SUMMARY
Melanoma cells are resistant to Transforming Growth Factor-β (TGFβ)-induced cell cycle arrest. In this study, we investigated a mechanism of resistance involving a regulatory domain, called linker region, in Smad2 and Smad3, main downstream effectors of TGFβ. Melanoma cells in culture and in tumor samples exhibited constitutive Smad2 and Smad3 linker phosphorylation. Treatment of melanoma cells with the MEK1/2 inhibitor, U0126, or the two pan-CDK and GSK3 inhibitors, Flavopiridol and R547, resulted in decreased linker phosphorylation of Smad2 and Smad3. Overexpression of the linker phosphorylation-resistant Smad3 EPSM mutant in melanoma cells resulted in an increase in expression of p15INK4B and p21WAF1, as compared with cells transfected with wild-type Smad3. In addition, the cell numbers of EPSM Smad3-expressing melanoma cells were significantly reduced compared to wild-type Smad3-expressing cells. These results suggest that the linker phosphorylation of Smad3 contributes to the resistance of melanoma cells to TGFβ-mediated growth inhibition.
Keywords: Melanoma, TGFβ/Smad, Linker domain, Mitogen-Activated Protein Kinases, Cyclin-Dependent Kinases, p15INK4B, p21WAF1
INTRODUCTION
While Transforming Growth Factor beta (TGFβ) suppresses growth and/or induces apoptosis of normal melanocytes, malignant melanoma (MM) cells appear to be refractory to TGFβ-mediated growth inhibition and apoptosis (Hoek et al., 2006; Krasagakis et al., 1999; Rodeck et al., 1994; Rodeck et al., 1999). Few studies have addressed the possibility of a direct inactivation of TGFβ signaling intermediates (loss of expression or mutation of TβR or Smads) to explain the resistance of melanoma cells to TGFβ inhibitory effects (Poser et al., 2005; Rodeck et al., 1999). The mechanisms of resistance of melanoma cells to TGFβ growth inhibition and apoptosis (Lasfar and Cohen-Solal, 2010) are unlikely to be related to a global defect of the TGFβ signaling system, since aggressive MM cells utilize TGFβ as a prooncogenic factor (Berking et al., 2001; Javelaud et al., 2005; Javelaud et al., 2007; Yang et al., 2002; Zhang et al., 2009). The TGFβ produced by melanoma cells (Berking et al., 2001; Javelaud et al., 2007; Krasagakis et al., 1999; Reed et al., 1994; Rodeck et al., 1994; Van Belle et al., 1996) not only modifies the tumor microenvironment (Berking et al., 2001; Liu et al., 2005) but also directly contributes to the aggressiveness of the melanoma cells through autocrine activation of Smads (Javelaud et al., 2005; Javelaud et al., 2007). One major idea emerging from these studies is that melanoma cells have inactivated the tumor suppressive arm of TGFβ, while the prooncogenic signals of TGFβ have been preserved (Chen et al., 2009; Hoek et al., 2006; Javelaud et al., 2008; Javelaud et al., 2007). Therefore, a potential model of resistance to TGFβ-mediated tumor suppression that has to be considered implies that not all Smad transcriptional activities have been disrupted in melanoma cells. One model would predict that Smad activity would be inhibited on promoters involved in tumor suppression, such as p15 and p21, but fully competent for regulating genes involved in TGFβ prooncogenic effects, such as Snail, PA1-1 (plasminogen activator inhibitor -1), vimentin and N-cadherin, important for invasion and metastasis.
It has become increasingly clear that the so-called linker region of Smads between the MH1 (N-terminal) and MH2 (C-terminal) domains represents a regulatory region for Smad activity (Wrighton and Feng, 2008). In Smad2 and Smad3 linker regions, 4 potential sites for proline-directed protein kinases, including the extracellular-signal-regulated kinases (ERKs), were suggested to be the target of phosphorylation induced by activated RAS. A Smad3 mutant with these four sites mutated (EPSM mutant) was able to rescue RAS-transformed epithelial cells to growth inhibition by TGFβ (Kretzschmar et al., 1999). The hypothesis supported by this pioneering work, was that linker phosphorylation of Smad2 and Smad3 can lead to the inhibition of Smad2 and Smad3 transcriptional activities. Other groups extended these results by showing that the linker region of Smad3 was not only the target of ERKs (Matsuura et al., 2005) but also of Cyclin-Dependent Kinases ((CDKs) (Alarcon et al., 2009; Matsuura et al., 2004)). CDK phosphorylation of Smad3 was shown to prevent its antiproliferative functions (Matsuura et al., 2004). The different studies on Smad linker phosphorylation (Wrighton and Feng, 2008) suggested to us that constitutive linker phosphorylation occuring in cancer cells due to aberrant activation of the mitogen-activated protein kinase (MAPK) pathways or the CDKs could potentially result in resistance to TGFβ-mediated growth suppression in MM. Interestingly, one of the major oncogenic events in the genesis of MM is the constitutive activation of the RAS-regulated RAF/MEK/ERK pathway, one of the MAPK pathways (Smalley, 2009). In addition, the activated ERK signaling pathway upregulates another MAPK, the c-Jun NH2-terminal kinase (JNK), in a so-called “rewiring program”, in melanoma (Lopez-Bergami et al., 2007). Adding to the complexity of the mitogenic signaling, the levels of CDK activities are high in melanoma due to the suppression of CDK inhibitors (CKI) and the upregulation of the CDK positive regulators, the cyclins (Bennett, 2008; Halaban, 2005).
The present study was designed to investigate whether constitutive linker phosphorylation of the Smads contributes to the resistance of melanoma cells to TGFβ-mediated tumor suppression. We demonstrate constitutive linker phosphorylation of Smad2 and Smad3 in human melanoma cell lines, in contrast to normal melanocytes. We also show that MAPK and CDKs/GSK3 kinases are involved in the phosphorylation of the Smad linker domain. We then investigated whether a high level of Smad3 linker phosphorylation found in melanoma cells, but not in melanocytes, could fulfill a repressive role on Smad3 activity and therefore result in resistance to TGFβ-mediated tumor suppression. Overexpression of the Smad3 EPSM mutant (unable to be phosphorylated in the 4 linker sites) in melanoma cells increased p15 and p21 expression and impaired their proliferation. Our results strongly suggest that constitutive linker phosphorylation of Smad3 plays a role in resistance to TGFβ, produced by melanoma cells or their microenvironment.
RESULTS
Resistance of human melanoma cells to TGFβ induced growth inhibition and apoptosis is not due to a defective Smad activation
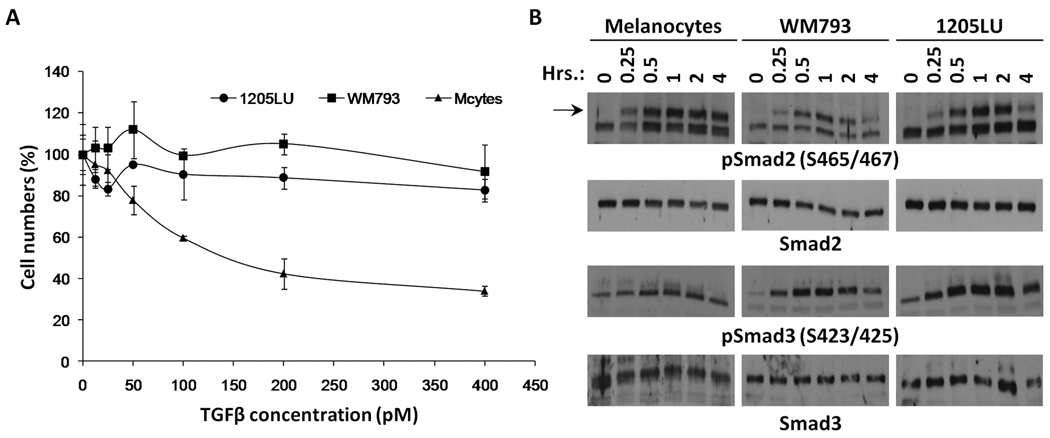
In order to test the biological response of melanoma lines and melanocytes to TGFβ, cells were treated with increasing concentrations of TGFβ for four days. At the end of this period, viable cells were counted. While melanocyte numbers dramatically decreased as a function of TGFβ concentration, melanoma lines were resistant to TGFβ treatment (Figure 1A).
Figure 1. Biological response of melanoma lines and melanocytes to TGFβ.
A. Melanoma cell lines are resistant to TGFβ treatment. 3–4 × 104 1205 LU, WM793 and melanocytes were seeded in 24-well plates. The following day, TGFβ was added at concentrations ranging from 12.5 to 400 pM. Each concentration was done in triplicate. Four days later, viable cells were counted using a Beckman Coulter. (cells with TGFβ/ cells without TGFβ) × 100 is expressed as a function of TGFβ concentration (pM). B. Phosphorylation of Smad2 and Smad3 induced by TGFβ treatment in melanoma lines, WM793 and 1205LU, and melanocytes. 4–5 × 105 cells were seeded in 35 mm dishes. The following day, cells were serum-starved for about 16 hours, before the addition of 100 pM of TGFβ for increasing periods of time, as indicated. At the end of the indicated period, whole cell extracts were made. Immunoblots were performed using antibodies against the C-terminal phosphorylated form of Smad2 (pSmad2(S465/467)) or Smad3 (pSmad3(S423/425)). Anti-total Smad2 and Smad3 antibodies were used for normalization.
In the classic model of the TGFβ signaling pathway, C-terminal serines at the SXS motif of Smad2 and Smad3 are phosphorylated by the TGFβ type I receptor following TGFβ activation. Considering the resistance of the melanoma lines to TGFβ growth inhibition and apoptosis, we examined whether Smad activation was intact in the melanoma lines. To analyze the induction of C-terminal phosphorylation of Smad2 and Smad3, cells were serum-starved for about 16 hours before the addition of TGFβ for increasing lengths of time (15, 30 minutes, 1, 2 and 4 hours). At each time point, whole cell extracts were prepared. Immunoblot experiments were performed, using antibodies recognizing phosphoSmad2 phosphorylated at Serines 465 and 467 and phosphoSmad3 phosphorylated at Serines 423 and 425. As shown in Figure 1B, the levels of phosphoSmad2 and phosphoSmad3 increased as early as 15 minutes following the addition of TGFβ in the melanoma lines. The peak of phosphorylation was reached at 1 hour, followed by a net decrease in phosphoSmad2 and phosphoSmad3 levels at 4 hours. The kinetics of activation were comparable for melanocytes and melanoma cells. However, phosphoSmad2 levels seemed to remain high even after 4 hours treatment of the melanocytes with TGFβ. The levels of total Smad2 and Smad3 were unchanged by TGFβ, indicating that TGFβ specifically increased the levels of Smad2 and Smad3 phosphorylation without modifying the expression level of the two proteins. These results suggest that the resistance of melanoma cell lines to growth inhibition and apoptosis is not related to an impairment of TGFβ-mediated phosphorylation of Smad2 and Smad3 at the C-terminal domain.
High constitutive level of phosphorylation of Smad2 and Smad3 linker regions in human melanoma lines
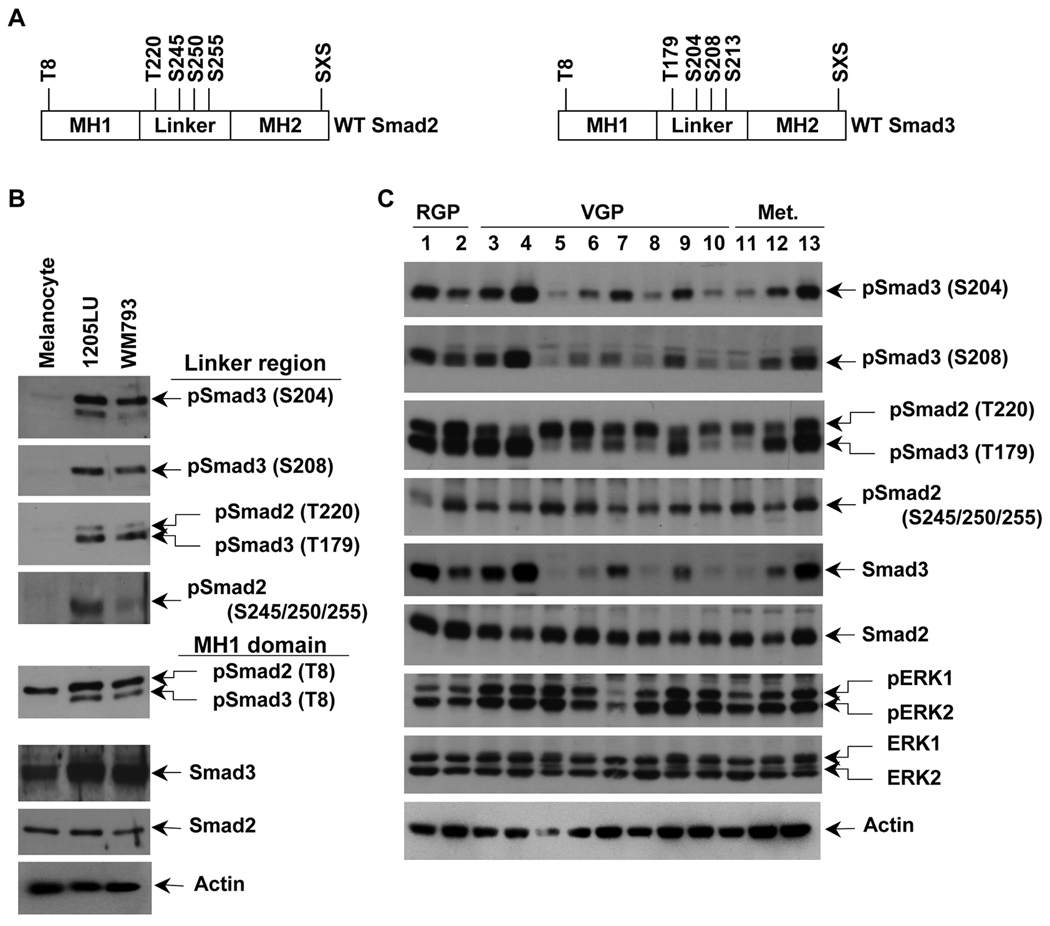
As previously mentioned, Smad2 and Smad3 are also regulated by phosphorylation at their linker region (in Figure 2A, the 4 sites for each Smad are represented). In order to test whether some differences in the levels of Smad2 and Smad3 linker phosphorylation could exist between melanoma lines and melanocytes, whole cell extracts from the serum-starved melanoma lines 1205LU and WM793, and the melanocytes were analyzed in immunoblot using polyclonal antibodies against specific phospho-amino acids in the linker region of Smad2 (phosphoSer245/250/255) and Smad3 (phosphoThr179, phosphoSer204 and phosphoSer208). As shown in Figure 2B, in the absence of any stimulation, melanoma lines have a high level of Smad2 phosphorylated at Ser245/250/255 and Smad3 phosphorylated at Thr179, Ser204 and Ser208. In contrast, normal melanocytes did not exhibit such phosphorylation events at the Smad2 and Smad3 linker sites. The antibody against phosphoSmad3 Thr179 also recognizes Smad2 phosphorylated at the corresponding Thr220, in melanoma lines but not in melanocytes. These results demonstrate that a high level of linker phosphorylated Smad2 and Smad3 exist in the melanoma lines but not in melanocytes. Thr8 in Smad3, previously identified as a major site for CDK phosphorylation (Matsuura et al., 2004) was also shown to be constitutively phosphorylated in the two melanoma lines but not in melanocytes (Figure 2B). We detected significant levels of constitutive linker phosphorylation for both Smad2 and Smad3 in a panel of human cell lines derived from melanoma at different levels of progression. Interestingly, we observed marked differences in the level of total Smad3 between the cell lines, suggesting that the expression level of Smad3 might be subject to downregulation in some melanoma cells. In addition, all these lines exhibit high levels of constitutively phosphorylated ERK1 and ERK2, reinforcing the hypothesis that these kinases might be involved in the Smad linker phosphorylation events (Figure 2C).
Figure 2. Constitutive linker phosphorylation of Smad2 and Smad3 in human melanoma lines.
A. Schematic representation of wild type Smad2 and Smad3. Smad2 has one phosphorylation site, threonine 8, in the MH1 domain (N-terminal domain), four phosphorylation sites in the linker region, threonine 220, serines 245, 250 and 255, and the SXS motif in the MH2 (C-terminal) domain. Smad3 has one phosphorylation site, threonine 8, in the MH1 domain (N-terminal domain), four phosphorylation sites in the linker region, threonine 179, serines 204, 208 and 213, and the SXS motif in the MH2 domain. B. High constitutive phosphorylation of Smad2 and Smad3 at the linker region in WM793 and 1205LU melanoma lines and not in melanocytes. The two melanoma lines, 1205LU and WM793, and the melanocytes were serum-starved overnight, before protein extraction. Immunoblot experiments using polyclonal antibodies against: Both phosphoSmad3 (Thr179) and phosphoSmad2 (Thr220); phosphoSmad2 (Ser245/250/255); phosphoSmad3 (Ser204); phosphoSmad3 (Ser208); both phosphoSmad3 (Thr8) and phosphoSmad2 (Thr8); total Smad3; total Smad2. C. Constitutive linker phosphorylation of Smad2 and Smad3 in a panel of human melanoma cell lines from different melanoma stages. The human melanoma lines were derived from Radial Growth Phase (RGP): 1: WM35; 2: 1552c; Vertical Growth Phase (VGP): 3: WM793; 4: WM278; 5: A2058; 6: SKMEL2; 7: SKMEL5; 8: SKMEL28; 9: SKMEL31; 10: HT144; primary melanomas and metastatic (Met.) melanomas: 11: WM1617; 12: C8161; 13: 1205LU. Cells were serum-starved overnight, before protein extraction as in B. p: phospho. S: Ser; T: Thr. Actin expression was used as a control.
Partial decrease of phosphorylation in Smad2 and Smad3 linker region in the presence of the MEK1/2 inhibitor, U0126
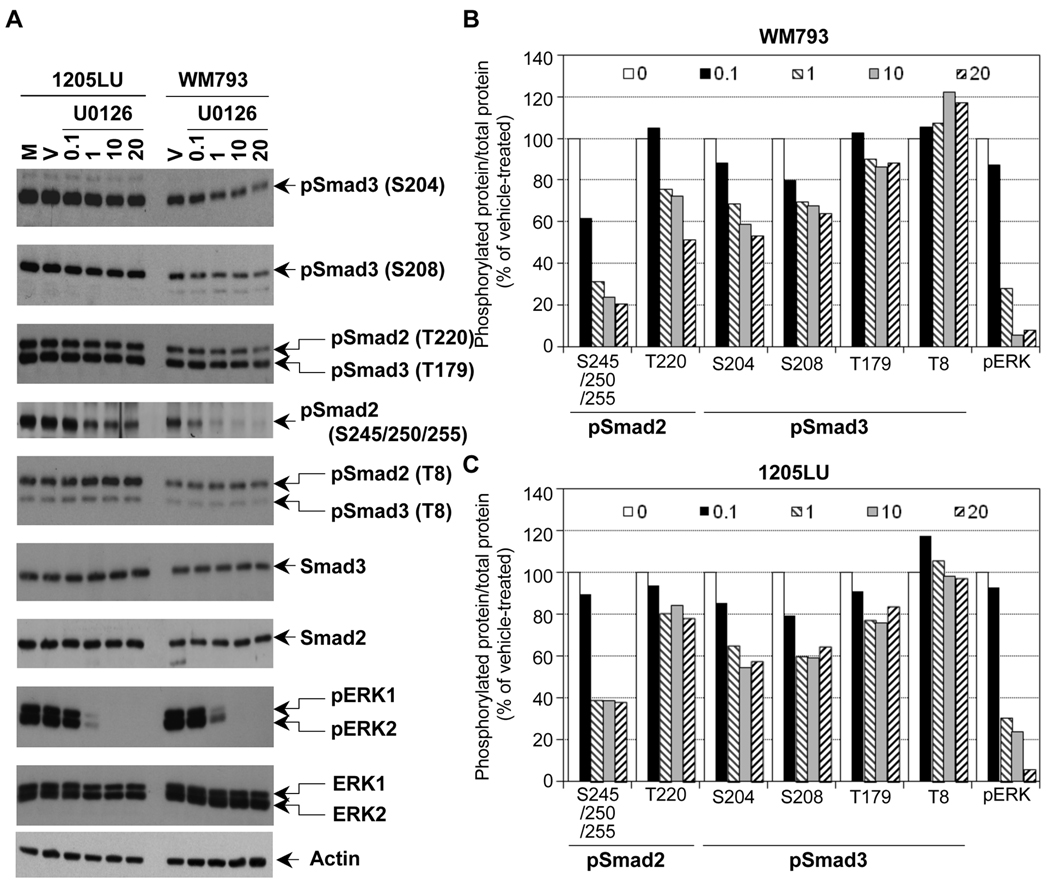
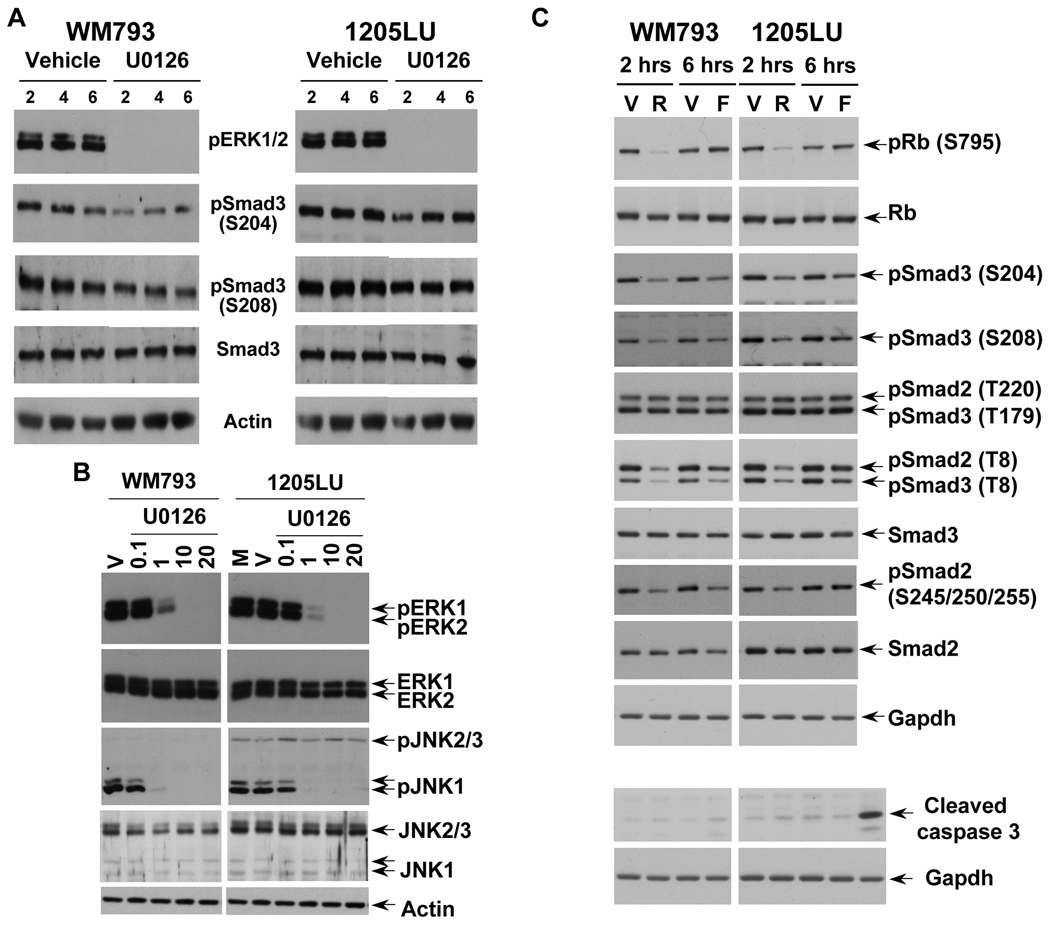
In order to assess the role of ERK kinases in the phosphorylation of Smad2 and Smad3 linker regions in the melanoma lines, we treated the same lines with the MEK1/2 inhibitor U0126. As shown in Figure 3A, inhibition of MEK1/2 by 1 µM of U0126 resulted in a dramatic inhibition of ERK phosphorylation. However, a longer exposure showed that a high level still remained within the melanoma lines at this concentration of U0126. At 10 and 20 µM, U0126 completely abolished ERK phosphorylation. In these conditions (total inhibition of ERK phosphorylation), we observed a significant reduction of phosphorylation at the cluster of serines 245/250/255, and to a lesser extent at Thr220 in Smad2 linker domain, and at Ser204, Ser208 and Thr179 in Smad3 linker region. Thr8 in Smad3, which is not a direct target of ERK (Matsuura et al., 2004; Matsuura et al., 2005), does not seem to be affected by U0126. The densitometric analysis of the bands (Figure 3B and 3C) confirmed that the phospho-sites that were the most affected by U0126 treatment were the cluster of serines and Thr220 for Smad2, and Ser204 and 208 in Smad3. Phosphorylation of Thr 179 in Smad3 was less affected by U0126, and Thr8 phosphorylation was not modulated. The fact that the inhibition of phosphorylation at Ser204, Ser208 and Thr179 of Smad3 is not complete while no activity of ERK kinases exist within the melanoma lines can have two explanations: 1) Because the time of treatment with U0126 is two hours, it is possible that the dephosphorylation of Smad3 is not completed within these two hours. 2) The phosphorylation sites Ser204, Ser208 and Thr179 could be the target of other kinases, such as JNK or CDK. In order to test the first possibility, WM793 and 1205LU melanoma cell lines were incubated in the presence of vehicle or U0126 for 2, 4 or 6 hours. For both lines, 6 hours treatment with U0126 did not result in complete dephosphorylation at Serines 204 and 208 of the Smad3 linker region, while resulting in complete disappearance of pERK1 and pERK2 (Figure 4A). 24 hours treatment with U0126 gave the same result and some toxicity was noted with this longer treatment (data not shown). These results suggest that, in addition to ERK, other kinases may be involved in the phosphorylation of the Smad3 linker region at these two sites.
Figure 3. The linker phosphorylation of Smad2 and Smad3 in human melanoma lines involves ERK.
A. Partial inhibition of Smad2 and Smad3 linker phosphorylation in the presence of the MEK1/2 inhibitor, U0126. 24 hours post seeding, cells were serum-starved for about 16 hours, before the addition of the MEK1/2 inhibitor, U0126, at the indicated concentrations for 2 hours. Cells left two hours in serum-free medium (M) or treated with the vehicle DMSO (V) for two hours were used as controls. Whole cell extracts were then prepared. Immunoblots were performed with polyclonal antibodies against: Phosphorylated ERK1 (pERK1) and ERK2 (pERK2); total ERK1 and ERK2; Smad2 and Smad3 and the phosphorylated forms of Smad2 and Smad3, as in Figure 2. Densitometric analysis of the immunoblots in B and C. B. Ratio of phosphorylated protein/total protein for pSmad2, pSmad3 and pERK is represented as a function of U0126 concentration (0.1; 1; 10 and 20 µM as indicated in the legend; 0 is for vehicle) for the WM793 melanoma cell line. C. Ratio of phosphorylated protein/total protein for pSmad2, pSmad3 and pERK is represented as a function of U0126 concentration for the 1205LU melanoma cell line.
Figure 4. JNK1 and CDK/GSK3 are involved in Smad linker phosphorylation.
A. Longer treatment with U0126 did not result in complete dephosphorylation of the Smad3 linker region at Serines 204 and 208. 24 hours post seeding, cells were serum-starved for about 16 hours, before the addition of 20µM of the MEK1/2 inhibitor, U0126 for 2, 4 and 6 hours. Cells treated with the vehicle DMSO for 2, 4 and 6 hours were used as controls. Immunoblots were performed with polyclonal antibodies against: Phosphorylated ERK1 (pERK1) and ERK2 (pERK2); phosphoSmad3 (Ser204); phosphoSmad3 (Ser208); total Smad3; Actin. B. U0126 also inhibits the constitutive phosphorylation of JNK1 on the canonical Thr183/Tyr185 MKK4/7 sites in WM793 and 1205LU human melanoma lines. Cells were serum-starved for about 16 hours, before the addition of the MEK1/2 inhibitor, U0126, at the indicated concentrations for 2 hours. Cells left two hours in serum-free medium (M) or treated with the vehicle (V) for two hours were used as controls. Immunoblots were performed with polyclonal antibodies against: Phosphorylated ERK1 (pERK1) and ERK2 (pERK2); phosphorylated JNK1 (pJNK1) and JNK2/3 (pJNK2/3); total ERK1/2 and total JNK1 and JNK2/3. C. Treatment with pan-CDK and GSK3 inhibitors inhibit phosphorylation at Serine 204 and 208 in Smad3 and at the cluster of serines 245/250/255 in Smad2 linker regions. 24 hours post seeding, cells were serum-starved for 16 hours, before adding R547 (R; 600 nM) for 2 hours or Flavopiridol (F; 300 nM) for 6 hours. For the controls, vehicle (V) was added either 2 hours (for R547) or 6 hours (for flavopiridol). Immunoblots were performed using the antibodies previously described in Figures 2 and 3. In addition, antibodies against total Rb and Rb, phosphorylated at serine 795, were used to monitor R547 and Flavopiridol effects on a known substrate of CDK2 and CDK4. Gapdh was used as an internal control. Lower part of panel C: Treatment with R547 (600 nM) or Flavopiridol (300 nM) does not increase caspase 3 cleavage. The last lane on the right is a positive control for cleaved caspase 3.
JNK1 may be implicated in Smad2 and Smad3 linker phosphorylation
In melanoma, sustained ERK activation increases JNK activity. This increased JNK activity requires the phosphorylation of JNK on Thr183/Tyr185 by the canonical MKK4/7 pathway (Lopez-Bergami et al., 2007). Therefore, if JNK activity is high in the melanoma lines, we should observe constitutive phosphorylation on these two MKK4/7 canonical sites. As shown in Figure 4B, 1205LU and WM793 had constitutively hyperphosphorylated JNK (pJNK) at the basal state (M or V lanes): pJNK1 only in the case of WM793, and both pJNK1 and pJNK2/3 for 1205LU. Interestingly, we observed that U0126 not only inhibited ERK phosphorylation, but also resulted in inhibition of JNK1 phosphorylation specifically, in both lines. The concentration necessary to completely inhibit phosphorylation of ERK1 and ERK2 (10 µM) was sufficient to totally inhibit JNK1 phosphorylation in both lines. From these experiments, we can conclude that in the case of WM793, while depriving the cells of any ERK or JNK1 activity, U0126 did not result in complete dephosphorylation of the Smad3 linker region. In the case of 1205LU, U0126 did not affect the levels of phosphorylated JNK2/3. If we hypothesize that these kinases are still active in the presence of U0126, they could be involved in the remaining linker phosphorylation of the Smads. Presently, the lack of a specific JNK inhibitor ((Bain et al., 2007); Dr. P. Cohen, Dundee, Scotland, UK, personal communication) represents an obstacle to the direct and definitive involvement of JNK1/2/3 activities in Smad2 and Smad3 linker phosphorylation.
CDK/GSK3 are involved in the linker phosphorylation of Smad2 and Smad3
In order to determine whether CDKs played a role in the linker phosphorylation of Smad2 and Smad3, we used two different types of pan-CDK/GSK3 inhibitors: Flavopiridol, a flavone derivative (Carlson et al., 1996; Rizzolio et al., 2010; Sedlacek, 2001), and R547, a diaminopyrimidine derivative (Chu et al., 2006; DePinto et al., 2006; Rizzolio et al., 2010). The effect of R547 and flavopiridol was assessed first on Rb phosphorylation at serine 795, a CDK2 and CDK4 site (Halaban, 2005). Rb phosphorylation was inhibited at 600 nM R547, but not at 300 nM Flavopiridol. We verified that at these concentrations, apoptosis was not occuring, as assessed by cleaved caspase 3 (Figure 4C) and cleaved PARP analysis (data not shown). Phosphorylation at threonine 8, in Smad2 and Smad3, was inhibited by R547 and Flavopiridol, confirming that this threonine was a good substrate for CDKs in vivo (Baughn et al., 2009; Matsuura et al., 2004). In the linker domain of Smad3, Serine 204 and S208 phosphorylation decreased as a result of R547 and flavopiridol treatments. Threonine 179 of Smad3 and threonine 220 of Smad2 were insensitive to R547 and flavopiridol. The phosphorylation of Smad2 at the cluster of serines (245/250/255) was reduced in the presence of R547 in both lines and in the presence of flavopiridol only in WM793 melanoma cell line. These results suggest that some of the linker phosphorylation sites in Smad2 (cluster of serines) and Smad3 (serines 204 and 208) might be the targets of CDKs and/or GSK3. Even in the event specific inhibitors of CDK2 and CDK4/6 were commercially available, well described compensatory mechanisms operating in the absence of a particular CDK have been demonstrated and constitute a technical challenge to precisely define the identity of the CDK(s) mediating the Smad linker phosphorylation events (Wang et al., 2009).
Expression of a linker phosphorylation mutant of Smad3 into melanoma cells impairs their proliferation
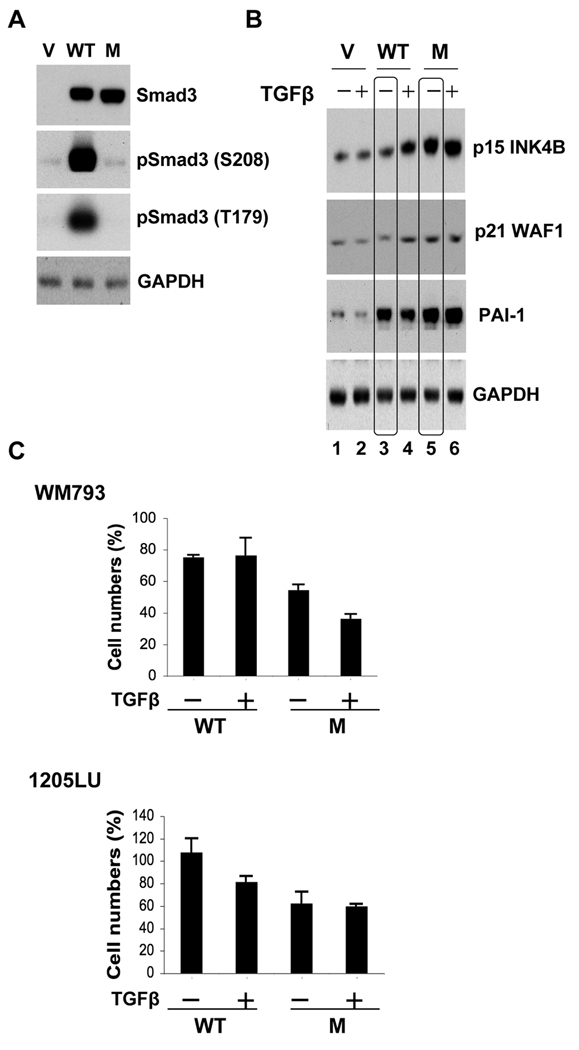
Studies performed in mouse embryonic fibroblasts from Smad2 or Smad3 deficient mice, as well as in HaCaT cells and few other epithelial systems suggest that Smad3 might have a more important role in TGFβ-mediated cell cycle arrest than Smad2 (Massague, 2008), while a more recent study proposed a role for Smad2 in apoptosis mediated by TGFβ in undifferentiated, stem cell-like, pluripotent prostate epithelial cells (Yang et al., 2009). Although the contribution of Smad3 and Smad2 in the cytostatic response has not been rigorously investigated in melanoma cells, we chose to investigate whether constitutive Smad3 linker phosphorylation could impair the sensitivity of melanoma cells to TGFβ-mediated growth inhibition and/or apoptosis. We used a Smad3 linker phosphorylation mutant called Smad3 EPSM, which has a threonine to valine substitution at position 179 and serine to alanine substitutions at positions 204, 208 and 213 (Kretzschmar et al., 1999; Matsuura et al., 2010; Sekimoto et al., 2007). Therefore, this mutant cannot be phosphorylated in the linker region. If constitutive linker phosphorylation of Smad3 inhibits its activity as an effector in the cytostatic and/or proapoptotic effects of TGFβ in melanoma cells, introducing the EPSM mutant into these cells should lead to their resensitization to TGFβ. In order to test this hypothesis, melanoma cells were transfected with the wild-type (WT) Smad3 or EPSM Smad3 expression vectors. As shown in Figure 5A, WT Smad3 and EPSM Smad3 were expressed at similar levels in melanoma cells. By cotransfecting a GFP (Green Fluorescent Protein) expression vector with either the empty vector, the WT Smad3 or the EPSM Smad3 expression vectors, we verified that the transfection efficiencies were comparable between conditions (data not shown). The high level of expression achieved for both WT Smad3 and EPSM Smad3 explains why we had to do a low exposure, preventing us from seeing the endogenous Smad3 in the vector-transfected cells. However, a longer exposure allowed us to observe the endogenous Smad3 in the vector-transfected cells, and to verify that the EPSM Smad3-transfected cells had a level of phosphorylation at serine 208 and threonine 179 identical to the vector-transfected cells (not shown). As expected, analysis of the linker phosphorylation sites showed that the WT Smad3-transfected cells exhibited a higher level of phosphorylation at serine 208 and threonine 179 than the vector-transfected cells (Figure 5A).
Figure 5. Expression of a linker phosphorylation mutant of Smad3 into melanoma cells impairs their proliferation.
WM793 and 1205LU cells were transfected with the vector (V), WT Smad3 (WT) or the linker phosphorylation mutant of Smad3, EPSM Smad3 (M) expression vectors. A. 24 hours post transfection, whole cell lysates were prepared for the analysis of Smad3 expression and the linker phosphorylation at serine 208 and threonine 179 in the WM793 melanoma cells. GAPDH expression was used as a control. (B). In parallel, 24 hours post transfection, WM793 melanoma cells were incubated in the absence (−) or presence (+) of 200 pM of TGFβ for 48 hours and extracted for protein expression analysis of p15INK4B, p21WAF1 and PAI-1. (C). 24 hours post transfection, the transfected WM793 and 1205LU melanoma cells were incubated in the absence (−) or presence (+) of TGFβ for 72 hours and counted. Cell numbers (%) have been normalized with the number obtained for the vector-transfected cells. Each experiment is representative of 4 experiments done in triplicate.
Some of the genes involved in the tumor suppressive transcriptional responses to TGFβ under the control of TGFβ-activated Smad complexes have been characterized. These include the cyclin-dependent kinase inhibitors, p15INK4B and p21WAF1. The induction of p15 and p21 expression results in CDK inhibition and cell cycle arrest (Massague, 2008). To determine whether EPSM Smad3 was more effective than WT Smad3 in inducing the expression of p15 and p21 in the presence of TGFβ, transfected cells were incubated in the absence or presence of TGFβ for 48 hours (Figure 5B). In the vector-transfected cells, TGFβ was unable to induce the expression of p15 or p21, which may be one of the reasons explaining the resistance of these cells to TGFβ. In the WT Smad3 transfected cells, TGFβ induced a moderate increase in the levels of p15 and p21. However, the expression of EPSM Smad3 was sufficient to induce a significant increase in p15 and to a lesser extent in p21 levels, even in the absence of TGFβ (compare lane 3 to lane 5). These results suggest that the activity of EPSM Smad3 is increased on the endogenous p15 and p21 promoters as compared with WT Smad3.
The plasminogen activator inhibitor-1 (PAI-1), a member of the urokinase plasminogen activator (uPA) system, plays a causal role in invasion and metastasis and a high level of PAI-1 in the primary tumor is one of the most informative markers of a poor prognosis in several types of human cancer (Andreasen, 2007; Ulisse et al., 2009). In order to test whether the linker phosphorylation of Smad3 inhibits the expression of PAI-1, involved in TGFβ prooncogenic effects, we analyzed the levels of PAI-1 in the transfected melanoma cells. As shown in Figure 5B, the over-expression of the WT Smad3 (linker phosphorylated) resulted in an increase in PAI-1 levels, even in the absence of TGFβ, as compared with the vector-transfected cells. When the linker phosphorylation was disrupted in EPSM Smad3-transfected cells, we observed an increase in PAI-1 levels comparable to that in the WT Smad3-transfected cells. Therefore, the linker phosphorylation (in WT Smad3) does not prevent the expression of PAI-1.
WM793 and 1205LU transfected cells (WT Smad3 or EPSM Smad3) were incubated for three days in the presence or absence of TGFβ and the number of viable cells was counted (Figure 5C). For both lines, the expression of EPSM Smad3 resulted in reduced numbers of cells as compared with the WT Smad3-expressing melanoma cells, even in the absence of TGFβ. These results indicate that the overexpression of a Smad3 mutant which cannot be phosphorylated in the linker region, impairs the proliferation ability of melanoma cells, likely through increased p15 and p21 expression. Therefore, the linker phosphorylation of Smad3 might contribute to the resistance of melanoma cells to TGFβ-mediated growth inhibition, but does not affect the expression levels of PAI-1.
Linker phosphorylation of Smad2 and Smad3 in human melanoma samples: A tissue microarray study
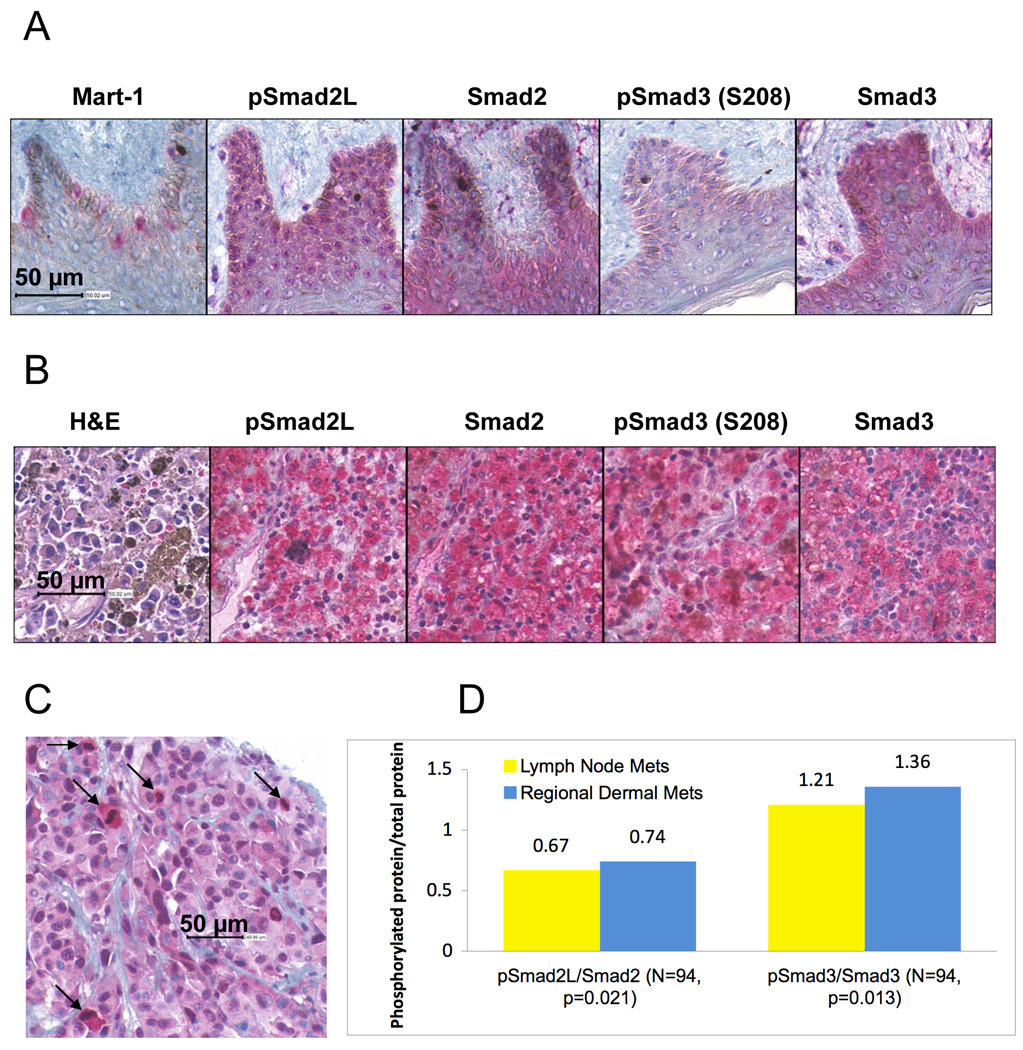
In order to examine the Smad linker phosphorylation in human melanoma samples, we performed an immunohistochemical analysis of phosphoSmad2 (S245/250/255) and phosphoSmad3 (S208), Smad2 and Smad3 using a melanoma tissue microarray (TMA). We simultaneously examined normal human skin sections for the expression of linker phosphorylated Smads and Smad2 and Smad3 (Figure 6). In Figure 6A, immunostaining with an antibody against Mart-1 highlights the presence of melanocytes in the basal layer of the epidermis. In the skin, light positive staining for phosphoSmad2 (S245/250/255) in the melanocytes was undistinguishable from that of adjacent keratinocytes, in contrast to negative staining for phophoSmad3 (S208) in the melanocytes and keratinocytes. Positive staining was observed for Smad2 and Smad3 in both melanocytes and keratinocytes. Positive staining for phosphoSmad2 (S245/250/255) and phosphoSmad3 (S208) was seen primarily in the cytoplasm of melanoma cells (Figure 6B). Figure 6C shows one area with high phosphoSmad2 (S245/250/255) staining predominantly in melanoma cells undergoing mitosis. For each of the melanoma samples (47 regional dermal metastases and 47 lymph node metastases), we determined the ratio of effective staining intensity between phosphoSmad2 and Smad2 and between phosphoSmad3 and Smad3. As shown in Figure 6D, we found that these two ratios were slightly increased in the regional dermal metastases as compared with lymph node metastases in a statistically significant way.
Figure 6.
Smad linker phosphorylation and Smad expression in normal human skin sections and melanoma samples. 4-µm sections of normal skin (A) and of the melanoma tissue microarray containing human melanoma samples (B), were stained using antibodies against Mart-1 (only normal skin), phosphoSmad2 (S245/250/255; pSmad2L), phosphoSmad3 (S208), Smad2 and Smad3. (C) One area of a melanoma core showing an association between high staining for phosphoSmad2 (S245/250/255) and mitotic figures (arrows). (D). Comparison of the ratios phosphoSmad2 (S245/250/255)/Smad2 (pSmad2L/Smad2) and phosphoSmad3 (S208)/Smad3 (pSmad3/Smad3) between regional dermal metastases (Mets) and lymph node metastases. N is the total number of samples submitted to one-tailed t-test. P value was generated through one-tailed t-test. The images were sub-samples from the whole slide images at 40X equivalent.
DISCUSSION
The development and progression of malignant melanoma appears to be characterized by resistance to TGFβ tumor suppressive effects on one hand (Lasfar and Cohen-Solal, 2010), and the autocrine/paracrine activation of the TGFβ pathway on the other hand. This suggests that high levels of TGFβ may provide a selective advantage to invasive and metastatic melanomas (Javelaud et al., 2008), as proposed for carcinomas (Massague, 2008). Therefore, the resistance to TGFβ tumor suppression represents a critical step in melanoma cell aggressiveness. The two TGFβ resistant melanoma cell lines used in our study still exhibit intact Smad activation, as evidenced by an increase in the C-terminal phosphorylation of the main downstream effectors, Smad2 and Smad3, with kinetics comparable to those observed in normal human melanocytes. This is in accordance with previous published results demonstrating functional TGFβ receptor complexes in melanoma cell lines (Chen et al., 2009; Javelaud et al., 2007). We tested the idea that the Smad linker region whose regulatory role on Smad2 and Smad3 transcriptional activity has been documented (Liu, 2006; Wrighton and Feng, 2008), could be differentially phosphorylated between normal melanocytes and melanoma cells. This hypothesis was reinforced by studies conducted in epithelial systems and showing the presence of phosphorylation sites for MAPK (ERK, JNK, p38; (Kamaraju and Roberts, 2005; Matsuura et al., 2005; Sekimoto et al., 2007) and CDK (Alarcon et al., 2009; Matsuura et al., 2004; Matsuzaki et al., 2009; Millet et al., 2009; Wang et al., 2009) in the Smad linker domain. In contrast to melanocytes, both MAPK and CDK, which are proline-directed serine-threonine kinases, are aberrantly activated in melanoma (Bennett, 2008; Halaban, 2005; Smalley, 2009). In the absence of treatment, melanoma cell lines demonstrated constitutive linker phosphorylation at threonine 179, serines 204 and 208 in Smad3 and at threonine 220 and at a cluster of serines (245/250/255) in Smad2. Basal (constitutive) phosphorylation at serines 250/255 and 208/213 in Smad2 and Smad3 linker regions was reported recently in two melanoma cell lines but no analysis of the kinases was performed (Chen et al., 2009). Immunohistochemical analysis of Smad phosphorylation at the cluster of serines (245/250/255) for smad2 and at serine 208 for Smad3 in melanoma samples confirmed the existence of constitutive linker phosphorylation of these two transcription factors in human tumors (Figure 6B, C and D). Some level of Smad2 linker phosphorylation in melanocytes of the skin (Figure 6A) might be achieved through paracrine activities originating from the melanocytes environment, such as the keratinocytes. By contrast, primary cultures of melanocytes were found negative for both Smad2 and Smad3 linker phosphorylation (Figure 2B). Interestingly, another group described the absence of Smad2 and Smad3 linker phosphorylation, in primary cultures of melanocytes (Chen et al., 2009).
Our next step was to investigate whether MAPK and CDK/GSK3 could be involved in the constitutive phosphorylation of Smad2 and Smad3 linker regions. The involvement of MAPK, in particular ERK1/2 and/or JNK, in these phosphorylation events, was revealed by using U0126, which inhibits both ERK1/2 and JNK1 constitutive phosphorylation/activation in the two melanoma cell lines (Figures 3 and 4). The so-called JNK inhibitor SP600125, routinely used by numerous investigators, is not a specific JNK inhibitor, since it inhibits 13 of 30 kinases tested with similar or greater potency than JNK isoforms ((Bain et al., 2007) and references therein). The authors of these studies recommend to discontinue the use of SP600125 as a JNK inhibitor in cell-based assays (Bain et al., 2007). In addition, it appears that no specific pharmacological inhibitor of JNK is available commercially yet (Dr. P. Cohen, personal communication). Therefore, it is difficult at the present time to definitely involve JNK isoforms, as kinases phosphorylating the Smad linker region. Our data suggest that they might be involved. As soon as specific pharmacological inhibitors of JNK become available, we will investigate whether these kinases participate directly in Smad linker phosphorylation.
The two pan-CDK inhibitors, R547 and flavopiridol, used in our study are currently in clinical trials for solid tumors (Lapenna and Giordano, 2009). R547 appears to decrease the linker phosphorylation of Smad2 at the cluster of serines and Smad3 at serines 204 and 208 more efficiently than Flavopiridol in our conditions. This difference is likely due to the fact that R547 exhibited a greater inhibitory activity for CDK2 and CDK4 as compared to flavopiridol (Lapenna and Giordano, 2009). For the 1205LU line, a CDK4 mutation at codon 22 (K22Q) (Smalley et al., 2008; Yang et al., 2005) was demonstrated, a mutation resulting in loss of p16 binding (Ceha et al., 1998). This p16-binding deficient CDK4 is therefore insensitive to p16 inhibition in the 1205LU melanoma line. This observation could partially explain that in our conditions, inhibition of Smad linker phosphorylation by the two pan-CDK inhibitors was less efficient in 1205LU than in WM793. Serines 204 and 208 seem to be good substrates of ERKs and/or JNK1 and CDKs/GSK3 in the studied melanoma cells. Further studies are required to determine the nature of the kinases phosphorylating threonine 179, in addition to ERK and/or JNK. Threonine 220 of Smad2 is probably also the target of other kinases in addition to ERKs and/or JNK, while the cluster of serines (for which we could not distinguish between the sites) responded well to U0126 and R547 treatments, suggesting that combined treatment with U0126 and R547 could result in complete dephosphorylation at this cluster. The use of antibodies against each of the sites in the cluster would allow us to test the possibility that differences in kinase specificity exist among the sites.
Interestingly, TGFβ was revealed as a major inducer of Smad linker phosphorylation in a wide variety of cellular systems. The different kinases involved in each of these studies include JNK, CDKs, GSK3β, depending on the phosphorylation site and the cellular context (Alarcon et al., 2009; Chen et al., 2009; Gao et al., 2009; Matsuzaki et al., 2009; Millet et al., 2009; Mori et al., 2004; Sekimoto et al., 2007; Wang et al., 2009). TGFβ-induced activation of JNK (independently of Smads) has been well documented (Zhang, 2009) and could explain the TGFβ-induced linker phosphorylation of Smads mediated by JNK. However, the mechanism by which TGFβ results in CDKs or GSK3β phosphorylation of Smad3 linker region remains uncertain. As previously mentioned, the TGFβ-based autocrine loop often described in melanoma (Javelaud et al., 2008) could constitute one of the many aberrant stimulations resulting in the constitutive linker phosphorylation observed in Smad2 and Smad3. As expected, the major cellular dysregulated pathways and kinases causing constitutive Smad linker phosphorylation will depend on the genetic and epigenetic contexts characterizing each melanoma cell. Therefore, the precise kinases, ERK1, ERK2, JNK1, JNK2, JNK3, the different CDKs and GSK3 isoforms participating in Smad linker phosphorylation, may very well differ between different melanoma cells, but a sufficient level of Smad linker phosphorylation would be maintained by any of these activated kinases, or a combination of these kinases to disrupt Smad antiproliferative activity.
An additional level of Smad2 and Smad3 regulation is achieved by phosphatases that dephosphorylate the linker region of the Smads. Using different mammalian systems, two independent studies showed that the so-called Small C-terminal Domain Phosphatases SCP1, SCP2 and SCP3 (serine/threonine phosphatases) dephosphorylate the Smad2 and Smad3 linker region in vitro and in vivo (Sapkota et al., 2006; Wrighton et al., 2006). RNA interference-mediated depletion of SCPs resulted in an increase in Smad2 and Smad3 linker phosphorylation. Another way for melanoma cells to maintain a high level of Smad2 and Smad3 linker phosphorylation would be to down regulate the expression of these specific linker phosphatases. Interestingly, microarray analysis of phosphatase gene expression in melanoma showed that 7 out of 47 serine/threonine phosphatases were down regulated, but the identity of these kinases was not the primary interest of this study (McArdle et al., 2005). The down regulation of the SCP proteins in melanoma could therefore reinforce the effects of activated pathways maintaining a high level of linker phosphorylation. The analysis of this hypothesis is underway.
To address the role of the linker phosphorylation of Smad3 in the resistance to TGFβ-mediated growth inhibition, we overexpressed a Smad3 linker phosphorylation mutant (EPSM Smad3, unable to be phosphorylated in the linker domain, Figure 5A) in melanoma cells, and analyzed the effects on p15INK4B and p21WAF1 expression and on the numbers of melanoma cells, in the absence or presence of TGFβ. We observed that EPSM Smad3 overexpression induced an increase in p15INK4B and p21WAF1 expression, even in the absence of exogenous TGFβ (Figure 5B). The two melanoma lines WM793 and 1205LU secrete active TGFβ (Berking et al., 2001; Javelaud et al., 2007) likely responsible for constitutive C-terminal phosphorylation of Smad3 (Javelaud et al., 2007) and Smad-dependent transcription (Rodeck et al., 1999) in these lines. In addition, the TGFβ secreted by 1205LU melanoma cells was able to induce paracrine responses in WI-36 human lung fibroblasts (Javelaud et al., 2007). Based on these data, we can suggest that the active TGFβ secreted by these melanoma cells could be sufficient to induce endogenous p15 and p21 expression when Smad3 activity has been derepressed by the mutation at the four linker sites. The increase in p15 and p21 expression, observed in the WT Smad3-transfected cells only in the presence of TGFβ (Figure 5B), could be explained by the high expression level of the exogenous WT Smad3. The high expression of WT Smad3 could then overwhelm the MAPK and/or CDK activities (or others) present in the melanoma cells, resulting in some unphosphorylated Smad3 (in the linker region), competent for p15 and p21 induction. However, overexpression of EPSM Smad3 in WM793 and 1205LU melanoma cells resulted in significantly reduced cell numbers as compared with WT Smad3-expressing cells. Our study furthers a recent published work reporting that in contrast to melanocytes, melanoma cell lines demonstrated constitutive linker phosphorylation of Smad2 and Smad3 on serines 250/255 and serines 208/213 respectively. The presence of the protooncogene Ski was required for a further increase of Smad3 linker phosphorylation by TGFβ (Chen et al., 2009). Therefore, TGFβ-induced linker phosphorylation has been observed in melanoma, and in the context of autocrine TGFβ signaling, could play a role in inhibiting Smad3 antiproliferative activity in a sustained way. In addition, in the melanoma cell line A375, the TGFβ-dependent increase in Smad3 linker phosphorylation was associated with resistance to c-myc downregulation and repression of p21 on one hand and TGFβ-mediated induction of the plasminogen activator inhibitor -1 (PAI-1) one the other hand, (Chen et al., 2009), suggesting the possible dual function of the Smad3 linker phosphorylation in the resistance to tumor suppression by TGFβ and promotion of TGFβ pro-oncogenic effects. Our results suggest that while Smad3 linker phosphorylation might contribute to the resistance to TGFβ-mediated cell growth inhibition in melanoma, it does not seem to inhibit the expression of PAI-1. According to our model, Smad activity would be inhibited on promoters involved in cell growth inhibition, such as p15 and p21, but fully competent for regulating some of the genes involved in TGFβ prooncogenic effects, such as PA1-1. This model is in accordance with the well-demonstrated fact that not all Smad transcriptional activities have been disrupted in melanoma cells.
Finally, we could envision that the complete identification of the kinases involved in the Smad3 linker phosphorylation will be rewarding, since dephosphorylating the Smad3 linker region, using agents targeting these kinases, could potentially reactivate Smad3 and resensitize the melanoma cells to their own secreted TGFβ or the TGFβ produced in the tumor microenvironment, thereby interfering with melanoma development.
METHODS
Cell lines
Primary cultures of melanocytes kindly provided by Dr M. Herlyn (Wistar Institute, Philadelphia, PA) were maintained in MCDB153 medium containing 2% FBS, 10% chelated FBS, 2 mM L-Glutamine, 20 pM cholera toxin, 60 pM basic FGF (Fibroblast Growth Factor), 100 nM Endothelin 3 and 10 ng/ml SCF (Stem Cell Factor).
WM1552c, WM793, WM278, WM1617 and 1205LU were kindly provided by Dr M. Herlyn. WM35 was purchased from ATCC. These lines were cultured in MCDB153/L-15 (4/1 ratio) medium containing 2% FBS, 5 µg/ml Insulin and 1.7 mM Calcium Chloride.
Cell count experiments
2–4 × 104 cells were seeded in 24-well plates. The following day, increasing concentrations of TGFβ (25 to 400 pM, in triplicate) were added to the plates. Four days following the addition of TGFβ, cells were washed in phosphate-buffered saline, trypsinized and counted, using a Beckman Coulter Vi-CELL 1.00. For the cell count experiments following transfection (see below), 200 pM TGFβ was used.
Transfection and TGFβ treatment
The CS2 vector and the wild type (WT) Smad3, and Smad3 EPSM (four sites mutated in the Smad3 linker region) expression vectors were previously described (Matsuura et al., 2010). Using the Nucleofector™ technology (Amaxa, Lonza), WM793 and 1205LU cells were transiently transfected with CS2, WT Smad3 or Smad3 EPSM expression vectors (4 µg per 106 cells). 24 hours post-transfection, cells were left untreated or treated with 200 pM TGFβ for 48 hours (immunoblot for p15, p21 and PAI-1 expression levels) or 72 hours (cell counts).
Basal phosphorylation of the Smad linker domains and treatments for immunoblot analysis
To analyze the basal phosphorylation of Smad2 and Smad3 linker domain, cells were seeded in 35 mm dishes or 6-well plates (4–5 × 105/ dish or well). The following day, cells were serum-starved for about 16 hours before being extracted. For TGFβ treatment, serum-starved cells were incubated for 15, 30 minutes, 1, 2 and 4 hours in the presence of 100 pM TGFβ and extracted at the end of each time point. For U0126 treatment, serum-starved cells were treated for 2, 4 or 6 hours, in the presence of 0.1, 1, 10 or 20 µM of U0126, depending on the experiments. For the treatments with R547 (600 nM; Hoffman-La-Roche) and Flavopiridol (300 nM; Sigma-Aldrich, St Louis, MO), serum-starved cells were incubated for 2 and 6 hours respectively.
Immunoblotting
Cells were harvested, washed with phosphate-buffered saline, and extracted in the presence of protease and phosphatase inhibitors (Roche) as previously described (Ge et al., 2004). Equal amounts of protein were subjected to polyacrylamide gel electrophoresis. After transfer onto nitrocellulose membranes, immunoblots were performed using antibodies against: Both phosphoSmad3 (Thr179) and phosphoSmad2 (Thr220); phosphoSmad3 (Ser204); phosphoSmad3 (Ser208); both phosphoSmad3 (Thr8) and phosphoSmad2 (Thr8) (Matsuura et al., 2004); phosphoSmad2 (Ser245/250/255); total Smad2; phosphorylated ERK1 (pERK1) and ERK2 (pERK2); phosphorylated JNK1 (pJNK1) and JNK2/3 (pJNK2/3); total ERK1/2 and total JNK1 and JNK2/3; phosphoRb (Ser 795); Rb; p15INK4B; p21WAF1; Actin; GAPDH; cleaved caspase 3; (purchased from Cell Signaling); total Smad3 (Zymed); PAI-1 (BD Transduction Laboratories), depending on the experiments.
Description of the Melanoma Tissue Microarray (TMA)
The TMA was built from de-identified non-linked melanoma tissue samples obtained by the Tissue Analytical Services of the Cancer Institute of New Jersey, in an IRB-approved and HIPAA-compliant fashion. The TMA contains 47 regional dermal metastases, 49 lymph nodes metastases and 4 distant skin metastases, with four fold redundancy (four cores/sample). The melanoma TMA has been validated and stained for the melanocyte specific markers, Mart-1 and tyrosinase.
Detection of Smad linker phosphorylation by immunohistochemistry
Normal skin control slides and the melanoma TMA were stained simultaneously for each of the antibodies tested. Human melanoma TMA and normal skin slides were deparaffinized and antigen retrieval was performed using extended CC1 treatment (Cell Conditioning Solution, Ventana Medical Systems). Rabbit polyclonal antibodies against pSmad2 (Ser 245/250/255. Cell signaling), pSmad3 (Ser 208; Abgent), Smad2 (Abcam) and Smad3 (Abcam) were applied and incubated at 37°C for 1 or 2 hours. Donkey anti-Rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was then applied and incubated at 37°C for 60 min, followed by chromogenic detection using the RedMap kit (Ventana Medical Systems). Slides were counterstained with Hematoxylin and dehydrated and cleared before coverslipping from Xylene. The antibody against Mart-1 was a mouse monoclonal (Ventana) and the secondary antibody (Ventana's Universal Secondary Antibody) was applied for 12 minutes before detection.
Imaging and sub-sampling (cropping)
Glass slides were digitized under 20× objective on a Trestle MedScan Whole Slide Scanner. The resulting image archives were viewed and sub-sampled via the Trestle web service hosted on a Windows 2003 Server R2 server.
Quantification and statistics
A mixture of three colors is present on each specimen prepared for this experiment: red from the immunohistochemical stain, brown from the presence of melanin and blue from hematoxylin counter stain. In order to isolate the immunohistochemical signal for quantification, three representative color vectors, also called the principal color vectors (PCVs) (Chen et al., 2004), were extracted from strategically prepared specimens immunostained but not counterstained with hematoxylin or just stained with hematoxylin.
Using our previously reported algorithm and software package (Chen et al., 2004), the scanned TMA specimens were first registered to correctly cross-reference spatial coordinates of detected TMA cores. The PCVs were subsequently used to decompose all core images into three constituent staining maps. The immunohistochemical signal, the red map, from each core image was analyzed to generate three distinct measurements: the integrated staining intensity (ISI), the effective staining area (ESA), and the effective staining intensity (ESI), among which the ESI is a reliable measure of staining intensity and was shown to be closely correlated with expert interpretation (Goodell et al., 2010). After consolidating scores coming from redundant TMA cores, the ratios of pSmad2/Smad2 and pSmad3/Smad3 were generated for 47 patients in the regional dermal metastases group and 47 patients in the lymph node metastasis group, respectively.
SIGNIFICANCE.
Resistance to Transforming Growth Factor β (TGFβ)-mediated tumor suppression represents a crucial step in melanoma aggressiveness, since it is coupled with the ability of TGFβ to drive the oncogenic process. We demonstrated the MAPK and CDK/GSK3-mediated constitutive linker phosphorylation of Smads in melanoma. We strongly suggested that constitutive Smad3 linker phosphorylation contributed to resistance to TGFβ-mediated cell cycle arrest likely by interfering with p15 and p21 expression. Restoring sensitivity to TGFβ by inhibiting phosphorylation at the Smad linker region may represent a new therapeutic avenue in the treatment of melanoma.
AKNOWLEDGEMENTS
This work was supported by a National Health Institute grant 3RO1CA041556-20S1 (M.R/K.C.S.), a Research Development Award (K.C.S.) from the Cancer Center Support Grant CCSG P30CA072720, and a Research Scholar Grant from the American Cancer Society 116683-RSG-09-087-01-TBE (K.C.S.). This research was funded also in part, by grants from the National Institutes of Health through contracts 5R01LM009239-04 and 3R01LM009239-04S2 from the National Library of Medicine and contracts 9R01CA156386-05A1 and SAIC/NCI#29XS154 from the National Cancer Institute (D.F.). We thank Joseph L.-K. Chan for some lysates from melanoma cell lines (A2058, SKMEL2, SKMEL5, SKMEL28, SKMEL31, HT144 and C8161), and Dr. Khanh G. Dinh for preliminary experiments with flavopiridol. We thank Allen Lovey from Hoffmann-La Roche for R547. We thank Lei Cong and the Tissue Analytical Services of the Cancer institute of New Jersey for the immunohistochemical staining of the control and melanoma tissue microarray slides. We are grateful to Dr. Marina Chekmareva in the Department of Pathology at UMDNJ for assessing the quality and specificity of the immunohistochemical stainings.
REFERENCES
- Alarcon C, Zaromytidou AI, Xi Q, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen PA. PAI-1 - a potential therapeutic target in cancer. Curr. Drug Targets. 2007;8:1030–1041. doi: 10.2174/138945007781662346. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn LB, Di Liberto M, Niesvizky R, Cho HJ, Jayabalan D, Lane J, Liu F, Chen-Kiang S. CDK2 phosphorylation of Smad2 disrupts TGF-beta transcriptional regulation in resistant primary bone marrow myeloma cells. J. Immunol. 2009;182:1810–1817. doi: 10.4049/jimmunol.0713726. [DOI] [PubMed] [Google Scholar]
- Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell Melanoma Res. 2008;21:27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- Berking C, Takemoto R, Schaider H, Showe L, Satyamoorthy K, Robbins P, Herlyn M. Transforming growth factor-beta1 increases survival of human melanoma through stroma remodeling. Cancer Res. 2001;61:8306–8316. [PubMed] [Google Scholar]
- Carlson BA, Dubay MM, Sausville EA, Brizuela L, Worland PJ. Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK) 2 and CDK4 in human breast carcinoma cells. Cancer Res. 1996;56:2973–2978. [PubMed] [Google Scholar]
- Ceha HM, Nasser I, Medema RH, Slebos RJ. Several noncontiguous domains of CDK4 are involved in binding to the P16 tumor suppressor protein. Biochem Biophys Res Commun. 1998;249:550–555. doi: 10.1006/bbrc.1998.9183. [DOI] [PubMed] [Google Scholar]
- Chen D, Lin Q, Box N, et al. SKI knockdown inhibits human melanoma tumor growth in vivo. Pigment Cell Melanoma Res. 2009;22:761–772. doi: 10.1111/j.1755-148X.2009.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Reiss M, Foran DJ. A prototype for unsupervised analysis of tissue microarrays for cancer research and diagnostics. IEEE Trans. Inf. Technol. Biomed. 2004;8:89–96. doi: 10.1109/titb.2004.828891. [DOI] [PubMed] [Google Scholar]
- Chu XJ, Depinto W, Bartkovitz D, et al. Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-diflu oro-6-methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity. J. Med. Chem. 2006;49:6549–6560. doi: 10.1021/jm0606138. [DOI] [PubMed] [Google Scholar]
- Depinto W, Chu XJ, Yin X, et al. In vitro and in vivo activity of R547: a potent and selective cyclin-dependent kinase inhibitor currently in phase I clinical trials. Mol. Cancer Ther. 2006;5:2644–2658. doi: 10.1158/1535-7163.MCT-06-0355. [DOI] [PubMed] [Google Scholar]
- Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massague J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Rajeev V, Subramanian G, et al. Selective inhibitors of type I receptor kinase block cellular transforming growth factor-beta signaling. Biochem. Pharmacol. 2004;68:41–50. doi: 10.1016/j.bcp.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Goodell L, Chen W, Javidian P, Chekmareva M, Hu J, Foran DJ. Use of computer assisted analysis to facilitate tissue microarray interpretation. United States and Canadian Academy of Pathology 2010 Annual Meeting; March 20–26, 2010; Washington, D. C.. 2010. [Google Scholar]
- Halaban R. Rb/E2F: a two-edged sword in the melanocytic system. Cancer Metastasis Rev. 2005;24:339–356. doi: 10.1007/s10555-005-1582-z. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Javelaud D, Alexaki VI, Mauviel A. Transforming growth factor-beta in cutaneous melanoma. Pigment Cell Melanoma Res. 2008;21:123–132. doi: 10.1111/j.1755-148X.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- Javelaud D, Delmas V, Moller M, Sextius P, Andre J, Menashi S, Larue L, Mauviel A. Stable overexpression of Smad7 in human melanoma cells inhibits their tumorigenicity in vitro and in vivo. Oncogene. 2005;24:7624–7629. doi: 10.1038/sj.onc.1208900. [DOI] [PubMed] [Google Scholar]
- Javelaud D, Mohammad KS, Mckenna CR, et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- Kamaraju AK, Roberts AB. Role of Rho/ROCK and p38 MAP kinase pathways in transforming growth factor-beta-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J. Biol. Chem. 2005;280:1024–1036. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- Krasagakis K, Kruger-Krasagakes S, Fimmel S, Eberle J, Tholke D, Von Der Ohe M, Mansmann U, Orfanos CE. Desensitization of melanoma cells to autocrine TGF-beta isoforms. J. Cell Physiol. 1999;178:179–187. doi: 10.1002/(SICI)1097-4652(199902)178:2<179::AID-JCP7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- Lasfar A, Cohen-Solal KA. Resistance to transforming growth factor beta-mediated tumor suppression in melanoma: are multiple mechanisms in place? Carcinogenesis. 2010;31:1710–1717. doi: 10.1093/carcin/bgq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Smad3 phosphorylation by cyclin-dependent kinases. Cytokine Growth Factor Rev. 2006;17:9–17. doi: 10.1016/j.cytogfr.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhang F, Lee J, Dong Z. Selective induction of interleukin-8 expression in metastatic melanoma cells by transforming growth factor-beta 1. Cytokine. 2005;31:241–249. doi: 10.1016/j.cyto.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Lopez-Bergami P, Huang C, Goydos JS, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Chiang KN, Lai CY, He D, Wang G, Ramkumar R, Uchida T, Ryo A, Lu K, Liu F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J. Biol. Chem. 2010;285:1754–1764. doi: 10.1074/jbc.M109.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Wang G, He D, Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry. 2005;44:12546–12553. doi: 10.1021/bi050560g. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Kitano C, Murata M, Sekimoto G, Yoshida K, Uemura Y, Seki T, Taketani S, Fujisawa J, Okazaki K. Smad2 and Smad3 phosphorylated at both linker and COOH-terminal regions transmit malignant TGF-beta signal in later stages of human colorectal cancer. Cancer Res. 2009;69:5321–5330. doi: 10.1158/0008-5472.CAN-08-4203. [DOI] [PubMed] [Google Scholar]
- Mcardle L, Rafferty MM, Satyamoorthy K, Maelandsmo GM, Dervan PA, Herlyn M, Easty DJ. Microarray analysis of phosphatase gene expression in human melanoma. Br. J. Dermatol. 2005;152:925–930. doi: 10.1111/j.1365-2133.2005.06454.x. [DOI] [PubMed] [Google Scholar]
- Millet C, Yamashita M, Heller M, Yu LR, Veenstra TD, Zhang YE. A negative feedback control of transforming growth factor-beta signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J. Biol. Chem. 2009;284:19808–19816. doi: 10.1074/jbc.M109.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Matsuzaki K, Yoshida K, et al. TGF-beta and HGF transmit the signals through JNK-dependent Smad2/3 phosphorylation at the linker regions. Oncogene. 2004;23:7416–7429. doi: 10.1038/sj.onc.1207981. [DOI] [PubMed] [Google Scholar]
- Poser I, Rothhammer T, Dooley S, Weiskirchen R, Bosserhoff AK. Characterization of Sno expression in malignant melanoma. Int. J. Oncol. 2005;26:1411–1417. [PubMed] [Google Scholar]
- Reed JA, Mcnutt NS, Prieto VG, Albino AP. Expression of transforming growth factor-beta 2 in malignant melanoma correlates with the depth of tumor invasion. Implications for tumor progression. Am. J. Pathol. 1994;145:97–104. [PMC free article] [PubMed] [Google Scholar]
- Rizzolio F, Tuccinardi T, Caligiuri I, Lucchetti C, Giordano A. CDK inhibitors: from the bench to clinical trials. Curr. Drug Targets. 2010;11:279–290. doi: 10.2174/138945010790711978. [DOI] [PubMed] [Google Scholar]
- Rodeck U, Bossler A, Graeven U, Fox FE, Nowell PC, Knabbe C, Kari C. Transforming growth factor beta production and responsiveness in normal human melanocytes and melanoma cells. Cancer Res. 1994;54:575–581. [PubMed] [Google Scholar]
- Rodeck U, Nishiyama T, Mauviel A. Independent regulation of growth and SMAD-mediated transcription by transforming growth factor beta in human melanoma cells. Cancer Res. 1999;59:547–550. [PubMed] [Google Scholar]
- Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- Sedlacek HH. Mechanisms of action of flavopiridol. Crit. Rev. Oncol. Hematol. 2001;38:139–170. doi: 10.1016/s1040-8428(00)00124-4. [DOI] [PubMed] [Google Scholar]
- Sekimoto G, Matsuzaki K, Yoshida K, Mori S, Murata M, Seki T, Matsui H, Fujisawa J, Okazaki K. Reversible Smad-dependent signaling between tumor suppression and oncogenesis. Cancer Res. 2007;67:5090–5096. doi: 10.1158/0008-5472.CAN-06-4629. [DOI] [PubMed] [Google Scholar]
- Smalley KS. Understanding Melanoma Signaling Networks as the Basis for Molecular Targeted Therapy. J. Invest. Dermatol. 2009;130:28–37. doi: 10.1038/jid.2009.177. [DOI] [PubMed] [Google Scholar]
- Smalley KS, Lioni M, Dalla Palma M, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol. Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulisse S, Baldini E, Sorrenti S, D'armiento M. The urokinase plasminogen activator system: a target for anti-cancer therapy. Curr. Cancer Drug Targets. 2009;9:32–71. doi: 10.2174/156800909787314002. [DOI] [PubMed] [Google Scholar]
- Van Belle P, Rodeck U, Nuamah I, Halpern AC, Elder DE. Melanoma-associated expression of transforming growth factor-beta isoforms. Am. J. Pathol. 1996;148:1887–1894. [PMC free article] [PubMed] [Google Scholar]
- Wang G, Matsuura I, He D, Liu F. Transforming growth factor-beta-inducible phosphorylation of Smad3. J. Biol. Chem. 2009;284:9663–9673. doi: 10.1074/jbc.M809281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Feng XH. To (TGF)beta or not to (TGF)beta: fine-tuning of Smad signaling via post-translational modifications. Cell Signal. 2008;20:1579–1591. doi: 10.1016/j.cellsig.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J. Biol. Chem. 2006;281:38365–38375. doi: 10.1074/jbc.M607246200. [DOI] [PubMed] [Google Scholar]
- Yang G, Rajadurai A, Tsao H. Recurrent patterns of dual RB and p53 pathway inactivation in melanoma. J. Invest. Dermatol. 2005;125:1242–1251. doi: 10.1111/j.0022-202X.2005.23931.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Wahdan-Alaswad R, Danielpour D. Critical role of Smad2 in tumor suppression and transforming growth factor-beta-induced apoptosis of prostate epithelial cells. Cancer Res. 2009;69:2185–2190. doi: 10.1158/0008-5472.CAN-08-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Dukhanina O, Tang B, et al. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. J. Clin. Invest. 2002;109:1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang F, Tsan R, Fidler IJ. Transforming growth factor-beta2 is a molecular determinant for site-specific melanoma metastasis in the brain. Cancer Res. 2009;69:828–835. doi: 10.1158/0008-5472.CAN-08-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]