ingenuity Pathways Analysis (IPA; ver. 5.5., Ingenuity Systems, Redwood City, CA) was applied to identify the relevant genes associated with molecular and cellular functions, canonical pathways and biological functions, as well as the biological interaction networks among significant genes. As a means to potentially identify the association between differentially expressed genes in the mammary gland tissue affected by unprotected dietary UFA, and the resulting milk FA composition, the relationship between milk FA groups and mRNA expression of affected genes was calculated using the mixOmics package of R (release 2.7). The significance of the correlations described was tested by linear model using limma package of R.
Results and discussion
Supplementation of dietary UFA decreased (P < 0.01) the concentration of short chain fatty acids (SCFA), 16-carbon FA (C16), and saturated fatty acids (SFA), and increased the concentration of conjugated linoleic acid (CLA) isomers (Table 1), suggesting that supplementing dairy cows with dietary UFA may help to improve the health and nutrition quality aspects of dairy milk. However, the omega-3 (n-3) and omega-6 (n-6) FA concentration decreased, and the concentration of trans- FA in milk increased with UFA supplementation (Table 1). Applying a statistical cut-off of FDR <0.05 together with an absolute FC threshold of 1.3, we identified a total of 972 genes as being differentially expressed in the mammary gland tissue when supplementing dairy cows with UFA compared with control diet. The expression of genes in mammary gland was not significantly affected by the different dietary unprotected UFA-sources. The genes affected by UFA supplementation were involved in molecule transport, lipid and protein metabolism, cell growth and proliferation, remodelling, and defense, inflammatory and immune response. A specific examination of the canonical pathways involved in remodelling and immune system using IPA revealed that UFA supplementation down-regulated a total of 78 genes associated with cell signalling pathways, including mTOR, JAK/Stat, prolactin and integrin signalling, as well as cellular and humoral immune responses, pathogen-induced signalling and cellular stress and injury. Ranking of the affected biological networks identified the P53 protein, a key transcription factor that controls mammogenesis in ruminants, as the major traffic controller.
Table 1.
Milk fatty acid concentration (mg/L) when comparing dairy cows fed with unsaturated fatty acids (UFA) enriched-diet relative to the same cows fed control diet
| Diet supplementation | ||||
| Non-UFA | UFA | SEM | P- value1 | |
| Item2 | UFAL | |||
| Short Chain Fatty Acids, mg/L | 11,343 | 8,035 | 418.5 | <0.001 |
| 316C, mg/L | 13,200 | 9,780 | 416.7 | <.0001 |
| Long-Chain Fatty Acids, mg/L | 15,282 | 15,412 | 467.9 | 0.84 |
| Unsaturated Fatty Acids, mg/L | 12,023 | 12,397 | 349.6 | 0.45 |
| Saturated Fatty Acids, mg/L | 28,393 | 21,160 | 938.6 | <.0001 |
| Polyunsaturated Fatty Acids, mg/L | 1,393 | 1,388 | 43.1 | 0.93 |
| 4n-3 Fatty Acids, mg/L | 269.3 | 201.7 | 9.28 | <.0001 |
| 5n-6 Fatty Acids, mg/L | 721.1 | 652.46 | 20.01 | 0.01 |
| Trans-octadecenoic Fatty Acids, mg/L | 1,798 | 2,900 | 72.5 | <0.001 |
| cis-9, trans-11-CLA, mg/L | 263.9 | 368.8 | 18.80 | 0.002 |
| trans-10,cis-12-CLAf, mg/L | 3.56 | 6.4 | 0.46 | <0.0001 |
1P- value = effect of unsaturated fatty acids enriched-diet supplementation
2Included n= 28 cows
316C = sum of 16:0 and 16:1,cis-9
4Sum of n-3 fatty acids (18:3n-3, 20:3n-3, 20:5n-3, and 22:5n-3)
5Sum of n-6 fatty acids (18:2n-6, 18:3n-6, 20:2n-6, 20:3n-6, 20:4n-6, and 22:2n-6)
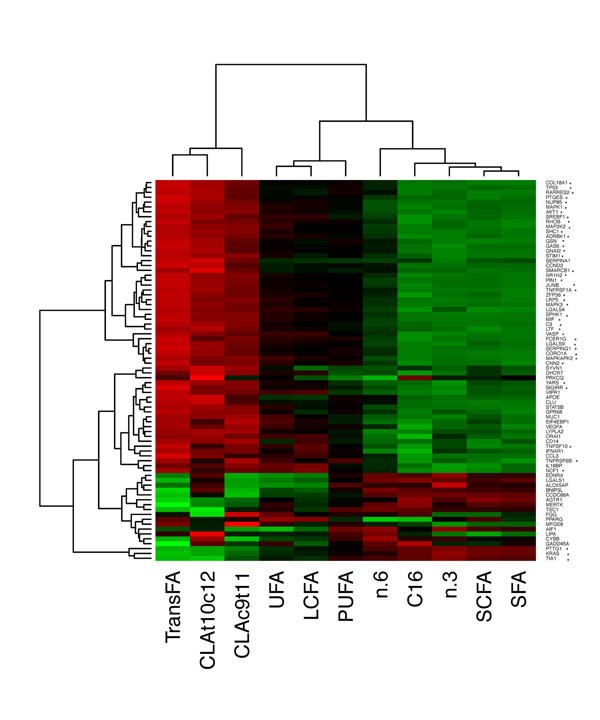
Little is known about remodelling, defense, and inflammatory and immune-related genes in response to dietary UFA supplementation in dairy cows. However, results presented here suggest that dietary UFA supplementation may affect mammary gland tissue integrity and cell adhesion, as well as immune functions, and thus may modify the susceptibility to mastitis in lactating cows and the resulting quality of milk. To determine whether the expression of these genes correlated with specific milk FA, we then studied the association between the set of 78-remodelling and immune response genes and different groups of FA. As depicted in Figure 1, we detected that the characterized FA groups could be divided into two main clusters, each with a distinctive correlation value to remodelling and immune response genes. A novel finding was that the first main cluster included the milk trans-FA and CLA isomers, and the second cluster the other groups of FA (SFA, C16, SCFA, n-3, n-6, and LCFA, UFA, and polyunsaturated FA (PUFA)). Furthermore, whereas trans-FA and CLA isomers negatively correlated with remodelling and immune response gene expressions (Figure 1), the other groups of FA showed a positive correlation with the level of remodelling and immune response gene expressions. This results are supported by the discovery that t10,c12 CLA isomer modulated the regulation of gene expression associated with immune system [3]. However, evidence for the effects of trans-FA on remodelling and immune-gene expression in the mammary gland of dairy cows is currently lacking. Remarkably, the regulation of gene expression by these FAs seems to be due to changes in the expression of different cell signalling and transcription factors related to a wide range of metabolic and developmental functions, among them, P53, ZFP36 (zinc finger protein 36) and SREBP1 (sterol regulatory element binding protein 1; Figure 1). ZFP36 has anti-inflammatory activity by binding to and destabilizing pro-inflammatory mRNAs, such as tumor necrosis factor-alpha (TNF-α) mRNA [4]. Further, P53 is a transcription factor regulating DNA repair, apoptosis, cell cycle arrest, and senescence [5], and SREBP1 is a key regulator of intracellular lipid homeostasis. Notably, recent results suggest that the immune system, through Lymphotoxin-beta receptor (LTBR) signalling, directly influences the enzymatic regulation of lipid homeostasis and thus the SREBP1 activation [6]. Knowledge of the mechanisms by which FA control specific gene expression may provide insight into the development of new nutrition strategies for a better management of milk quality and mammary gland health status.
Figure 1.
Milk fatty acids correlated to remodelling and immune response related-genes. Heat map of the correlations between the 78 significantly expressed genes involved in remodelling and immune response-related genes when comparing dairy cows supplemented with unsaturated fatty acid relative to the same cows fed a control diet, and the milk fatty acids content. The 78 immune function-related genes were identified by Ingenuity Pathways Analysis and presented a Fold Change (FC) ≥ 1.3 and a false discovery rate (FDR) q-values < 0.05. The horizontal rows of the map represent genes, whereas the columns represent the different groups of fatty acids. Each pixel represents the correlation value between each gene and group of fatty acids: the colours depict the coefficient of correlation from green (large negative) to red (large positive). * indicates gene signatures that presented a significant correlation with the FA in milk
Conclusions
The results of this study show that supplementing grazing dairy cows with different unprotected unsaturated fatty acids reduced the concentration of short chain fatty acids, C16 and saturated fatty acids in the milk, whereas that of trans-fatty acids increased. Dietary unsaturated fatty acid supplementation affected 78 remodelling and immune response-related genes, suggesting an effect on mammary gland tissue integrity and health. A novel finding was that the milk trans-FA presented a significant and negative correlation profile to these genes, therefore it is tempting to speculate on an active role of this FA-group in the regulation of mammary gland gene expression associated with immune system and remodelling components. Further functional knowledge by which fatty acids control these genes will be of great importance in understanding the resulting mammary gland health and integrity, as well as the milk composition and quality.
Competing interests
The author(s) declare that they have no competing interests.
Contributor Information
Núria Mach, Email: nuria.mach@wur.nl.
Jürgen van Baal, Email: Jurgen.vanBaal@wur.nl.
Leo Kruijt, Email: Leo.Kruijt@wur.nl.
Antoon Jacobs, Email: Antoon.Jacobs@wur.nl.
Mari Smits, Email: Mari.Smits@wur.nl.
Acknowledgements
The authors gratefully acknowledge the financial support of WUR investment funds (BAS code KB-05-003-040-ASG-V-2) and the financial support of the “Comissionat per a Universitats i Recerca (CUR) del DIUE” of the Government of Catalonia.
This article has been published as part of BMC Proceedings Volume 5 Supplement 4, 2011: Proceedings of the International Symposium on Animal Genomics for Animal Health (AGAH 2010). The full contents of the supplement are available online at http://www.biomedcentral.com/1753-6561/5?issue=S4.
References
- Bauman D, Perfield J, Harvatine K, Baumgard L. Regulation of fat synthesis by conjugated linoleic acid: lactation and the ruminant model. J Nutr. 2008;138:403–409. doi: 10.1093/jn/138.2.403. [DOI] [PubMed] [Google Scholar]
- Bionaz M, Loor J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366–381. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers R, van der Wielen R, Brink E, Hendriks H, Dorovska-Taran V, Mohede I. Effects of cis-9, trans-11 and trans-10, cis-12 conjugated linoleic acid (CLA) isomers on immune function in healthy men. Eur J Clin Nutr. 2003;57:595–603. doi: 10.1038/sj.ejcn.1601585. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai W, Blackshear P. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Jerri D, Dickinson E, Roberts A, Said T. Regulation of apoptosis during mammary involution by the p53 tumor suppressor gene. J Dairy Sci. 2002;85:1103–1110. doi: 10.3168/jds.S0022-0302(02)74171-4. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Szustakowski JD, Nanguneri NR, Mickanin C, Labow MA, Nohturfft A, Dev K, Sivasankaran R. Identification of Novel Genes and Pathways Regulating SREBP Transcriptional Activity. PLoS ONE. 2009;4:e5197. doi: 10.1371/journal.pone.0005197. [DOI] [PMC free article] [PubMed] [Google Scholar]