Abstract
Mitochondria and plastids have their own DNAs and are regarded as descendants of endosymbiotic prokaryotes. Organellar DNAs are not naked in vivo but are associated with basic proteins to form DNA-protein complexes (called organelle nuclei). The concept of organelle nuclei provides a new approach to explain the origin, division, and inheritance of organelles. Organelles divide using organelle division rings (machineries) after organelle-nuclear division. Organelle division machineries are a chimera of the FtsZ (filamentous temperature sensitive Z) ring of bacterial origin and the eukaryotic mechanochemical dynamin ring. Thus, organelle division machineries contain a key to solve the origin of organelles (eukaryotes). The maternal inheritance of organelles developed during sexual reproduction and it is also probably intimately related to the origin of organelles. The aims of this review are to describe the strategies used to reveal the dynamics of organelle division machineries, and the significance of the division machineries and maternal inheritance in the origin and evolution of eukaryotes.
Keywords: mitochondria, plastids, division machinery, organelles, maternal inheritance, Cyanidioschyzonmerolae
Introduction
Howard and Pelc (1953) proposed the concept that the cell cycle is classified into gap 1 (G1), DNA synthesis period (S) and gap 2 (G2) on basis of chromosomes’ behaviors.1) Since then, the cell cycle has been taken to mean a mitotic cycle that considers only the dynamics of cell nuclei. Molecular biological studies of cell cycles using yeast cells and cultured animal cells have accelerated this assumption.2) However, basic eukaryotic cells contained at least a minimal set of double-membrane-bound compartments; the cell nucleus, mitochondria, and plastids (chloroplasts), and single-membrane-bound compartments; for example, the microbody (peroxisome), the Golgi apparatus, the endoplasmic reticulum and lysosomes (vacuoles). The original cells increased the number of compartments (organelles), except the nucleus, during the evolution of eukaryotes into Amoebozoa, Bikonta and Opisthokonta.3) All life depends on photosynthesis by plastids in plants for food and oxygen, while mitochondria are important organelles for energy supply in all organisms. Despite their importance, very little information is known about origin, division, and heredity of mitochondria and plastids. One hundred years ago, Correns and Baur (1909) independently discovered that different plastid phenotypes were inherited in a non-Mendelian fashion in higher plants.4) If their discoveries are taken as the first step, the history of the study of the inheritance of mitochondria and plastid genomes can be classified into four steps. 1) the discovery of non-Mendelian plastid phenotypes. 2) the discovery of plastid and mitochondrial DNA as carriers of non-Mendelian genes. 3) sequencing of entire genomes of mitochondria and plastids, and 4) the discovery of a nucleus in mitochondria and plastids (the nucleoid; a complex of DNA and proteins).4)
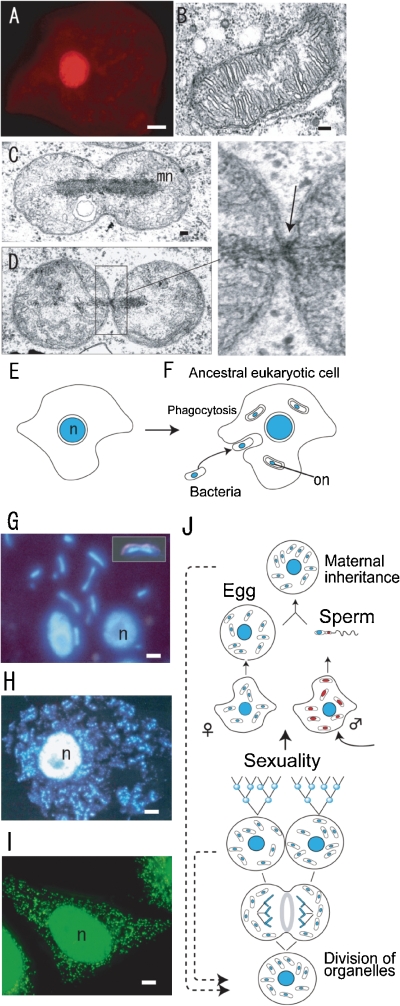
The discovery of mitochondrial (mt-) and plastidal (pt-) nuclei was important for the study of organelle division and inheritance. After the discovery of non-Mendelian phenotypes, a new phase in the study of traits inherited in a non-Mendelian manner began with genetic studies of microorganisms. However, these studies could not identify the non-Mendelian factor in the heredity of the cells. Electron microscopy demonstrated that plastids of alga5) and the mitochondria of animals contained DNA-like fibrils in electron transparent regions.6) Although molecular biological studies on the structure of organellar DNA and the sequencing of organelle genomes has progressed,7) it has proved difficult to observe DNA in mitochondria and plastids in vivo (Fig. 1A, B). Thus, it was thought that organelle DNAs, as well as bacterial DNAs, were naked in cells. In 1973–1974, we discovered that mitochondria in the slime mould Physarum polycephalum contained a large amount of mt-DNA, which was organized with proteins to form a rod-shaped electron dense mt-nucleus (Fig. 1C, D).6,8) The mitochondria divided according to a simple sequence, including spherule-, ovoid- and dumbbell-shaped structures, along with mt-nuclear division (Fig. 1C, D).6,9) The process of mt-division can be clearly classified into two main events: mt-nuclear division and mitochondriokinesis (also called mitochondrial division). These events in the division of the Physarum mitochondria seemed to be similar to those in the division of Rickettsiella melolonthae,9) suggesting that mitochondria could be regarded as descendants of endosymbiotic prokaryotes (Fig. 1E, F). The hypothesis is confirmed by the genome sequencing of Rickettsiella.10)
Figure 1.
Fluorescence (A, G, H, I) and Nomarski/fluorescence (insert in G) microscopy, electron micrographs (B, C, D) and diagrammatic representations of cell division (E, F, J). A. A human cell stained with acridine orange. Only the cell nucleus (red) was visualized. B. The mitochondrion in Chlamydomonas reinhardtii is composed of many cristae around an electron transparent region. C and D. In Physarum polycephalum, the mitochondria contain electron dense rod-shaped mitochondrial nuclei (mt-nuclei) and mitochondriokinesis occurs using a small ring (indicated by an arrow in the enlarged image of D) after mt-nuclear division (mn in C). E and F. Change of concept of the cell. In addition to the cell nucleus (n), the DNAs in organelles (mitochondria and plastids) that are regard as ancestors of bacteria can be visualized as organellar nuclei (on in F). G, H, and I. The fluorescence images demonstrate mt-nuclei, pt-nuclei (rod-shaped or small spots) and the cell nucleus (n) in P. polycephalum (G), Nicotiana tabacum (H), and Homo sapiens (I). J. Organellar dynamics during life cycle of eukaryotes with non-sexual and sexual reproduction stages. Scale bars: 10 µm (A, H), 5 µm (I), 1 µm (G), and 0.1 µm (B,C). C and D are from Ref. 6, H is from Ref. 13, I is from Dr. Sasaki-Higashiyama, N.
High-resolution fluorescence microscopy was developed, which enabled visualization of mt-nuclei, containing a small amount of DNA, in the mitochondria of slime mould P. polycephalum (Fig. 1G), Nicotiana tabacum (BY2) (Fig. 1H), and Homo sapiens (Hela cell) (Fig. 1I). Furthermore, the concept of mt-nuclei expanded to plastids.11) In plastids, the DNAs are also organized into pt-nuclei. The HU-like protein (histone-like protein of plastid) in pt-nuclei has a similar role to “Glom (a protein inducing agglomeration of mitochondrial chromosome)” in mt-nuclei.11,12) Thus, all eukaryotic cells contain two types of nuclei; cell nuclei and mt-nuclei, with plants also containing a third type; pt-nuclei.6) Statistical and quantitative observations of individual mt- and pt-nuclei made it possible to study the dynamic behavior of organelle division during the cell cycle of non-sexual reproduction and to study cytoplasmic (maternal, paternal and biparental) inheritance of organelle genomes (DNAs) during sexual reproduction (Fig. 1J).13) Thus, they could also be used to study the mechanisms of organelle division and cytoplasmic inheritance. We also hypothesized that their mechanisms would provide an important key for understanding the evolutionary origin of eukaryotic cells.4,14) In this review, I describe the mechanisms and machineries of organelle division and briefly summarize the mechanism of organelle inheritance, as recently reviewed.4) Finally, the implications of the mechanisms of organelle division and inheritance regarding the evolutionally origin of eukaryotic cells are discussed.
Strategies for studying the mechanism of mitochondrial and plastid division during non-sexual reproduction
Strategy 1. Identifying the most suitable organism for studying organellar division.
The mitochondria and plastids in almost all eukaryotes divide after organelle-nuclear division, as in slime mould. When daughter mitochondria were pinched off at the final stage of division, a small ring-like structure (Fig. 1D) and a smooth bridge between daughter mitochondria were observed in P. polycephalum and Nitella flexilis,6) respectively. These structures have been found to be universal in eukaryotes.15) However, it proved very difficult to study the cellular and molecular mechanisms of organelle division in eukaryotes for the following reasons. In Bikonta (plantae), Opisthokonta (animalia and fungi) and Amoebozoa (amoebae, slime mould), 1) the cells contain a large number of mitochondria, 2) mitochondrial shapes are irregular, 3) mitochondrial division occurs randomly (sometimes mitochondrial fusion also occurs), and 4) mitochondria move actively in whole cells (Figs. 1J and 2A).
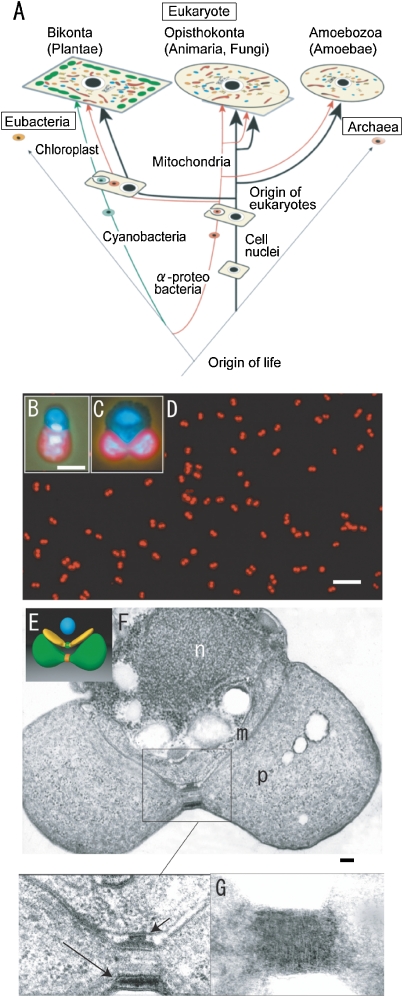
Figure 2.
Schematic representation of the eukaryotic phylogenic tree (A) and the model cell (E), fluorescence microscopy (B–D) and electron micrographs (F, G) of C. merolae cells. A. Mitochondria and plastids originated from α-proteobacteria and cyanobacteria of eubacteria, respectively, and increased in number during evolution. B and C. The photographs show a cell nucleus (top), mitochondrial nucleus (middle), and plastid nucleus (bottom) in interphase (B) and dividing cells (C) after staining with DAPI. The plastids emit red autofluorescence. D. The synchronized dividing plastids show a dumbbell-shape. E. Model of the dividing cell showing a spherical cell nucleus (blue), v-shaped mitochondrion with division machinery (green), and a dumbbell-shaped plastid with division machinery (red). F. Dividing cell shows cross sections of cell nucleus (n), mitochondrion (m), plastid (p), dividing electron dense mt- (short arrows in enlarged image) and pt-division machineries (long arrows in enlarged image) at the division sites. G. The contracted outer pt-division machinery looks like a bundle of fine filaments, 5–7 nm in diameter, after negative staining. Scale bars: 10 µm (D), 1 µm (B, C), and 0.1 µm (F). E is from Dr. Yoshida, Y., F is from Ref. 3, G is from Ref. 23.
As a basis for study for multi-organelle organisms, we searched for the simplest suitable organism for the study of organellar division under several conditions using the eukaryotic phylogenetic tree shown in Fig. 2A. Tamiya (1966) achieved synchronous culture of the green alga Chlorella ellipsoidea using light/dark cycles.16) Plastids seemed to be an essential organelle as a sensor of light for synchronization. Lim et al. (1986) showed that red alga are the most primitive eukaryotes by phylogenic analysis of 5S ribosomal RNA.17) Primitive eukaryotes live in hot extremophile environments.18) Therefore, we searched for simpler hot spring red algae among the Cyanidiophyceae (Fig. 2A). This group comprises of three genera, Cyanidioschyzon merolae, Cyanidium caldarium, and Galdieria sulphurarium. C. caldarium (18.5 Mbp) and G. sulphurarium (24 Mbp), which were isolated from a Kusatsu hot spring in Japan, have four endospores and 16 endospores, respectively.14) In 1986, we discovered electron dense plastid division (PD) rings (machineries) in C. caldarium, but could not isolate the rings from the cells because the cell had rigid cell walls.14) We then turned our attention to the simplest alga C. merolae (13 Mbp)14) and purified completely the alga from a mixture with C. caldarium.19) C. merolae is a small (1.5–2 µm diameter) unicellular organism that inhabits sulfate-rich hot springs (pH 1.5, 42 ℃). One cell division of C. merolae is equivalent to reproduction-an entirely new organism is created. The cells offer unique advantages for studies of mitochondria and plastid division because they do not have a rigid cell wall and contain just one nucleus, one mitochondrion, and one plastid (Fig. 2B, C).3,20) The alga also has the smallest genome of all photosynthetic eukaryotes, and contains a minimal set of small membrane-bound compartments; for example, a microbody (peroxisome), a single endoplasmic reticulum, a single Golgi apparatus, and a few lysosomes (vacuoles).21)
Strategy 2. The cell cycle is composed of a mitotic cycle, a mitochondrial division cycle, and a plastid cycle, which could be synchronized by light/dark cycles.
To elucidate the molecular mechanisms of mt- and pt-division, one would usually create and analyze many mutants by molecular biological techniques. However, we challenged to isolate the very small organellar division machineries that are directly responsible for organelle division. When cells in stationary phase were transferred into a new medium under a 12:12 h light/dark cycle, plastids, mitochondria, and cell nuclei divided synchronously, in that order, soon after the initiation of the dark period (Fig. 2D). This synchronized system proved useful for studying the cellular and molecular mechanisms of organelle divisions (Fig. 2D).3) The cell cycle C. merolae comprises three division cycles: the mitotic cycle (G1, S, G2, M), the mitochondrial division cycle (mitochondrial (mt) G1, mtS, mtG2, mtM) and plastid division cycle (plastid (pt) G1, ptS, ptG2, and ptM).3,20) Plastid division could be visualized in living cells. Large electron-dense mt-division rings (outer and inner membrane MD-rings) were observed and, as predicted by the discovery of PD rings in C. caldarium,13,14) electron dense pt-division rings (outer and inner membrane PD-rings) were discovered at the division sites (Fig. 2E–G). As the outer PD- and MD-rings contracted, they grew thicker and maintained a constant volume, while the thickness of the inner PD-ring did not change, and its volume decreased at a constant rate as it contracted. Thus, the outer PD- and MD-rings seemed to generate the motive force for contraction at the dividing site. Although these MD-rings become smaller in size in higher animals and plants, and the PD-rings also become simpler in higher plants, they are universal structures in eukaryotes.3,15) Mitochondria and plastids divided using their own division machineries. The pt-division machineries in C. merolae are 5 µm in circumference, which is larger than the mt-division machineries (1 µm in circumference); therefore, we first isolated the pt-division machineries from these cells.22,23)
Strategy 3. Isolation and fine structural analysis of plastid division machineries.
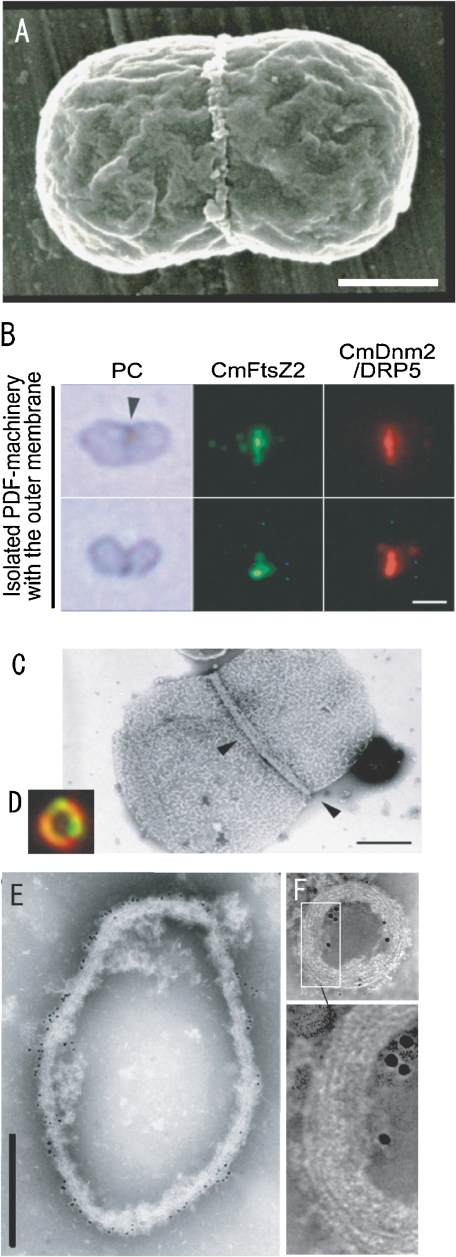
We isolated dividing plastids with their pt-division machineries from a synchronized culture of C. merolae (Figs. 2G and 3A) and treated them with nonionic detergent Nonidet P-40 to extract stroma and thylakoids.22) The inner rings and outer ring remained intact after the treatment. Negative staining revealed that the outer ring consists of a bundle of 5-nm filaments (Fig. 2G), in which globular proteins are spaced 4.8 nm apart.22) Sliding of the fine filaments seemed to generate the motive force for contraction of the machineries.13,22) Moreover, the structure and properties of the filaments are unlike those of known cytoskeletal filaments. We also examined dividing phase-specific proteins of plastids as candidate components of the rings. However, as the outer membranes could not be solved completely by NP-40 treatment and as the architectural proteins in the pt-division machineries were very small or were present in very low amounts, we could not determine the candidate proteins.
Figure 3.
Electron micrograph (A: SEM, C, E, F: TEM) and phase contrast (PC in B) and fluorescence micrograph (cmFtsZ, cmDnm in B, D) of isolated plastids with pt-division machineries (A–C) and isolated pt-division machineries (D, E, F). A. The image shows a dividing plastid with pt-division machinery at the center of the plastid. B and C. The pt-division machineries with the outer membrane are a chimera of bacterial FtsZ (CmFtsZ), dynamin (CmDnm2), and PD rings (arrow head in C). D, E, and F. Isolated pt-division machineries are composed of a bundle of fine filaments (enlarged image in F) and contained dynamin (small gold particles in F) and FtsZ (large gold particles in F). Scale bars: 1 µm (B), 0.5 µm (A, C, E), and 0.1 µm (F). A is from Ref. 3, B, C, D and F are from Ref. 28, E is from Ref. 3.
Strategy 4. Identification of proteins in plastid division machineries using the full genome sequence of C. merolae.
To identify the small proteins and their genes in the pt-division machineries, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was used. However, the genome information of C. merolae was a prerequisite before the use of the TOF MS. The full genome sequences of the cell nucleus of 20 chromosomes (16,546,747 bp), mitochondrion (32,211 bp), and the plastid (149,987 bp), were determined,21,24) for the first time in eukaryotes. A total of 5,335 genes were identified. Almost all the genes were expressed. The unique characteristics of these genomic structures included: a lack of introns in all but 26 genes; only three copies of ribosomal DNA units that maintain the nucleolus; and two dynamin genes that are involved only in the division of mitochondria and plastids. The lack of a myosin gene, in addition to an unexpressed actin gene, suggested a simpler system of cytokinesis (Table 1).21,24) During the sequencing of the genome, comparative genome analysis identified five novel genes, including FtsZ, dynamin, and Mda1 (mitochondrial division apparatus 1) gene, which are related to mt- and pt-division machineries.23,25–27) The division of mitochondria and plastids involves a dynamic trio: an FtsZ ring of bacterial origin, electron dense MD/PD rings, and eukaryotic mechanochemical dynamin rings. After the complete sequencing of the genomes, the identities of genes related to organelle division were clarified. Four genes representing mitochondrial FtsZ (FtsZ2-1 and FtsZ2-2) and plastid FtsZ (FtsZ1-1 and FtsZ1-2) were identified. A large gene family of dynamins, comprising of more than 10 members encoding functionally diverse proteins with a wide range of membrane pinching roles, has been found in other organisms. However, in C. merolae, only two dynamin genes (Dnm1 and Dnm2) are present, with a role in the later stages of the mitochondrion and plastid division, respectively.21,23,25) These findings suggest that plastids and mitochondria divide in a similar way, using common systems that are an amalgamation of bacterial and eukaryotic rings. This dynamic trio of plastid division is conserved in lower algae to higher plants.
Table 1.
Nuclear and organellar genomes of C. merolae and comparison to other genomes
| Feature | E.coliO157 | C.merolae | S.pombe | S.cerevisiae | A.thaliana |
|---|---|---|---|---|---|
| Cell Nucleus | |||||
| No. of chromosomes | 1 | 20 | 3 | 16 | 5 |
| Sequenced length (bp) | 5,528,445 | 16,546,747 | 12,462,637* | 12,495,682* | 115,409,949* |
| G+C content (%) | 50 | 55 | 36 | 38.3 | 34.9 |
| CpG occurence (Obs/Exp) | 1.151 | 0.886 | 0.803 | 0.724 | |
| No. of genes | 5,472 | 5,335 | 5,950 | 8,069 | 39,640 |
| Mean gene length† (bp) | 1,552 | 1,416 | 1,424 | 1,310 | |
| Gene density (bp per gene) | 3,102 | 2,508 | 2,088 | 4,526 | |
| Per cent coding | 87 | 44.8 | 57.5 | 70.5 | 28.8 |
| Genes with introns (%) | 0.5 | 45.9 | 5 | 79 | |
| No. of introns | 27 | 4,730 | 272 | 107,784 | |
| Mean length of intron (bp) | 248 | 81 | NA | 170 | |
| Mean length of exons (bp) | 1,540 | ND | ND | 170 | |
| No. of tRNA genes | 10 | 30 | 174 | 274 | 620 |
| No. of 5S rRNA genes | 8 | 3 | 30 | 100–150 | 1,000 |
| No. of 5.8S, 18S and 28S rRNA units | 7 | 3 | 200–400 | 100–150 | 700–800 |
| Mitochondrion | |||||
| Genome size (bp) | 32,211 | 19,431 | 85,779 | 366,924 | |
| G+C content (%) | 27.1 | ||||
| No. of protein genes | 34 | 10 | 29 | 58 | |
| Density (bp per protein genes) | 947 | 1,943 | 2,958 | 6,326 | |
| Plastid | |||||
| Genome size (bp) | 149,987 | NA | NA | 154,478 | |
| G+C content (%) | 37.6 | ||||
| No. of protein genes | 208 | NA | NA | 79 | |
| Density (bp per protein genes) | 721 | NA | NA | 1,955 | |
| Genes with introns (%) | 0 | NA | NA | 18 |
Data are from Refs. 21 and 24. ND, not determined; NA, not applicable.
*Ribosomal DNA repeats were not sequenced.
To identify further novel proteins in the pt-division machineries, we improved the method for isolating intact pt-division machineries. The outer membrane (associating dynamin and PD rings) of the plastids could not be completely solved by high concentrations of NP-40 (Fig. 3A–C).28) Instead, dividing plastids were obtained from synchronized cells and treated with n-octyl-β-D-glucopyranoside (OG) after Nonidet P-40 treatment. The purity of the isolated pt-division machineries was examined by fluorescent and electron microscopy, SDS-polyacrylamide gel electrophoresis, MALDI-TOF MS, and immunoblotting. The isolated division machineries showed super twist, spiral, and ring structures (Fig. 3D–F),28) suggesting that pt-division machineries included structures and molecules that generate motive force for contraction at the dividing site. The isolated pt-division machinery was smooth, and comprised a bundle of fine filaments 5–7 nm in diameter (Fig. 3F) that was composed of an FtsZ ring (inside) and the PD ring and dynamin rings (outside) through the membranes (Fig. 3F).28) The existence of membrane-free pt-machineries suggested that there is a linking structure through the membrane between the inner (FtsZ and inner PD) and outer (outer PD and dynamin) rings (Fig. 4C).
Figure 4.
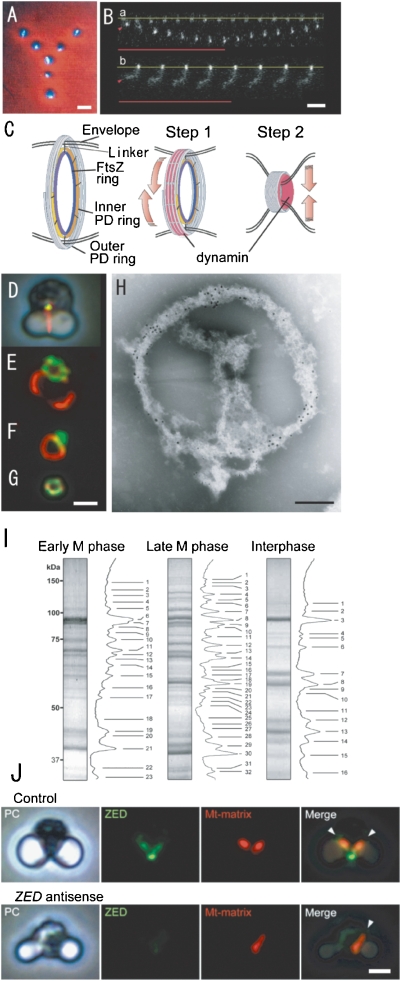
Phase contrast/fluorescence (A, D, J). phase-contrast (J), fluorescence micrographs (B, E. F, G, J) of isolated mitochondria (A), pt-division machineries (B), complexes of mt- and pt-division machineries (D, E, F), mt-division machinery (G), mitochondria and ZED (J), contraction model of pt-division machineries (C), an electron micrograph of the complex of isolated mt- and pt-division machineries (H), and SDS-PAGE (I). A. Physarum mitochondria with m-nuclei (bright rods) were drawn up in a “Y” image using optical tweezers. B. Manipulation of intact PD division machinery of C. merolae (a) and a dynamin-released PD division machinery (b) with the optical tweezer. The base line of one end of each of the pt-division machineries is fixed to the cover glass (yellow lines), while the other end is trapped by the optical tweezers (arrowheads) and an infrared laser (red lines). C. The schema shows a contraction model of pt-division machineries. In the first step, the dynamin molecules (red) drive the sliding of the fine filaments. In the second step, dynamin moves from the surface to the inside and pinches off the narrow bridge between daughter plastids. D. Mt-division machinery (yellow) is associated with pt-division machinery (red) in intact dividing cells. E and F. The bulk of isolated mt-division machinery (green) adheres to the pt-division machinery (red). G. Isolated mt-division machinery is a small ring. H. Immuno-electron micrograph showing the distributions of Mda1 and dynamin (Dnm2) in isolated mt- (large gold particles) and pt- (small gold particles) division machineries after negative staining. I. SDS-PAGE images of isolated PD machineries were compared with the interphase fraction (right). Proteins of isolated mitochondrial and plastid division machineries from cells in early M-phase (left), later M-phase (middle), and inter-phase (right) were separated by SDS-PAGE. J. In cells with transient DNA introduction and expression of ZED promoter without antisense-ZED, mitochondrial division (red) occurred normally (control). Plastid division occurred normally (phase contrast, PC), but mitochondrial division did not occur in ZED (blue)-downregulated cells (ZED antisense). Scale bars: 1 µm (A, B, G, J), 0.2 µm (H). B and C are from Ref. 28, D–J are from Ref. 30.
Using electrophoresis, proteins of the isolated pt-division machineries were classified into about 30 bands. The dense bands were examined by MALDI-TOF MS and compared with fractions from interphase cells. Fifty proteins and their genes became candidates to form the pt-division machineries. The most obvious protein was the dynamin-like protein, as predicted. Therefore, we first analyzed the structure and function of this protein. Pt-division machineries involve PD, dynamin, and FtsZ rings.28) To examine the function of an individual organelle and organellar division machinery, we developed an micro-optical tweezer that emitted a small beam (Fig. 4A, B). To check the power of the tweezer, a “Y” was drawn with seven isolated Physarum mitochondria, each of which contained one intact bright mt-nucleus (Fig. 4A). By the trapping method, it was easy to stretch the isolated pt-division machineries of C. merolae (Fig. 4B). After individual intact pt-division machineries were stretched to four times their original lengths with optical tweezers, they spontaneously returned to their original sizes. Dynamin released pt-division machineries did not retain the spiral structure and could not be stretched (Fig. 4B).28) Thus, dynamin might generate the motive force for contraction by filament sliding in dividing plastids, in addition to pinching-off the membranes. We hypothesize that the function of dynamin in the operation of pt-division machineries is two-fold: first, as a mediator of filament sliding at the early phase of plastid division, and then as a “pinchase”29) to pinch off the neck of the dividing plastid at the late phase (Fig. 4C).28) It was thought that contraction of the PD ring through sliding of the PD-ring fine filaments is caused by myosin-like proteins, but no genes encoding myosin or myosin-like proteins were found in the C. merolae genome.21,24,28) Thus, it is likely that dynamin, rather than myosin-like proteins, drives the sliding of the PD-ring fine filaments and causes the contraction required for plastid division. Thus, dynamin molecules in the plastid division machinery appeared to function as cross-bridges that undergo microscopic movement to drive the sliding of the microtubule filaments in the PD ring during the early phase of plastid division. At the late phase of plastid division, the dynamin molecules move from the surface of the contracted pt-division machinery to the inside of the machinery (Fig. 4C).28) When the dynamin molecules are associated with the membrane at the final stage of plastid division, and interact with the lipid membranes, they would act as a pinchase to pinch off the membranes at the bridge between the daughter plastids.
Strategy 5. Isolation, structure, and function of mitochondrial division machineries.
Using information gained from genome sequencing, we searched for novel proteins related to mt-division machineries. As a result, three proteins (FtsZ-like, dynamin-like, and Mda1-like) that function during contraction of mitochondrial division machineries were identified.25–27) To identify all the proteins related to mt-division machineries, the machineries must be isolated. However, mt-division machineries are smaller and more fragile than pt-division machineries and during isolation, the machineries were broken. To date, the mt-division machineries have not been directly isolated. It might be possible to isolate the mt-division machineries using their association with the pt-division machineries: previous electron microscopic studies showed that the mt-division machineries are attached tightly to the pt-division machinery during early M phase, but are separated by late M phase (Fig. 4D). Therefore, we first isolated the complex of mt- and pt-division machineries from the early M phase (Fig. 4E, F, H).30) Thus, the small mt-division machinery was isolated as part of the complex fraction (Fig. 4G).
To identify novel mt-division proteins, we performed proteomic analyses of the isolated complex of mt- and pt-division machineries from cells in early M phase, isolated pt-division machineries from cells in late M phase, and background proteins in this fraction from cells in interphase by SDS-PAGE and MALDI-TOF MS, respectively.30) The unique spots in plastid division machineries have been analyzed and the hypothetical components of the mt- and pt-division machineries were determined by subtracting background proteins from those of the isolated mt- and pt-division machineries fraction. Consequently, 76 mt- and pt-division machinery proteins from the cells in early M phase, 57 isolated pt-division machinery proteins from the cells in late M phase, and 29 background proteins in this fraction from the cells in interphase were identified, respectively (Fig. 4I). We identified a bacterial ZapA (Z ring associated protein A)-like protein, ZED (Z ring extended device), that is part of the inner membrane mt-division machinery in the division machinery proteins. ZED contains a predicted mitochondrial transit signal and two coiled-coil regions and has partial homology with the bacterial division protein ZapA. Cytological studies revealed that ZED accumulates to form a ring structure that colocalizes with FtsZ beneath the inner membrane. ZED proteins are expressed just before mitochondrial division and are required for mitochondrial division. We also demonstrated compelling functional similarity between bacterial ZapA and mitochondrial ZED, suggesting that the bacterial cell division system was incorporated into the MD machinery with remodeling of bacterial division proteins during evolution.30) The similarity between ZED and Zap is significant for eukaryote evolution as described later.
Strategy 6. Immunofluorescent microscopy and gene targeting of unknown proteins.
Gene targeting is the most effective technique for analyzing the function of genes, and is especially suitable in C. merolae cells as they contain each single organelle and a simple gene composition. Recently, gene targeting techniques were developed in C. merolae.30–32) Immunofluorescence images were obtained from C. merolae cells targeted with antisense-ZED. In cells with transient DNA introduction and expression of the ZED promoter without antisense-ZED, mt-division occurred normally (Fig. 4J). In ZED-downregulated cells, pt-division occurred normally but mt-division did not occur (Fig. 4J). FtsZ1 did not form ring structures at the division site, and Mda1, which is part of the outer membrane mt-division machinery, was not recognized in ZED-downregulated cells.30) These results showed that ZED is part of the inner membrane mt-division machineries, promotes formation of the FtsZ ring in the inner membrane mt-division machineries, and that the presence of Mda1 in the outer membrane mt-division machineries is dependent on ZED/FtsZ1 function. Gene targeting techniques will lead to the elucidation of the functions of the novel unknown proteins.
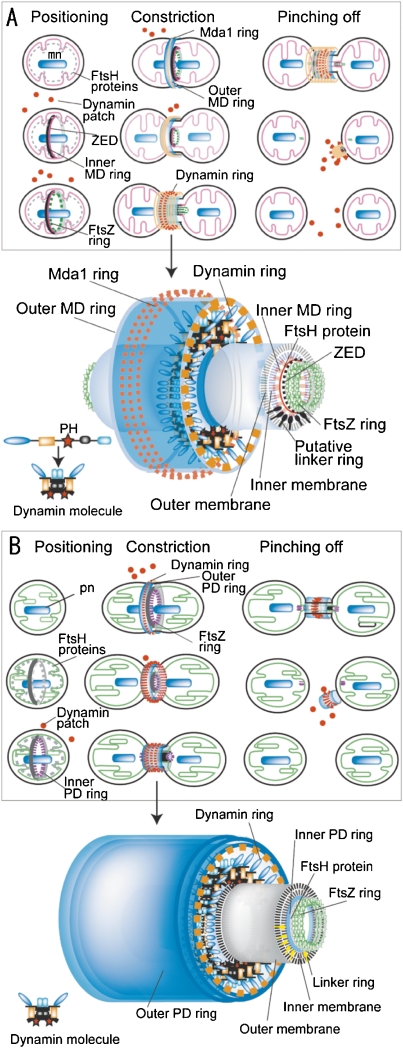
Model of mitochondrial and plastid division machineries
Mt- and pt-division machineries have very precise architectures at the nanolevel, although they appear very smooth and simple. Fig. 5A shows a model of mt-division by the mt-division machinery.3) In the positioning phase of the mt-division machineries, the inner ZED ring, inner MD ring and FtsZ ring are formed on the matrix side, under the control of FtsH, and the division site is determined. The electron dense outer MD ring is located at the cytoplasmic side and then the dynamin ring is located around the MD ring in the cytoplasm. Finally, the outer membrane mt-division machinery is composed of the Mda1 ring, the outer MD ring, the dynamin ring, and unknown rings. The inner membrane mt-division machinery is composed of the ZED ring, the FtsZ ring, the inner MD ring and unknown rings. The existence of membrane-free mt-division machineries suggests that there is a linking structure through the membrane between the inner and outer mt-division machineries.30) In the constriction phase, dynamin is recruited from patches in the cytoplasm to form the outer membrane mt-division machinery, with Mda1 and the outer MD ring.30) Then dynamin drives the sliding of the MD ring fine filaments and causes the contraction required for mitochondrial division. At the late phase of mitochondrial division, the dynamin molecules move from the surface of the outer membrane mt-division machinery to the inside of the machinery (Fig. 5A).30) Praefcke and McMahon (2004) reviewed the superfamily of dynamins and the domain structure and function.33) Classical dynamins contain a GTPase domain, middle domain, pleckstrin-homology (PH) domain, GTPase effector doman (GED) and proline-rich domain. Dynamin probably pinches off the membranes at the bridge between the daughter plastids by a biochemical reaction between the PH domain and the double membrane. After mitochondrial division, the remnants of the mt-division machinery then adhere to one side of the daughter mitochondria and the dynamin ring is broken into small patches.26)
Figure 5.
Models of mitochondrial division by mt-division machinery (A) and molecular model of mt-division machinery (enlarged image in A) and plastid division by pt-division machinery (B) and molecular model of pt-division machinery (enlarged image in B). A. In the positioning phase, the inner MD ring and FtsZ ring are formed on the matrix side, and the division site is determined. In the constriction phase, the Mda1 ring and outer MD ring appear on the cytoplasmic side of the outer membrane. The inner MD, FtsZ, Mda1, and outer MD rings begin to constrict the equator of the dividing mitochondrion. When the mitochondrion has constricted at the division site, dynamin is recruited from the patches in the cytoplasm to form the mt-division machinery, with Mda1 and the outer MD ring, and finally migrates to a space inside the thickened outer MD ring and outside the outer membrane. In the pinching-off phase, the inner MD ring and the FtsZ ring are split to form a patch in each matrix. The dynamin ring pinches off the membrane of the bridge between the daughter mitochondria. The remnants of the mt-division machineries are broken into small patches. Putative molecular model of pinching-off at the bridge between daughter mitochondria. Inner MD and FtsZ rings are split to form two divided matrices and then the dynamin ring pinches off the outer and inner membranes simultaneously, at the center of the bridge, by a biochemical reaction between the PH domain and the double membranes under the control of Mda1 and the outer MD ring. B. Events similar to the mt-division machineries occur in the pt-division machineries. mn, mitochondrial nuclei; pt, plastid nuclei. Dynamin molecules is improved from Ref. 33, A is from Ref. 3.
The mechanism of plastid division is very similar to that of mitochondrial division (Fig. 5B).3) The outer membrane pt-division machinery is composed of the outer PD ring, dynamin ring and unknown rings. The inner membrane pt-division machinery is composed of the FtsZ ring, the inner PD ring and unknown rings. There is a linking structure through the membrane between the inner and outer mt-division machineries.28) In Arabidopsis thaliana, ARC6 (accumulation and replication of chloroplasts), PDV1 (plastid division 1) and PDV2 were found as linker-like proteins.34) The constriction model at the division site shows two steps involving dynamin molecules in the pt-division machinery. In the first step, the dynamin GTPase molecules in the pt-division machinery function as cross-bridges that undergo microscopic movements to drive the sliding of the 5 nm filaments in the electron dense outer PD ring during the early phase of plastid division (Figs. 4C and 5B). The dynamin ring generates the driving force for contraction with the aid of the other PD rings. In the second step, the dynamin molecules move from the surface of the pt-division machinery to the inside of the machinery, and play a role in pinching off the narrow bridge between daughter plastids during the late phase of plastid division (Figs. 4C and 5B).3) After the inner PD and FtsZ rings are split to the stroma of the daughter plastids, the dynamin ring then simultaneously pinches off the outer and inner membranes at the center of the bridge, by a biochemical reaction between the PH domain and the double membranes, under the control of the outer PD ring.23,28)
In almost all eukaryotes without sexual reproduction and eukaryotes with sexual reproduction, mitochondria and plastids are thought to divide according to the methods as described above. Why an original eukaryotic cell requires the outer membrane organelle division machineries for organelle divisions will be discussed later.
Mechanisms of maternal inheritance of mitochondrial and the plastid genome (DNA) during sexual reproduction
In almost all organism after the multiplication of cells and organelles during the non-sexual reproduction phase, the cells and organelles enter the sexual reproduction phase (Fig. 1J). The main processes of sexual reproduction are fertilization, involving the fusion of gametes and meiosis, involving the halving of the number, and recombination between the cell-nuclear chromosomes. However, very little is known about the molecular mechanism of cytoplasmic inheritance of organelles and its implications for the early evolution of eukaryotic cells. Recently, the mechanism of maternal inheritance of mitochondrial and plastid genomes (DNAs) has been summarized in a special issue.4) Therefore, I will describe the mechanisms briefly.
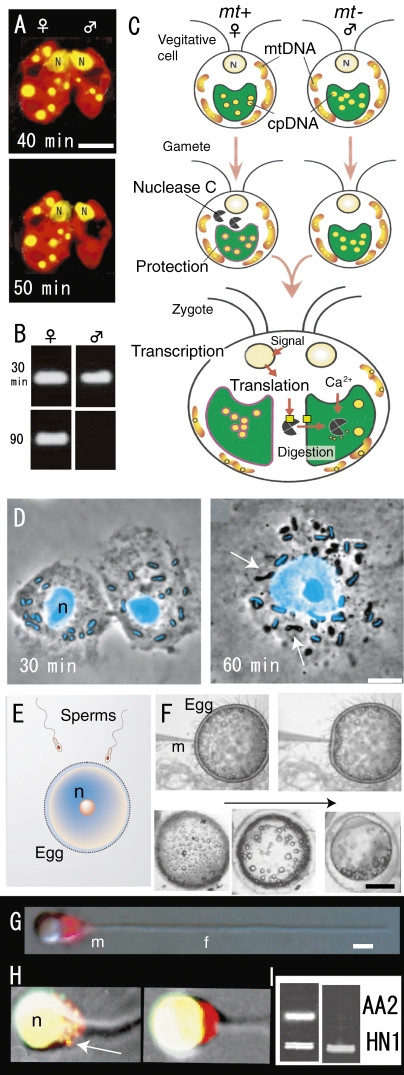
Maternal inheritance of organelle genomes was thought to result from dilution of the paternal contribution. This is because the paternal gametes (sperm) are much smaller than maternal gametes (egg) and contribute only a small amount of cytoplasm to the progeny. However, if this idea is correct, maternal transmission of organelle DNA should not occur in isogamous organisms where there are no size differences between male and female gametes. Therefore, to obtain direct evidence for sexual transmission of organelle DNAs, we examined their behaviors in gametes and young zygotes of the isogamous alga Chlamydomonas reinhardtii using high resolution fluorescence microscopy. The preferential digestion of male (mt−) gamete-originated plastid (pt-) DNA occurred, while pt-DNA originating from female (mt+) gametes remained 50 min after mating of female and male gametes (Fig. 6A, B).35,36) They were observed in living cells (Fig. 6A) and confirmed by nested PCR (Fig. 6B). Thereafter, the maternal (uniparental) transmission of organelles that resulted from “the active digestion of male organelle nuclei (genomes) in young zygotes” was observed in almost all algae and land plants.4,13,37) The mechanism was analyzed by molecular and cellular biological techniques (Fig. 6C). A model for the molecular mechanism of uniparental inheritance in C. reinhardtii was proposed on the basis of the active digestion of male plastid DNA by the Ca2+-dependent nuclease MDN (mt+-specific DNase, 140 kDa).4,37) In vegetative cells, MDN is absent or inactivated in both mating types. During gametogenesis, MDN is synthesized or activated only in female cells.36) At the same time, female pt-DNA becomes resistant to the action of MDN. During gamete fusion, MDN obtains access to unprotected male plastids and digests the male pt-DNA, leading to the uniparental inheritance of pt-DNA. Several factors might mediate the successful digestion of male plastid DNA after zygote formation: (1) entry of MDN into male plastids; (2) efficient access of MDN to plastid DNA molecules; and (3) an increase in Ca2+ concentration inside male plastids. Zygote-specific gene expression might be crucial to these processes (Fig. 6C).37)
Figure 6.
Fluorescence micrographs of living young zygotes of C. reinhardtii (A), gametes and a zygote of P. polycephalum (D), sperm mitochondria of Oryzias latipes (G, H), patterns of nested PCR after mating (fertilization) of C. reinhardtii (B) and O. latipes (I) and models of active digestion of male plastid DNA (C), fertilization of vertebrate (E) and process of microinjection and the development of injected eggs (F). A. Preferential disappearance of male plastid nuclei (yellow) visualized in a SYBR Green I stained living zygote 50 min after mating. B. Male gamete plastid DNA is digested within 90 min after mating. C. During gametogenesis, nuclease C is synthesized or activated only in female (mt+) cells. At the same time, female plastid DNA becomes resistant to the action of nuclease C. During gamete fusion, nuclease C obtains access to unprotected male plastids and digests the male plastid DNA, leading to maternal inheritance of plastid DNA. Several factors might mediate the successful digestion of male plastid DNA after zygote formation. D. Each amoeba contained about 12 mitochondria and each mitochondrion was characterized by the rod shaped mt-nucleus at its center (blue in D). 1 hr after nuclear fusion, mt-nucleus fluorescence completely disappeared in about half of the mitochondria in the zygote (arrows). E. A large egg fertilizes with a sperm. F. After injection, the eggs remained intact and progressed through normal developmental stages. G and H. Phase-contrast and SYBR green I-MitoTracker double-stained images showed mitochondria (red) in sperm before (G) and sperms in fertilized egg (H) after fertilization. Sperm mitochondrial nuclei (yellow) disappeared completely 60 min after fertilization (right). I. Single sperm with or without mitochondrial nuclei was selectively extracted from fertilized eggs using optical tweezers and analyzed by nested PCR. Sperm and eggs were derived from AA2, and the HNI PCR product was added to each reaction as an internal control. Active digestion of sperm mitochondrial DNA (AA2) occurred 60 min after injection into eggs (right). N and n, cell nuclei. Scale bars: 0.5 mm (F), 5 µm (A, D), and 1 µm (G). Scale bars = 1 µm. A and B are from Ref. 36, C is from Ref. 37, D is from Ref. 39, F-I are from Ref. 40.
Furthermore, similar preferential digestion of male organellar DNA was observed in mitochondria in zygotes of an isogamous slime mould after mating (Fig. 6D).38,39) Male and female gametes contained about ten mitochondria with rod-shaped mitochondrial nuclei (nucleoids). Soon after mating, mitochondria originating from both sexes are mixed in a zygote and the male mt-nuclei are selectively digested by the nuclease (Fig. 6D). The empty male mitochondria were digested 70 h after mating.39) Furthermore, in a vertebrate (Japanese medaka Oryzias latipes), selective digestion of sperm-originated mitochondrial DNA also occurred in the fertilized egg 60 min after fertilization (Fig. 6E–I).40) Nishimura et al. (2006) visualized the selective digestion of sperm mt-nuclei in living sperm using highly sensitive SYBR green I vital staining (Fig. 6G, H) and nested PCR of one cell by test tube fertilization (Fig. 6I).40) The elimination of sperm mt-DNA upon fertilization is achieved in two stages: (i) gradual decrease of mt-nuclei numbers during spermatogenesis and (ii) rapid digestion of sperm mt-DNA just after fertilization.40) This “active digestion of organelle nuclei from uniparental gametes by nucleases” is probably universal among isogamous, anisogamous, and oogamous organisms.4) In higher plants, it has been difficult to analyze maternal inheritance of organellar genomes at the molecular level in vivo because of the large number of opaque ovular cells that usually enclose them. Recently, an in vitro system for observing the guidance of pollen tubes was established using the naked embryo sac of T. fournieri,41,42) and hopefully the molecular mechanism of maternal inheritance will be elucidated.
In addition to the active digestion of male organelle nuclei by the nuclease, the difference in size between male and female gametes in anisogamous and oogamous organisms indicates that the contribution to the progeny of male organellar genomes (DNA) is small, while that of female gametes is large, suggesting that anisogamy and oogamy reinforce maternal inheritance.4)
Implications of organelle division machineries and organelle inheritance regarding the origin of eukaryotic cells
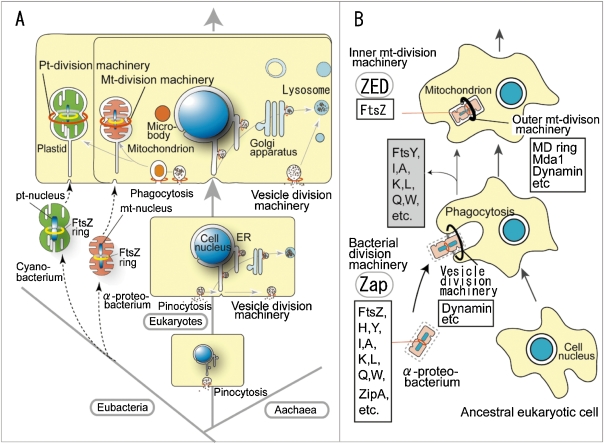
Cellular and molecular data has suggested that mitochondria and plastids are descendants of endosymbiotic prokaryotes. However, there is little information as to how free-living autonomous bacteria became semi-autonomous organelles during endosymbiosis in early eukaryotic evolution. Semi-autonomous organelles were probably generated from the bacteria in four steps: 1) the host cell (organism) captured and engulfed bacteria by endocytosis (phagocytosis);3,15,43) 2) the cell controlled the division and multiplication of endosymbiotic bacteria or pre-organelles by division machineries;3,15) 3) part of endosymbiotic bacterial genome and genes was transferred to the host’s own genome;2,44) and 4) with development of sexuality, the host cell genome regulated the evolution of the organellar genome by uniparental (maternal) inheritance.3,4,14,15)
The first two steps are particularly interesting. Vesicle division rings around the neck of vesicle buds seem to play a role in phagocytosis (Fig. 7A, B).3,43) The formation of clathrin-coated vesicles could serve as a guide, as the use of dynamin-related proteins during the severing of vesicles at the plasma membrane and during division of the mitochondrial outer membrane suggests a common evolutionary origin.3,4,43) van del Bliek (2000) imagined that the first endosymbiotic bacteria used dynamin of the host cell during the entry at the plasma membrane and used it again during division of its outer membrane, before eventually giving rise to modern day mitochondria.43) A small (40 nm diameter) electron-dense ring was observed around the neck of vesicle buds formed from the plasma membrane.44) Since then, it has been generally believed that this electron-dense ring, or spiral, consists of dynamin recruited to the vesicle necks. This hypothesis was supported by the fact that the endocytic protein, GTPase dynamin, binds directly to liposomes, deforming them into tubules in vitro, and plays critical roles in membrane fission and curvature during clathrin-mediated endocytosis. However, several problems remain to be solved.3,15) There is no direct in vivo evidence that the vesicle division rings contain dynamin. Using immunoelectron microscopy with antibodies to dynamin, gold particles indicating dynamin signals were not located directly on the filaments of the vesicle division ring in vivo, but only near the vesicle ring. The dynamin rings in mt- and pt-division machineries in C. merolae were 20 or 100 times larger in diameter and more than 1,000 times greater in volume than those of the vesicle division machinery. However, even when the dynamin rings were observed clearly by immunoelectron and immunofluorescent microscopy, they were never observed directly using electron microscopy. Thus, the vesicle division machineries were considered to be analogous to the mt- and pt-division machineries were considered: the collar is composed of vesicle division rings, dynamin rings, and putative rings, and is divided by a biochemical reaction between one domain in the vesicle division machinery and the constricted plasma membrane (Fig. 7A, B).3,15) It is presumed that the vesicle division ring is similar to prototypes of the outer mt- and pt-division machineries; therefore, it seemed logical to hypothesize that they evolved into these outer membrane organelle division machineries (Fig. 7A, B).3,15,43)
Figure 7.
Schematic representation of the origin of eukaryotic cells (A) and dynamic events of bacterial division machineries and mitochondria during endosymbiosis with phagocytosis and vesicle-division machineries (B). A. Origin and evolution of eukaryotic cells with emphasis on mt- and pt-division machineries, including plasma membrane vesicle division, and cytoplasmic vesicle division. Basic eukaryotic cells are composed of double-membrane-bound organelles (cell nucleus, mitochondrion and plastid and single-membrane-bound organelles (ER, Golgi apparatus, lysosomes (vacuoles), microbodies). With phagocytosis, microbodies, mitochondria, and plastids are generated from Eubacteria. The formation of eukaryotic cell organelles consists of two phases: before and after mitochondria and plastids are generated from phagocytosis. B. Schematic representation of the bacterial division machinery, the vesicle division machinery, the inner membrane mt-division machinery and the outer membrane mt-division machinery. Eubacteria divided using bacterial division proteins. Almost all bacterial proteins were lost during endosymbiosis and FtsZ and ZED remained in inner mt-division machineries in eukaryotes. A is from Ref. 3.
On the other hand, how did the inner membrane organelle division machineries originate from bacterial division machineries? It is thought that eubacteria divided using division machineries that were composed of more than 10 proteins, including filamentous temperature-sensitive (fts) proteins FtsA, FtsB, FtsI, FtsK, FtsL, FtsN, FtsQ, FtsZ, ZipA, and ZapA (Fig. 7B).3,45) However, although almost all bacterial proteins (FtsA, FtsB, FtsI, FtsK, FtsL, FtsN, FtsQ, FtsZ, and ZipA) were lost during endosymbiosis,3,30) with FtsZ and ZED (Zap-like protein) remaining in the inner mt-division machineries in eukaryotes, it was unknown how the bacterial division proteins were lost (Fig. 7A). The discovery of ZED gave us a hint. Coiled-coil structures similar to those of ZED are also found in the bacterial FtsZ associated protein ZapA, which is widely conserved among bacteria. Sequence identity between ZED (excluding insertions) and ZapA was 25.8% (48.3% positive) (Fig. 7),30) suggesting that the host genome incorporated the gene encoding bacterial division protein Zap to form its own gene encoding division protein ZED. It is probable that the other lost proteins were similarly transformed from bacterial division proteins to host division proteins. Finally, bacterial division proteins were lost while the nuclear genome of the host cell must have evolved genes for the outer membrane mt- and pt-division machineries to control the division of organelles (Fig. 7B).
In the third step, the host cell promoted the transfer of more than 80% of the symbiont genes into the host’s cell genome during the early stage of endosymbiosis.2,3,44) Thereafter, small amounts of DNA continued to move from the organelle genome to the host genome.15,44) As a result, endosymbiont bacteria lost their autonomy.
In the last step, a new theory concerning the significance of maternal inheritance of organelle genomes in eukaryotes has been proposed.3,4,14) The “active digestion of male organelle nuclei (genomes) during sexual reproduction from uniparental gametes by nucleases” is now thought to be universal among isogamous, anisogamous, and oogamous organisms.4,40) In addition, the difference in size between male and female gametes in anisogamous and oogamous organisms showed that contribution to the progeny of the male organellar genome (DNA) is small, while that of female gametes is large, suggesting that anisogamy and oogamy reinforce maternal inheritance. It is well known that during sexual reproduction, recombination and independent assortment of homologous chromosomes in cell nuclei allows for a diversity of genotypes in the population. As a result, this produces genetic variation in gametes that promote genetic variation in a population of offspring. On the other hand, maternal (uniparental) transmission of organellar genomes to progeny seemed to be an effective method to avoid the evolution of organelles through recombination of the organellar genes of both parents.
Perspective
Using genome information, liquid-MALDI-TOF MS instead of gel-MALDI-TOF MS, cell biological technique, and gene targeting of molecular biology in C. merolae, future studies will elucidate the architecture of mt- and pt-division machineries, which are composed of more than 30–40 proteins, and will reveal their functions. Furthermore, the mechanism of mt- and pt-division must be elucidated by structural biology and atomic biology. Elucidation of the mechanisms at the atomic level will require the crystal structure of organelle division machineries and organelles for X-ray analysis and NMR analyses. C. merolae inhabits hot springs; therefore, its proteins are likely to be more thermostable than other organisms. Recently, the Center for Eukaryotic Structural Genomics (CESG) at Wisconsin University has reported that eukaryotic proteins generally are considered to be much more challenging targets than prokaryotic proteins, and will require technological innovations for structure determination by X-ray crystallography or NMR spectroscopy. During the first three years of PSI-2, CESG selected crystal targets representing 601 proteins from Homo sapiens, 33 from mouse, 10 from rat, 139 from Galdieria sulphuraria, 35 from Arabidopsis thaliana, 96 from C. merolae, 80 from Plasmodium falciparum, 24 from yeast, and about 25 from other eukaryotes.46)
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (No. 19207004 to T.K.), and by grants from the Japan Society for Frontier Project “Adaptation and Evolution of Extremophile” (to T.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and from the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN, to T.K.).
Profile
Tsuneyoshi Kuroiwa was born in 1941 in Tokyo. In 1971, he graduated from the Faculty of Science, Tokyo Metropolitan University, and in the same year entered the graduate school of Sciences, University of Tokyo. He studied the cytogenetics in the Department of Biological Sciences, Faculty of Sciences, University of Tokyo. In 1971, he received his Ph.D. degree from University of Tokyo under the direction of the late Professor Nobunori Tanaka. In same year, he was promoted to a Researcher, the Tokyo Metropolitan Isotope Research Center. He was promoted to a Lecturer, Okayama University and an Associate Professor in 1973. He moved to the National Institute for Basic Biology as an Associate Professor in 1977, and to a Professor in 1983. He returned to University of Tokyo and was a Professor (Developmental Biology) of the Department of Biological Science in 1987–2002. After retirement from University of Tokyo in 2002, he became a Professor emeritus of University Tokyo. He moved to a Professor (Cell Biology) of the Department of Life Sciences, Rikkyo (St. Paul’s) University in 2003-present. He has discovered the division machinery of mitochondria and chloroplasts and clarified the molecular mechanisms of organelle division which had been wrapped in mystery. The DNA of mitochondria and chloroplasts is known to be maternally-inherited in most living organisms. He revealed that this is because of the selective degradation of paternal DNA. In order to develop these researches, he found out the primitive red alga (Cyanidioschyzon merolae), as the key model organism of eukaryotes, performed genome sequencing, and became the first researcher in the world who succeeded in 100% genome sequencing. Cyanidioschyzon is now used world-wide as an important model organism. He was elected a member of the Science Council of Japan in 2003-present. He was awarded the Purple Ribbon Medal 2008, won the 2008 BSJ Merit Award by BSP (Botanical Society of Japan) and the 2008 Charles Reid Barnes Life Membership Award by ASPB (American Society of Plant Biologists).
References
- 1).Howard A., Pelc S.R. (1953) Synthesis of deoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Heredity 6 (Suppl.), 261–273 [Google Scholar]
- 2).Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K. and Walter, P. (2002) Molecular Biology of the Cell (4th ed.). Garland Publishing, New York. [Google Scholar]
- 3).Kuroiwa T., Misumi O., Nishida K., Yagisawa F., Yoshida Y., Fujiwara T., et al. (2008) Structure, function, and origin of vesicle, mitochondrial and plastid division machineries with emphasis on dynamin rings and electron-dense rings. Int. Rev. Cell Mol. Biol. 271, 97–141 [DOI] [PubMed] [Google Scholar]
- 4).Kuroiwa T. (2010) Review of cytological studies on cellular and molecular mechanisms of uniparental (maternal or paternal) inheritance of plastid and mitochondrial genomes induced by active digestion of organelle nuclei (nucleoids). J. Plant Res. 123, 207–230 [DOI] [PubMed] [Google Scholar]
- 5).Ris H., Plaut W. (1962) Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 13, 383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Kuroiwa T. (1982) Mitochondrial nuclei. Int. Rev. Cytol. 75, 1–59 [DOI] [PubMed] [Google Scholar]
- 7).Gillham, N.W. (1994) Organelle Genes and Genomes. Oxford University Press, USA. [Google Scholar]
- 8).Kuroiwa T. (1974) Studies on mitochondrial structure and function in Physarum polycephalum III. Electron microscopy of a large amount of DNA released from a central body in mitochondria by trypsin digestion. J. Cell Biol. 63, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Kuroiwa T., Kawano S., Hizume M. (1977) Studies on mitochondrial structure and function in Physarum polycephalum V. Behavior of mitochondrial nucleoids throughout mitochondrial division cycle. J. Cell Biol. 72, 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Andersson S.G.E., Zomorodipour A., Andersson J.O., Sicheritz-Ponten T., Alsmark U.C.M., Podowski R.M., et al. (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–142 [DOI] [PubMed] [Google Scholar]
- 11).Kobayashi T., Takahara M., Miyagishima S., Kuroiwa H., Sasaki N., Ohta N., et al. (2002) Detection and localization of a chloroplast-encoded HU-like protein that may organize chloroplast nucleoids. Plant Cell 14, 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Sasaki N., Kuroiwa H., Nishitani C., Takano H., Higashiyama T., Kobayashi T., et al. (2003) Glom is a novel mitochondrial DNA packing protein in Physarum polycephalum and causes intense chromatin condensation without suppressing DNA function. Mol. Biol. Cell 14, 4758–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Kuroiwa T. (1991) The replication, differentiation and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128, 1–62 [Google Scholar]
- 14).Kuroiwa T. (1998) The primitive red algae Cyanidium caldarium and Cyanidioschyzone merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20, 344–354 [Google Scholar]
- 15).Kuroiwa T., Nishida K., Yoshida Y., Fujiwara T., Mori T., Kuroiwa H., et al. (2006) Structure, function and evolution of mitochondrial dividing apparatus. Biochimi. Biophys. Acta. Mol. Cell 1763, 510–521 [DOI] [PubMed] [Google Scholar]
- 16).Tamiya H. (1966) Synchronous cultures of algae. Ann Rev. Plant Physiol. 17, 1–27 [Google Scholar]
- 17).Lim B.L., Kawai H., Hori H., Osawa S. (1986) Molecular evolution of 5S ribosomal RNA from red and brown algae. Jpn. J. Genet. 61, 169–176 [Google Scholar]
- 18).Seckbach J. (1991) Systematic problems with Cyanidium caldarium and Galdieria sulphuraria, and their implications from molecular biology studies. J. Phycol. 27, 79–796 [Google Scholar]
- 19).Toda K., Takano H., Miyagishima S., Kuroiwa H., Kuroiwa T. (1998) Characterization of a chloroplast isoform of serine acetyltransferase from the thermo-acidiphilic red alga Cyanidioschyzone merolae. Biochim. Biophys. Acta 140, 72–84 [DOI] [PubMed] [Google Scholar]
- 20).Kuroiwa T., Kuroiwa H., Sakai A., Takahashi H., Toda K., Itoh R. (1998) The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181, 1–42 [DOI] [PubMed] [Google Scholar]
- 21).Matsuzaki M., Misumi O., Shin-i T., Maruyama S., Takahara M., Miyagishima S., et al. (2004) Genome sequence of the ultra-small unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–665 [DOI] [PubMed] [Google Scholar]
- 22).Miyagishima S., Takahara M., Kuroiwa T. (2001) Novel filaments 5nm diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Miyaghishima S., Nishida K., Mori T., Matsuzaki M., Higashiyama T., Kuroiwa H., et al. (2001) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15, 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Nozaki H., Takano H., Misumi O., Terasawa K., Matuzaki M., Maruyama S., et al. (2007) The first 100% complete eukaryotic genome sequences from the red alga Cyanidioschyzon merolae 10D. BMC Biol. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Nishida K., Takahara M., Miyagishima S., Kuroiwa H., Matsuzaki M., Kuroiwa T. (2003) Dynamic recruitment of Dynamin for final mitochondrial severance in a red alga. Proc. Natl. Acad. Sci. USA 100, 2146–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Nishida K., Yagisawa F., Kuroiwa H., Nagata T., Kuroiwa T. (2005) Cell cycle-regulated, microtubule-independent organelle division in Cyanidioschyzon merolae. Mol. Biol. Cell 16, 2493–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Nishida K., Yagisawa F., Kuroiwa H., Yoshida Y., Kuroiwa T. (2007) WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA 104, 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Yoshida Y., Kuroiwa H., Misumi O., Nishida K., Nanamiya H., Yagisawa F., et al. (2006) Isolated chloroplast division machinery can actively constrict after stretching. Science 313, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 29).Kozlov M. (1999) Dynamin: Possible mechanism of “Pinchase” action. Biophys. J. 77, 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Yoshida Y., Kuroiwa H., Hirooka S., Yoshida M., Fujiwara T., Misumi O., et al. (2009) Novel mitochondrial division protein ZED forms the inner complex structure of the mitochondrial division machinery with the FtsZ ring as revealed by isolated mitochondrial division machineries. Curr. Biol. 19, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 31).Kobayashi Y., Kanesaki Y., Tanaka A., Kuroiwa H., Kuroiwa T., Tanaka K. (2009) Tetrapyrole signal as a cell cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. USA 106, 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Imamura S., Kanesaki Y., Fujiwara T., Kuroiwa T., Tanaka K. (2009) R2R3-type MYB transcription factor, CmMYB1, controls the expression of nitrogen assimilation genes in response to nitrogen status in the unicellular red alga Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA 106, 12548–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Praefcke G.J.K., McMahon H.T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 [DOI] [PubMed] [Google Scholar]
- 34).Yang Y., Glynn J.M., Olson B.J.S.C., Schmitz A.J., Osteryoung K.W. (2008) Plastid division: across time and space. Curr. Opin. Plant Biol. 11, 577–584 [DOI] [PubMed] [Google Scholar]
- 35).Kuroiwa T., Kawano S., Nishibayashi S., Sato C. (1982) Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature 298, 481–483 [DOI] [PubMed] [Google Scholar]
- 36).Nishimura Y., Misumi O., Matsunaga S., Miyagishima S., Yokota A., Kuroiwa T. (1999) Active digestion of mt+ derived chloroplast DNA in individual zygotes of Chlamydomonas reinhardtii revealed by the optical tweezers. Proc. Natl. Acad. Sci. USA 96, 12577–12582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Nishimura Y., Misumi O., Kato K., Inada N., Momoyama Y., Kuroiwa T. (2002) mt+ gamete specific nuclease that targets mt− chloroplasts during the sexual reproduction in Chlamydomonas reinhardtii. Genes Dev. 16, 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Kawano S., Takano H., Kuroiwa T. (1995) Sexuality of mitochondria fusion, recombination and plasmid. Int. Rev. Cytol. 161, 49–110 [DOI] [PubMed] [Google Scholar]
- 39).Moriyama Y., Kawano S. (2003) Rapid, selective digestion of mitochondrial DNA in accordance with the matA hierarchy of multiallelic mating types in the mitochondrial inheritance of Physarum polycephalum. Genetics 164, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Nishimura Y., Yoshinari T., Naruse K., Yamada T., Sumi K., Mitani H., et al. (2006) Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc. Natl. Acad. Sci. USA 103, 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S., Kuroiwa H., et al. (2001) Pollen tube attraction by the synergid cell. Science 293, 1480–1483 [DOI] [PubMed] [Google Scholar]
- 42).Okuda S., Tsutsui H., Shiina K., Sprunck S., Takeuchi H., Yui R., et al. (2009) Defensin-like peptides LUREs are pollen tube attractants secreted from the synergid cell. Nature 458, 251–376 [DOI] [PubMed] [Google Scholar]
- 43).van der Bliek A.M. (2000) A mitochondrial division apparatus takes shape. J. Cell Biol. 151, F1–F4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Ohta N., Sato N., Kawano S., Kuroiwa T. (1994) The trp A gene on the plastid genome of Cyanidium caldarium strain RK-1. Curr. Genet. 25, 357–361 [DOI] [PubMed] [Google Scholar]
- 45).Bramhill D. (1997) Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13, 395–424 [DOI] [PubMed] [Google Scholar]
- 46).Markley J.L., Aceti D.J., Bingman C.A., Fox B.G., Frederick R.O., Makino S., et al. (2009) The Center for Eukaryotic Structural Genomics. J. Struct. Funct. Genom. 10, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]