Abstract
A genomic interval of ∼1–1.5 Mb centered at the MSR marker on 8p22 has emerged as a possible site for a tumor suppressor gene, based on high rates of allele loss and the presence of a homozygous deletion found in metastatic prostate cancer. The objective of this study was to prepare a bacterial contig of this interval, integrate the contig with radiation hybrid (RH) databases, and use these resources to identify transcription units that might represent the candidate tumor suppressor genes. Here we present a complete bacterial contig across the interval, which was assembled using 22 published and 17 newly originated STSs. The physical map provides twofold or greater coverage over much of the interval, including 17 BACs, 15 P1s, 2 cosmids, and 1 PAC clone. The position of the selected markers across the interval in relation to the other markers on the larger chromosomal scale was confirmed by RH mapping using the Stanford G3 RH panel. Transcribed units within the deletion region were identified by exon amplification, searching of the Human Transcript Map, placement of unmapped expressed sequence tags (ESTs) from the Radiation Hybrid Database (RHdb), and from other published sources, resulting in the isolation of six unique expressed sequences. The transcript map of the deletion interval now includes two known genes (MSR and N33) and six novel ESTs.
[The sequences described in this paper have been submitted to the GenBank data library under the accession nos. AF126202–AF126212.]
The short arm of chromosome 8 is a frequent site of allelic deletions in epithelial tumors as diverse as colon (Cunningham et al. 1993; Fujiwara et al. 1993; Chang et al. 1994), breast (Kerangueven et al. 1997; Anbazhagan et al. 1998), prostate (Vocke et al. 1996; Deubler et al. 1997; Prasad et al. 1998), head and neck (Wu et al. 1997; El-Naggar 1998), bladder (Takle et al. 1996; Wagner et al. 1997), and lung (Ohata et al. 1993). In prostate cancer, loss of heterozygosity (LOH) for markers on 8p is one of the most frequent somatic mutations, occurring in >60% of these tumors (Cunningham et al. 1996). Functional evidence that chromosome 8 contains tumor suppressor gene-associated activity is provided by microcell-mediated chromosome transfer studies, which demonstrate suppression by this chromosome of in vivo metastatic ability and in vitro invasiveness in the highly metastatic rat prostate cancer cell line AT6.2 (Nihei et al. 1996; Kuramochi et al. 1997), suppression of both tumorigenicity and in vitro invasiveness in the COKFu colon carcinoma cell line (Tanaka et al. 1996), reduction of tumorigenicity in the SW620 colon carcinoma cell line, and total suppression of soft agar clonicity in the HT29 colon carcinoma cell line (Gustafson et al. 1996). Together, these studies strongly suggest that 8p harbors a tumor suppressor gene involved in the genesis of epithelial malignancies.
Subregional deletion analysis of chromosome 8 in a wide spectrum of tumors demonstrates a high degree of LOH for probes within 8p22, including prostate (Bova et al. 1993; Macoska et al. 1995; Cunningham et al. 1996), colon (Cunningham et al. 1993; Fujiwara et al. 1994; Takanishi et al. 1997), lung (Fujiwara et al. 1994; Lerebours et al. 1999; Wistuba et al. 1999), liver (Fujiwara et al. 1994; Pineau et al. 1999), male breast (Chuaqui et al. 1995), breast (Patel et al. 1994; Kerangueven et al. 1997), ovarian (Wright et al. 1998), and urinary bladder (Ohgaki et al. 1999) cancers, although current evidence suggests that there are several discrete, nonoverlapping sites of LOH on chromosomal bands 8pter → p23, 8p22, 8p21, and 8p12 → p11. The general interpretation of these findings is that there is more than one tumor suppressor gene on 8p and additional data are needed to further narrow the critical region(s) to identify the genes (The Third International Workshop on Human Chromosome 8 mapping 1996).
The best evidence to date for a localized tumor suppressor gene interval on 8p is provided by Bova and colleagues (1993), who reported a homozygous deletion of macrophage scavenger receptor (MSR) sequences in a case of metastatic prostate cancer. Bookstein and colleagues assembled a YAC contig encompassing 9 cM of genomic sequence (Bookstein et al. 1994), which was used to develop a long-range restriction map of the 2-Mb interval around MSR by pulse-field gel electrophoresis and Southern blotting of YAC DNA (Bova et al. 1996). Genomic probes isolated from this region were then applied to the tumor sample, indicating that the homozygous deletion interval spans 730–970 kb, extending from N877-13 to E20. In addition, supporting evidence is provided by Levy and colleagues (1999), who recently reported another homozygous deletion in the pancreatic cancer cell line MIA PaCa-2, which overlaps with the prostate deletion and encompasses markers D8S549 and D8S1992.
So far only two genes have been located to the interval. One of them, MSR, is expressed in macrophages only and does not appear to be a plausible candidate for a tumor suppressor gene in epithelial neoplasm (Emi et al. 1993). Another gene, called N33, was isolated in the process of direct cDNA selection of YAC clones encompassing the interval (MacGrogan et al. 1996). N33 is silenced by aberrant methylation in most colon cancer cell lines and some primary colorectal tumors. However, SSCP analysis and sequencing of the 11 exons of N33 in 10 metastatic prostate tumor samples did not detect any mutations. Additionally, studies of N33 replacement in several colorectal lines failed to show evidence of tumor suppressor activity manifested as either in vitro inhibition of cell proliferation or suppression of tumorigenicity in nude mice (MacGrogan and Bookstein 1997). The significance of these findings with regard to oncogenesis remains uncertain.
Efforts are under way to identify additional candidate genes within this interval. Chinen and colleagues reported the results of exon amplification of YAC clones in 8p22, including the N877-13 to E20 region (Chinen et al. 1996). Two interesting candidates emerged from this study, including PDGF-receptor β-like tumor suppressor (PRLTS) and centrosome autoantigen PCM-1 (Ohata et al. 1994; Fujiwara et al. 1995), but both lie outside of the homozygous deletion interval. None of the remaining 43 exons have been evaluated further or mapped precisely.
Here we report the results of our efforts to identify additional candidate genes within the tumor suppressor gene interval. A high-resolution contig of bacterial clones has been prepared that spans the entirety of the deletion interval. The integration of this map with existing radiation hybrid (RH) databases and preparation of a RH breakpoint map facilitated identification of mapped and unmapped ESTs extracted from the Radiation Hybrid Database (RHdb) and provided a physical scale to the map. This physical map was used to localize and identify potential transcript units from published sources, databases, and exon amplification experiments. Our efforts resulted in the assignment of six novel transcription units within the N877-13 to E20 interval, of which five are expressed in epithelial tissues and cell lines and possibly represent novel genes. The complete contig of the deletion interval and the transcript map should prove useful for the further identification and evaluation of candidates for the 8p tumor suppressor gene.
RESULTS
Physical Map
Figure 1A illustrates the homozygous deletion interval that was the target of the physical map effort. Figure 1B shows the names and approximate extent of the selected YAC clones that encompass the deletion interval, of which YAC clones 946_c_9 and 932_e_9 had been used previously to develop a long-range restriction map of the deletion interval depicted in Figure 1C (Bookstein et al. 1994). To prepare a physical contig, PCR was used to screen human genomic BAC (Shizuya et al. 1992) and P1 (Pierce et al. 1992) pooled DNA libraries, as well as a cosmid library prepared from flow-sorted chromosome 8 (Deaven et al. 1986), using the primer pairs listed in Table 1. A total of 39 sequence tagged sites (STSs) were used for library screening: 20 are published, 2 were a gift from Dr. R. Bookstein (unpubl.), 6 were designed based on the published sequences of MSR (Emi et al. 1993) and N33 genes (MacGrogan et al. 1996), and the remaining 11 STSs were developed from the end sequences of BACs, P1, and cosmid clones isolated in this study (Table 1). Bacterial clone end sequences were determined not to contain repetitive elements by BLAST search against nucleotide databases accessible through the National Center for Biotechnology Information (NCBI at http://www.ncbi.nlm.nih.gov/BLAST/).
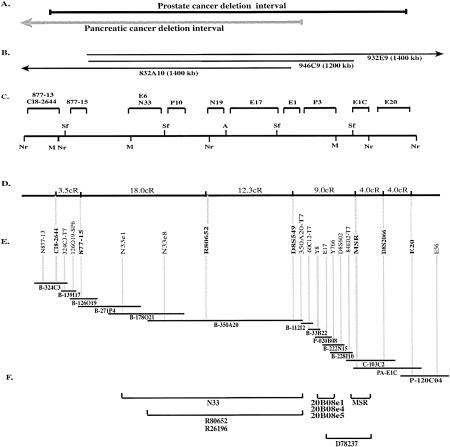
Figure 1.
Physical and transcript map of the 8p22 homozygous deletion interval. (A) Extent of the target interval (Bova et al. 1996; Levy et al. 1999). (B) Selected YAC clones that cover the deletion interval and their approximate extent (Bookstein et al. 1994). The numbers refer to the CEPH database YAC name; the approximate sizes of YAC clones are given in parenthesis. (C) Long-range restriction map of the YAC clones encompassing the interval (adapted from Bova et al. 1996). (D) Two-point distances between the indicated markers in cR based on the analysis with the Stanford G3 RH panel. (E) Minimal-tiling path contig of the deletion interval. Only selected markers are shown for simplicity. Those shown in boldface type have also been used for RH analysis. Vertical gray lines connect markers to the bacterial clones with which they tested positive. (Prefix P) PI clones; (PA) PAC clones; (B) BAC clones; (C) cosmid clones. (F) Location of known genes and partial cDNAs described in Tables 3 and 4. Horizontal bars indicate the clones to which transcripts have been localized.
Table 1.
Complete List of STSs Used for Contig Construction
| STS name | Primer | Annealing temp. (°C) | Product size (bp) | Source | |
|---|---|---|---|---|---|
| forward 5′ → 3′ | reverse 5′ → 3′ | ||||
| N877-13 | GGACCTAACCAGATCCAGTA | GCAGACCAAATATCATCTCTC | 55 | 135 | Bova et al. (1996) |
| D8S1731a | CCAAGCAAATCATGGAAATC | AGCAAACTTATCCCACAAGG | 55 | 220b | Stanford G3 RH Map |
| C18-2644 | CCTCTAAGTCCCTGTGTCAG | AAGCGCTAATGGGTCTCT | 55 | 186 | Bova et al. (1996) |
| 324T7 | GCTAAGATCACAGAGGGAACTGA | ATTGTGTTACGATTGCCGTG | 55 | 146 | end sequence of BAC 324C3c |
| 126SP6 | GCCCTGCTTCAGCCTTATG | GCAGTCCAGTTTATTAGGTCCTG | 55 | 416 | end sequence of BAC 126O19c |
| 150E3-T7 | TTATAGATTGAGGAGCTATGGTTGG | TGTATACATAGGTGTGTATACATAGGT | 55 | 148 | end sequence of BAC 150E3c |
| N877-15 | CACGTCTGGCCTTTATTATG | GTGAATGAACTGCTTTGAGG | 55 | 288 | Bova et al. (1996) |
| N33e1 | GGCCCGCCCCCGACCACAG | CCCGGGGCGCTCCTTCACG | 65 | 176 | N33, exon 1 sequenced |
| E6 | TCCGAGGGCTATATACTTCC | CATCTTGTCTTTGCTCCAAT | 55 | 101 | Bookstein et al. (1994) |
| N33e3 | ATGTGGCTGTGTCGCAATAAAAC | GGGCCATAGTAGAAACACATCAGT | 55 | 217 | N33, exon 3 sequenced |
| D8S1992b | TTCATCGTCTGAACCTGG | CAGGGTTGAAGCAATCTG | 55 | 300b | MacGrogan et al. (1996) |
| P10 | TTGCAGAGGTTCAGGACAG | CATTCAGACAGATGTCAGAGTC | 55 | 800b | Bova et al. (1996) |
| N33e8 | GCCAGGCTCAGTTTGTG | TGTAGCTTTAGTTTGCCCTTTTC | 55 | 94 | N33, exon 8 sequenced |
| 115A12-SP6 | TGCCCCGTAACTTGAAATCT | AGCCATCATCACCTTGCTCT | 55 | 160 | end sequence of P1 115A12c |
| J28 | AGCTTCATCTTCAGAGCAAG | GTACGATAAGGGTGACCATC | 55 | 222 | gift from R. Bookstein (unpubl.) |
| N33e11 | AGGGAACTGGATGAATGGT | TTGCTCGAAAATGGATAGTG | 50 | 229 | N33, exon 11 sequenced |
| D8S549a | AAATGAATCTCTGATTAGCCAAC | TGAGAGCCAACCTATTTCTACC | 55 | 170b | Gyapay et al. (1994) |
| 115A12-T7 | CAGAAACCTTTGTTAACTGTCAG | CTTACCTCATATGCTCACACTGT | 50 | 146 | end sequence of P1 115A12c |
| 350A20-T7 | AGTTTGGGGAGCTGTTTGTTTGAA | CGCGCCCGGCCTACTCTC | 55 | 194 | end sequence of BAC 350A20c |
| 60C12-T7 | GAGCTGCTCCGAGAACTTACAT | GTCAGGCCTTCTACCAACAACAT | 55 | 170b | end sequence of P1 60C12c |
| N19 | GTGAAGGAGGGCAGTCAT | CAGGGTTGAAGCAATCTG | 55 | 175 | MacGrogan et al. (1996) |
| 60C12-SP6 | ACTCTGGTATACACTGATGTAATACAG | TTAGCTGAGACATTTTCTGAAG | 55 | 125 | end sequence of P1 60C12c |
| Y8 | GGGAAGCTATGGTAAAACAT | GGCAAGTAGATGTTACAGAAG | 55 | 150b | gift from R. Bookstein (unpubl.) |
| E17 | CAAGGCATATCACAACTGC | GATAATTGAACTGTCACCTCTG | 55 | 121 | Bookstein et al. (1994) |
| YE766 | GACTCTTGCCACCTTGTAAA | ATCTCCAAACCTACTTCTCC | 55 | 89 | Bookstein et al. (1994) |
| E1 | TGACACACTTGCCATTTGAT | TTCCATTAGTCCCAGTTGTC | 55 | 131 | Bookstein et al. (1994) |
| D8S602a | GACTCTATTTTACTATCATTGACAG | GTTATCTTCTGCCTGGTGAAAAG | 55 | 300b | Takanishi et al. (1997) |
| 45G06-SP6 | GAGGAAAATTCTAAACAAAATGTT | AATTTGCCAGATCTTTAG | 50 | 143 | end sequence of P1 45G06c |
| E2d | TGAAGCCATCTGTGGG | GTAAGAACTGTTAAGTGGAAAG | 55 | 116 | Bookstein et al. (1994) |
| MSR10 | GTGGGAGCGGCCCTCACGAG | TGCACGGCTTGAACACCTGGGTAT | 60 | 139 | MSR, exon 10 sequenced |
| 84E02-T7 | CTACCTTGCCACCTTCCAAAACTA | GAGAAGCCAACACTGAGGGAACTA | 50 | 154 | end sequence of COS 84E02c |
| MSR5 | TTAGAGGAGCGTGTTTACAATGT | GAATGTTCCCAATCTTTCAGTCT | 55 | 143 | MSR, exon 5 sequenced |
| E3 | GCCTGTTTCATCGAACC | CCTGGCATTCTTTACCTAGA | 55 | 85 | Bookstein et al. (1994) |
| GATA29A08 | CCTGAGTCCAGGGAGGTC | CGGAAAAAACCCAAGTACCA | 55 | 332 | Stanford G3 RH Map |
| MSRa | TTCATCTATTGCATTCC | CAAAATTTCAGCATGACAACTG | 50 | 102 | Matsumoto et al. (1990) |
| D8S2066 | TTTTCTCCATCCGGTGACTC | CCAACTACGGCATGGTTTCT | 55 | 170b | Stanford G3 RH Map |
| 84E02-T3 | CCCTGGTGAGACAGCGAGAC | AACTCACTCTTCACCCCATTTAGG | 50 | 149 | end sequence of Cos 84E02c |
| E20 | TGAATTTGCATAGTCTGCAG | CAGCTCTAACAAGGCTCCTA | 55 | 107 | Bookstein et al. (1994) |
| E56 | TTTGTTGAGGACAAATACCC | TGTCACGATGAGGATTGTTA | 55 | 170 | Bookstein et al. (1994) |
Denotes polymorphic markers.
Approximate PCR product size.
Primers derived from end sequences of bacterial clones obtained in this study.
Primers designed based on published sequences.
Additional clones were isolated from the BAC and P1 libraries by hybridization-based screening of high-density gridded filters using an Alu PCR amplification product of DNA from the YAC clones shown in Figure 1C as a probe. A PAC clone, E1C, included in this contig, was kindly provided by Dr. R. Bookstein (unpubl.). This clone helped to close a gap that was not represented in the P1 and BAC libraries. Finally, hybridization-based screening of an additional BAC library (human, RPCI-11, Research Genetics) allowed isolation of the BAC clone 112I2, which bridged the last remaining gap between telomeric and centromeric portions of the contig between markers 350A20-T7 and 60C12-T7.
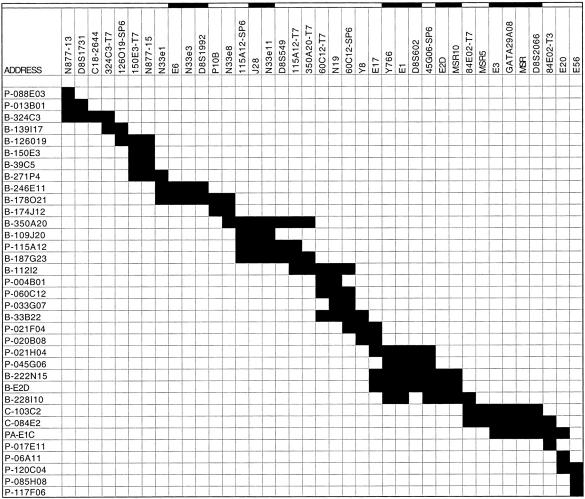
The completed physical map of the N877-13 to E20 interval is depicted in Figure 1E as a least-tiling path of selected bacterial clones and markers; a comprehensive map, showing all of the isolated clones and their STS content, is presented in Figure 2. Marker order in Figure 2 was based on STS content of the individual bacterial clones, as well as additional information given in Table 1 (such as derivation from the intronic or exonic sequences of MSR and N33 genes, and from the end sequences of bacterial clones). The contig is about 1.5 Mb in length, and the average marker density across the contig is approximately one per 40 kb of genomic sequence. The complete contig map represents at least twofold coverage of much of the interval by contiguously overlapping BAC, PAC, P1, and cosmid clones.
Figure 2.
Complete list of the isolated bacterial clones and their STS content. Marker names are listed across the top row. Markers who order is unresolved in relation to other markers that map to the same clone(s) are indicated by horizontal solid bars at top. Clone names are listed in the left column in order from telomere (top) to centromere (bottom). (Prefix P) PI clones; (PA) PAC clones; (B) BAC clones; (C) cosmid clones. STS content of each clone is indicated by a solid box next to the clone name.
RH Analysis
RH analysis was performed to define the position of selected markers on the physical map in relation to other markers on the Stanford G3 RH map, and to provide an independent verification of the orientation of the entire contig on the larger chromosomal scale. A total of 15 markers were analyzed, 12 of which were assigned to bins 2, 3, and 4 on the Stanford G3 RH map, including MSR (or WI-7228), D8S549, and R80652 markers. Three markers (E20, CI8-2644, and N877-15) were typed on the Stanford G3 RH panel (Stewart et al. 1997) in our laboratory. Note that markers E20 and CI8-2644 define the boundaries of the deletion interval. Markers were analyzed using the RHMAP 3.0 statistical package (Lange et al. 1995). Ten markers entered the same linkage group under an lod score criterion of 10.0; two point distances for these markers were calculated with lod scores >7 and are shown in Figure 1D for those located in the deletion interval. The calculated overall distance between markers CI8-2644 and E20 (opposite ends of the deletion interval) is ∼50.8 cR or 1.47 Mb, using the conversion of 1 cR = 29 kb for the G3 panel (Stewart et al. 1997). Markers entering the same linkage group were analyzed using the RHMAXLIK program. Although it was not possible to order loci with minimum support for the best locus order of >1.8 log10 units, the following marker order was calculated: TEL–D8S511–CI8-2644–N877-15–R80652–D8S549–MSR–D8S2066–E20–SHGC33312–D8S1715–CEN. This level of support reflects the limitations of the G3 panel; however, this order is consistent with the large-scale restriction map of the interval (Bova et al. 1996) and our bacterial contig data.
Placement of ESTs Identified from the Human Transcript Map
ESTs from the publicly available RH consensus transcript map (http://www.ncbi.nlm.nih.gov/SCIENCE96/) were integrated into the constructed physical map as follows. The interval to be searched was defined from D8S511 to D8S261, which includes the homozygous deletion interval. The search produced a total of 36 ESTs. BLASTN analysis reduced this number to 27 nonredundant ESTs. All 27 ESTs were then analyzed in more detail using Whitehead-MIT WC8.1 YAC contig assignment data (when available) and placement information from the publicly available RH maps. Based on this information, 19 mapped outside of the deletion interval and, hence, were not pursued further. The remaining eight markers were typed by PCR reactions using DNA from the YAC clones spanning the interval. Three of the eight produced positive hits and identified the following YAC clones: YAC 832_a_10 was positive for marker R80652 and YAC 946_c_9 was positive for markers stSG4991 and WI-17604. YAC-positive markers were then typed using DNA from the bacterial clones in the interval. Marker R80652 was localized to BAC 350A20 (Fig. 1F), but markers stSG4991 and WI-17604 could not be confirmed by PCR to localize to any bacterial clone in the contig.
Exon Amplification
Bacterial clones from the deletion interval were searched for coding sequences using exon amplification. Five clones covering approximately half of the 1.5-Mb interval were subjected to exon amplification. The resulting 300 clones were digested to release the insert, examined by gel electrophoresis, and subgrouped based on the insert size. A total of 32 clones, each of unique insert size, were sequenced and analyzed. After discarding those containing false splices or vector-derived sequences, five clones remained that contained unique inserts. All five inserts were isolated, labeled, and hybridized to a Southern blot containing BamHI digests of total genomic DNA and DNA from the bacterial clones in the interval (data not shown). Three inserts (20B08e1, 20B08e4, and 20B08e5) appeared to be single-copy genomic sequences, as they recognized single restriction digest bands. Although all three hybridized back to the same clone, P1 20B08 (Fig. 1F), each of them detected a different size band. Two inserts did not hybridize to either genomic DNA or bacterial clones and were not investigated further.
Placement of New ESTs Deposited in RHdb
The RHdb release 13 data set (http://www.ebi.ac.uk/RHdb/index.html) was used as a source of unmapped ESTs. The identification of those located either within or near the target interval was done based on the RH breakpoint map using methods similar to those described previously (Horrigan et al. 1999). The analysis produced 23 nonredundant EST markers that were located around the interval and required further confirmation. Two of them were localized independently to the interval by other methods: EST R80652 (RH70072) was obtained from the Human Transcript Map (see above), and EST RH36122 represented a fragment of the N33 coding sequence. Ten of the resulting 21 markers had genomic assignments to adjacent locations, and 11 were previously unmapped. All 21 markers were tested by PCR on the minimal tiling path of bacterial clones and YAC clones covering the interval. EST (RH36526 or R26196) mapped to YAC 932_e_9 and BAC 350A20 (Fig. 1F). EST RH36122 represented a segment of DLC-1 (deleted in human liver cancer), a gene reported recently that was suggested as a possible tumor suppressor gene (Yuan et al. 1998). On PCR analysis, this EST did not test positive with either the BAC or the YAC clones and has been reported recently to be located outside of the interval (P.J. Wilson and G. Chenevix-Trench, unpubl.). The remaining 20 ESTs did not represent any known genes and did not test positive with bacterial clones within the interval. Thus, the analysis resulted in the identification of one novel EST that had not been identified by other methods.
Other Sources of Expressed Sequences
Chinen and colleagues (1996) previously have reported results on the exon-trapping of three YAC clones, of which 931_a_1 and 946_c_9 cover the deletion interval. Among 45 exon-like fragments isolated, 4 represent cDNA fragments of two known genes (PCM-1 and PRLTS) and map outside of the interval. To ascertain which of the remaining 41 fragments lie within the deletion interval, PCR primers were designed for those that had sufficient sequence information (Table 2). A total of 18 primer pairs were successfully prepared, and these were typed on the bacterial contig and YAC clones. Primers designed from exon D78237 identified two overlapping bacterial clones on the contig: BAC 222N15 and BAC 228I10 (Fig. 1F). Two exons (D78242 and D78220) mapped to YAC 932_e_9, but not to any bacterial clone, and are thus thought to be outside of the interval.
Table 2.
PCR Primers Prepared for Evaluation of Expressed Sequences and Their Location to the Physical Map
| Sequence name | Primer | Ta (C)a | Prod size (bp) | YAC testingbc | Contig testingbc | Source | |
|---|---|---|---|---|---|---|---|
| forward 5′ → 3′ | reverse 5′ → 3′ | ||||||
| D78212 | TCCCAAAACAGAAAGAGCAGATGT | GTGGGGCAGGTGTTCCAATAAA | 50 | 121 | 932 e 9 | − | Chinen et al (1996) |
| D78215 | CCACCGGCAGGAATGAACAAC | CGCTCTAGTGCCGTCTGAGTCTGT | 60 | 165 | − | − | Chinen et al (1996) |
| D78216 | ATCAGTACAAGGGGATTCTAACT | ATGTTTTCTTAACGAGGAGACA | 50 | 82 | − | − | Chinen et al (1996) |
| D78221 | CCCCTCAGGATTGACTAT | TCACGTCACCTTTTGCAT | 50 | 104 | − | − | Chinen et al (1996) |
| D78225 | ACAGCCCCTCAGGATTGACTA | GAGGCATCATCGAAGGTAAGG | 55 | 88 | − | − | Chinen et al (1996) |
| D78226 | AGAAGCCCCGAAGAGACACCAA | GGCGCTTTGCCAGGGAAGA | 55 | 134 | − | − | Chinen et al (1996) |
| D78227 | GCACCGTTAACAACATGTCTATTT | GAAGAGCCAAGGAAGAGAAACTT | 50 | 143 | − | − | Chinen et al (1996) |
| D78228 | TTCAGGTCATTTTTCCAGTTT | CTTCCCTATAGAACGACTTG | 50 | 90 | − | − | Chinen et al (1996) |
| D78229 | ATGCGCTACCCCCTGAGAAAAC | GCAGGTGCTGAATCACAACTGTCA | 55 | 147 | − | − | Chinen et al (1996) |
| D78230 | TTTTTGAGCTAGAGTTCACATAGA | TGGCAAGCGAATTCACTTCT | 50 | 85 | − | − | Chinen et al (1996) |
| D78231 | AGAGGAAAGGCCACATGAGGACA | AGTGTTGGCAGGTCTGATTTCTCC | 60 | 90 | − | − | Chinen et al (1996) |
| D78233 | CCTAGGACCCCTGCCTCAG | CCAAAGGGTGGTCAAAGTAGGT | 60 | 81 | − | − | Chinen et al (1996) |
| D78234 | TCCTGGCCTAGAGGTCTTA | TGTTTTTGTAGGTCTCAATCTTA | 50 | 103 | − | − | Chinen et al (1996) |
| D78237 | TATCTTGAGCCAACCTCCTGTCTC | TCTTGATAGGGGCTGGGTAAAATA | 55 | 103 | 932 e 9 | B-222N15, B-222I10 | Chinen et al (1996) |
| D78239 | CTATCCATGCTAAGACATCC | TGTATAAAGGAAAGAAGTTGAATG | 50 | 98 | − | − | Chinen et al (1996) |
| D78242 | CGGCCTATGCGACGAGTCA | GTAATTTCGCTGCTTCCAGTCG | 50 | 111 | 932 e 9 | − | Chinen et al (1996) |
| D78246 | TTCTGCTTTTCCTGGACCTGTTCC | CACCGGGCTGGGAGGACTGT | 55 | 123 | − | − | Chinen et al (1996) |
| D78247 | CCCCAGTTGAGCCATGAGATAA | TTGGCAATTTCTGTAAGTCAGGAA | 55 | 123 | − | − | Chinen et al (1996) |
| R80652 | GTCTGATGTCACTCAGATTGAAGG | TGAGCAAAAAGTCTATGCCTAAA | 55 | 109 | 932 e 9 | B-350A20 | Chinen et al (1996) |
| WI-17604 | TGGGCCAAGAGCTTTCTG | TAGACTACTTTGGAGATGCAGCC | 50 | 139 | 932 e 9 | B-350A20 | Hum. Gene Map, RHdb |
| 20B08e1 | CAATGAGAGGGGCTTTGT | GTTTCCATGGGATGTTCA | 50 | 70 | 932 e 9 | P-20B08 | exon amplification |
| 20B08e4 | ACTGGCCACCACCAGCGTGAT | GAGCAAGATGGCAGGGGTGGTGTG | 55 | 102 | 932 e 9 | P-20B08 | exon amplification |
| 20B08e5 | GGGAATTTTAGGTGGTGGTGGTTC | AGGATTCAGGCATAATGGTTTAGC | 55 | 191 | 932 e 9 | P-20B08 | exon amplification |
| R26196 | GAATCGTTCCATCATATCCACA | TGACAGCAGGTAACCGATACC | 55 | 201 | 932 e 9 | B-33B22, P-20B08 | RHdb |
(Ta) Annealing T.
The YAC or BAC names indicate that the EST tested positive with this clone.
Negative signs indicate that the EST did not test positive with any YAC clone or bacterial contig.
Characterization of the Expressed Sequences within the Interval
All ESTs localized to the interval were evaluated for their expression profile by Northern blot analysis. Inserts from the exon amplification-isolated clones 20B08e1, 20B08e4, and 20B08e5 were gel-purified and labeled using random hexamer priming. For the remaining three ESTs, PCR amplification products of genomic DNA with EST primers were used as a probe. PCR products were labeled by random hexamer priming or kinase end-labeling reactions, depending on the size of the product. All probes were hybridized to a multiple tissue Northern blot (Clontech) containing the following tissues: spleen, thymus, prostate, ovary, small intestine, colon, heart, placenta, skeletal muscle, and peripheral blood lymphocytes (PBL). Probes 20B08e1 and 20B08e4 detected an RNA transcript of ∼1.2 kb in size, which is expressed in all tissues tested with a high abundance in spleen and PBLs. Probes 20B08e5, D78237, R80652, and R26196 did not detect any message.
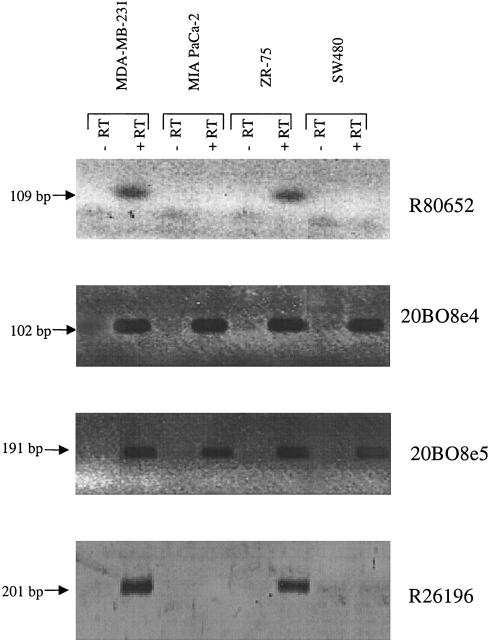
EST primers derived from all six sequences (Table 2) were used for RT–PCR analysis using total cellular RNA isolated from the epithelial cancer cell lines SW480 (colon), MDA-MB-231 and ZR-75 (breast), and MIA PaCa-2 (pancreas). Primers designed from sequence 20B08e1 tested negative in all cell lines studied. However, this probe clearly detects a 1.2-kb message on the Northern blot, thus confirming its expression in normal epithelial tissues. The remaining five ESTs showed differential patterns of expression, presented in detail in Table 3. Of particular interest is the fact that ESTs R80652 and R26196 did not show expression in the pancreatic cancer cell line MIA PaCa-2 (Fig. 3). This cell line recently has been reported to contain a homozygous deletion encompassing markers D8S549 and D8S1992 at the telomeric portion of the interval (Levy et al. 1999); therefore, the genomic sequences from which these ESTs are derived are expected to be deleted in this cell line. Representative pictures of the RT–PCR analysis are shown in Figure 3.
Table 3.
Expression Analysis of Candidate Sequences by Northern Hybridization and RT–PCR
| Sequence name | Clone IDa | Sequence available (kb) | Northern | RT–PCR | ||||
|---|---|---|---|---|---|---|---|---|
| band size (kb) | tissue specificityb | MB231 | PaCa-2 | ZR-75 | SW480 | |||
| R80652 | 146794 (3′) | 1.86c | none | N.A. | + | − | + | − |
| 20B08e1 | — | 0.1c | 1.2 | all tissues tested | − | − | − | − |
| 20B08e4 | — | 0.5c | 1.2 | all tissues tested | + | + | + | + |
| 20B08e5 | — | 0.35c | none | N.A. | + | + | + | + |
| D78237 | — | 0.154 | none | N.A. | + | − | − | − |
| R26196 | 133050 (3′) | 1.22c | none | N.A. | + | − | + | − |
IMAGE Consortium clone ID. Clones are available through Research Genetics.
Tissues used for analysis are: spleen, thymus, prostate, testis, ovary, small intestine, colon, PBL. (N.A.) Not applicable.
Figure 3.
Representative pictures of RT–PCR analysis on selected epithelial cancer cell lines. RT–PCR was performed on total cellular RNA. First-strand cDNA was synthesized using oligo(dT); PCR primers were designed from each partial cDNA clone. The lanes are as indicated. Control reactions without reverse transcriptase and reactions containing water were performed for every sample.
Only limited sequence information was originally available for the identified transcript units. Additional sequence information was obtained for exon-like fragments 20B08e1, 20B08e4, R80652, and R26196 by performing 3′ and 5′ RACE on a Marathon-Ready human fetal brain and placenta cDNA library (Clontech), as is shown in Table 3. The available sequence of all six transcribed units was then used to search the UniGene human sequence collection and The Institute for Genomic Research (TIGR) human gene index database (http://www.tigr.org/tdb/tdb.html) to identify all corresponding cDNA clones and detect possible overlap with other clones. The EST R26196 is represented by two cDNA clones, and EST R80652 represented single-clone isolates in the TIGR human gene index database. None of the other sequences (20B08e1, 20B08e4, 20B08e5, and D78237) identified cDNA clones in any of these databases.
Sequences of all six ESTs were used to BLAST search nucleotide and protein databases accessible through NCBI and TIGR. None of the expressed sequences appeared to be identical to any known genes. Exon 20B08e5 showed 95% nucleotide homology to the uncharacterized Homo sapiens cDNA clone (GB#AI142134). Some interesting homologies were revealed for the following sequences: 20B08e1 showed 76% homology with the TIGR cDNA clone contig for H. sapiens mRNA for phosphatidylinositol-4-phosphate 5-kinase, type II, isoform c (TGI#THC154497). Sequence 20B08e4 did not show any strong homologies at the nucleotide level using the BLASTN program, but the BLASTX search revealed two stretches of 69% and 84% protein homology with H. sapiens mRNA for UDP–GalNAc:polypeptide N-acetylgalactosaminyl transferase (GalNAc-T3) (GB#X92689). Finally, D78237 showed 88% nucleotide homology with sperm surface protein PH-20 mRNA (GB#L13779). BLAST search results for all six ESTs are shown in Table 4.
Table 4.
Results of BLAST Search with Expressed Sequences from the Deletion Interval
| Sequence name | Source | BLASTN | BLASTX | ||||||
|---|---|---|---|---|---|---|---|---|---|
| similarity | identity (%) | length (bp) | P(N) | similarity | identity (%) | length (bp) | P(N) | ||
| R80652 | Human Transcript Map, RHdb | none | none | ||||||
| 20B08e1 | exon trap | none | THC154497; PIP5K | 76.2 | 27 | 5e-22 | |||
| 20B08e4 | exon trap | none | X92689; H. sapiens mRNA | 84 | 26 | 2e-8 | |||
| for UDP–GalNAc: GalNac-T3 | 69 | 13 | 9e-12 | ||||||
| 20B08e5 | exon trap | AA278418; H. sapiens cDNA clone IMAGE: 703548 | 95 | 71 | 2e-25 | none | |||
| D78237 | published | L13779; sperm surface protein PH-20 mRNA | 52 | 88 | 1e-25 | ||||
| R26196 | RHdb | none | none | ||||||
DISCUSSION
An adequate delineation of a putative tumor suppressor gene site and preparation of a detailed physical map of the interval are the first critical steps in gene identification when using a positional candidate strategy. The 8p22 interval has been thought to contain a tumor suppressor gene, based on recurring LOH in many different types of epithelial neoplasia; however, different reports suggest different locations of the minimal deletion region. A small homozygous deletion at the MSR locus, reported in metastatic prostate carcinoma, provides supporting evidence for this site and also sets a logical starting point for the cloning effort. The deletion spans a genomic interval of ∼1–1.5 Mb in size from N877-13 to E20 encompassing D8S549 and MSR. Two genes previously have been mapped to the interval, but neither one appears to be a tumor suppressor gene.
Here we present the preparation of a high-resolution bacterial contig of the deletion region. The contig provides twofold or greater redundant coverage over much of the interval with contiguously overlapping BAC, PAC, P1, and cosmid clones. The approximate size of this contig is 1–1.5 Mb, and the average marker density across the contig is ∼1/40 kb of genomic sequence.
An unresolved contradiction in the literature leaves somewhat questionable the relative position of MSR and CI8-2644 in the context of adjacent markers on 8p22 (Fujiwara et al. 1994; Bova et al. 1996), which may potentially affect the relative placement of some key markers across the deletion interval and orientation of the entire contig on the larger chromosomal scale as well. To obtain independent data addressing this question, we attempted to position CI8-2644, MSR, and E20 in relation to each other and to other markers outside of the deletion boundaries, based on the Stanford G3 RH panel. Even though we could not position markers with a high level of support, the obtained order unequivocally agreed with the marker placement on the YAC-based long-range restriction map of the interval (Bova et al. 1996).
The bacterial contig and its integration with the existing EST mapping resources proved to be useful in the identification of the potential coding sequences. A combination of complementary approaches was used in this search, including database analysis, exon amplification of a portion of the interval (primarily excluding the N33 region), and an extensive search of published literature. ESTs from the D8S511 to D8S261 segment on the Human Transcript Map were identified and confirmed on the bacterial contig. This search produced EST R80652. Other ESTs from RHdb were assessed by means of a RH breakpoint panel, similar to an approach reported recently for a chromosome 5 analysis (Horrigan et al. 1999). This yielded three clones, two of which had been placed on the Human Transcript Map, and one, R26196, which was previously unmapped. Exon amplification provided three sequences, all of which are expressed, based on Northern blot and/or RT–PCR analysis; two of them show homology to known genes. Localization to the interval was confirmed for a fourth exon-like fragment, D78237, which was reported in a study by Chinen and colleagues (1996). Interestingly, neither we nor Chinen et al. isolated coding sequences of the MSR gene in this approach, even though this gene is present within the interval analyzed.
The transcripts described in Tables 3 and 4 represent six unique sequences localized to the interval, five of which appear to be expressed and therefore derived from cDNAs. Exon-like fragment D78237 did not show expression on either Northern blot or RT–PCR analysis and probably does not originate from coding sequence. None of the identified ESTs are known genes, although three show interesting homologies that will warrant further study.
Sequence 20B08e4 was found to be homologous with GalNAc-T3. It is an interesting finding, as GalNAc-transferases initiate mucin-type O-glycosylation, which is the formation of an O-glycosidic linkage between GalNAc and serine or threonine residues. Aberrant glycosylation and expression of mucins is associated with malignant transformation of epithelial cells from different sites (Hennebicq-Reig et al. 1997; Reis et al. 1998). O-glycosylation has also been shown to be important in the folding of proteins such as human chorionic gonadotropin and the binding of cell adhesion molecules such as selectins and selectin ligands (Clausen and Bennett 1996). GalNAc transferases are also required for the synthesis of ganglioside GM2, which is usually significantly elevated in human melanoma cells (Hoon et al. 1992) and is one of the major gangliosides on the cell surface of neuroectodermal tumors (Nakamura et al. 1994).
Sequence 20B08e1 revealed homology with cDNA for PIP5K. The phosphatidylinositol pathway is implicated in the regulation of numerous cellular functions and responses to extracellular signals. An important branching point in the pathway is the phosphorylation of phosphatidylinositol 4-phosphate by PIP5K to generate a second messenger, phosphatidylinositol-4,5-bisphosphate (PIP2). PIP5K and PIP2 have been implicated in signal transduction, cytoskeletal regulation, DNA synthesis, and vesicular trafficking. Some findings suggest that a subset of TNF responses may result from the direct association between PIP5Kβ (isoform of PIP5K) and TNF receptor p55 (Castellino et al. 1997).
Sequence D78237 revealed homology with a sperm membrane protein, PH-20. PH-20 has hyaluronidase activity that is required for sperm penetration through the cumulus cell layer that surrounds the oocyte. This gene was shown to be testis-specific by Lin et al. (1993) and thus is not a likely candidate for a tumor suppressor gene of epithelial origin.
Additional studies are under way to obtain the complete cDNA coding sequence for all of these isolated ESTs and to investigate tumor mutations and biologic function, to evaluate them as candidates for the 8p22 tumor suppressor gene.
METHODS
PCR-Based Library Screening
PCR amplification was performed on DNA superpools and plate pools of the BAC (Research Genetics) and P1 libraries (Research Genetics), according to the recommended procedures. Cosmid clones were obtained by PCR screening of 110 plates of the LA08Nco1 flow-sorted chromosome 8-specific cosmid library (Deaven et al. 1986) using a plate-pool strategy. PCR reactions were carried out in a 20-μl volume consisting of the following: 100 ng of DNA, 1× PCR buffer with 1.5 mm MgCl2 (PE Applied Biosystems), 0.1 mm dNTPs (Pharmacia), 1 μm of each primer, and 1 unit of AmpliTaq DNA polymerase (PE Applied Biosystems). Amplification was done in a PTC-100 Thermocycler (MJ Research) with an initial denaturation of 94°C for 3 min; then 35 cycles of denaturation at 94°C for 30 sec, annealing for 20 sec, and elongation at 72°C for 30 sec; followed by a 3 min elongation at 72°C. The annealing temperature for each PCR primer pair is listed in Table 1. PCR products were analyzed by electrophoresis on a 2% Nusieve agarose gel (FMC BioProducts) and visualized by ethidium bromide staining.
Hybridization-Based Library Screening
High-density gridded filters were hybridized using an inter-Alu repeat amplification product of YAC DNA as a probe. Probes were labeled by random decamer priming using the DECAprime II DNA labeling kit (Ambion). PCR amplification of YAC DNA and hybridization were performed as described by Horrigan and colleagues (Horrigan and Westbrook 1997). Primers used for amplification were previously described (Horrigan and Westbrook 1997). Identified positive clones were ordered from Research Genetics and checked by PCR on cultures for STS content.
DNA Purification
DNA was prepared using tip-500 columns (Qiagen) and the suggested manufacturer's protocol was modified as follows. Glycerol stocks were inoculated in 1 liter of LB media with 25 μg/ml chloramphenicol (for BAC clones) or 12.5 μg/ml kanamycin (for P1, PAC, and cosmid clones) and incubated overnight at 37°C. The culture was divided into two 500-ml preparations, precipitated, resuspended in P1 buffer containing 1 mg/ml lysozyme, and incubated at 37°C for 15 min. DNA was eluted from the columns by adding three 5-ml aliquots of QF buffer preheated to 65°C.
Generation of BAC, P1, and Cosmid End Sequences
P1 and cosmid clones were sequenced manually, using SequiTherm Cycle Sequencing kit (Epicentre Technologies). Five fmoles of template and 1.5 pmoles of kinase-labeled primer were used for each sequencing reaction; samples were prepared as described in the manufacturer's protocol. The following primers were used: P1 SP6 (5′-GGCCGTCGACATTTAGGTGACAC-3′) and P1 T7 (5′-CCGCTAATACGACTCACTATAGGG-3′) primers for P1 clones and P1 T7 and T3 (5′-ATTAACCCTCACTAAAGGGA-3′) primers for cosmid clones. PCR reactions were carried out in the Perkin-Elmer 480 thermocycler at the following cycle conditions: initial denaturation at 95°C for 3 min; 20 cycles of 95°C for 30 sec, 50°C for 30 sec, 70°C for 1 min; 10 cycles of 95°C for 30 sec, 70°C for 1 min. After the PCR reaction, loading dye was added to the samples and they were separated on a gel or stored at −20°C. Samples were analyzed on a 6% Long Ranger gel (FMC BioProducts) with 8 m urea at 80 V for 1 hr and 20 min. Dried gels were exposed to Kodak X-Omat film for 12 hr at −20°C with an intensifying screen.
BAC end sequencing was performed on the automated ABI 373 DNA sequencer. The ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) was used. Three micrograms of BAC DNA and 50 pmoles of primer were used in a total volume of 40 μl. The following primers were used: T7 (5′-TAATACGACTCACTATAGGG-3′) and SP6 (5′-ATTTAGGTGACACTATAG-3′) (Boysen et al. 1997). PCR reactions were carried out in the PTC-100 thermocycler (MJ Research) under the following cycle conditions: initial denaturation at 94°C for 4 min; 25 cycles of 96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min. After cycling, PCR reactions were purified by ethanol precipitation (protocol 1) as described in the manual.
Southern Hybridization
Genomic or bacterial DNA was digested with either BamHI or HindIII in the presence of spermidine. Restriction fragments were then fractionated through a 0.7% agarose gel and transferred to a Hybond N+ nylon membrane (Amersham) under alkaline conditions. Probes were labeled by random decamer priming using the DECAprime II DNA labeling kit (Ambion). Hybridization and washing were performed at 65°C as described in Hybridization-Based Library Screening (above), without the prehybridization step.
Northern Analysis
Human MTN (multiple tissue Northern) blots (Human II 7759-1 and Human 7760-1, Clontech) were hybridized in ExpressHyb hybridization solution (Clontech) according to the manufacturer's protocol.
RT–PCR
Total cellular RNA was isolated using Trizol reagent (BRL, Life Technologies) following the suggested protocol. First-strand cDNA synthesis was performed using SuperScript Preamplification System and oligo(dT) primer (BRL, Life Technologies) according to the supplied protocol. First-strand cDNA was amplified in the standard PCR reaction using 10% of the first-strand reaction and AmpliTaq DNA polymerase (PE Applied Biosystems). The sequences of primers used and annealing temperatures are indicated in Table 2. Control reactions without reverse transcriptase and reactions containing water instead of cDNA were performed for every sample. RT–PCR products were analyzed by electrophoresis on a 2% Nusieve agarose gel (FMC BioProducts) and visualized by ethidium bromide staining.
Exon Amplification
Exon amplification was performed using GIBCO BRL Exon Trapping System (BRL, Life Technologies) according to the manufacturer's and published protocols (Buckler et al. 1991; Church et al. 1994). Briefly, DNA from bacterial clones was restricted using a BamHI–BglII double digest. The resulting fragments were subcloned into the vector pSPL3. Individual transformants were pooled, and episomal DNA extracted from these pools was electroporated into COS-7 cells. Cytoplasmic RNA was extracted and used as a template for reverse transcription followed by PCR using vector-specific primers. Primary PCR amplification was BstXI-treated to reduce the recovery of vector and cryptic splicing products. The product of the secondary PCR amplification with nested primers was cloned into the pAMP10 vector. Individual colonies were transferred to 96-well plates, propagated, and stored as 20% glycerol stocks at −20°C. To minimize the number of false positives, colony filters were hybridized with kinase-labeled PCR product derived from the amplified HIV-intron portion of the vector pSPL3 with primers SD6 (5′-TCTGAGTCACCTGGACAACC-3′) and SA2 (5′-ATCTCAGTGGTATTTGTGAGC-3′). To size-select material for sequencing, 96-well plates with single clones were PCR amplified using primers designed from the cloning site flanking sequences of the vector pAMP10: EXO6 (5′-GCCAGTGAATTGAATTTAGGTG-3′) and EXO7 (5′-CAAGCTCTAATACGACTCAC-3′). PCR samples were run on a gel and inserts were grouped based on size. Hybridization-negative clones from different insert-size groups were chosen for sequencing. Clones were sequenced on both strands using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) following the manufacturer's instructions. Sequences were analyzed using BLAST programs (Altschul et al. 1990).
RACE analyses (5′ and 3′) were performed using Marathon-Ready cDNA (Clontech) and Advantage KlenTaq Polymerase Mix (Clontech) according to the manufacturer's instructions. First-amplification primers and nested primers were designed from the exon amplified and sequenced clones. The following primers were used for 20B08e1: sense ex1-RACE F (ATGGGATGTTCATGAGAAAGATTT), annealing temperature = 58°C, and antisense ex1-5′ RACE (CTGCCCAGTGAGAGGGGCTTTG), annealing temperature = 62°C; nested sense nex1-3′ RACE (ATGGACAGGAGACTGCAAACAAA), annealing temperature = 60°C; and nested antisense ex1-RACE R (ATGGGATGTTCATGAGAAAGATTT), annealing temperature = 62°C. For 20B08e4: sense ex4 F (ACTGGCCACCACCAGCGTGAT), annealing temperature = 65°C; antisense ex4-5′ RACE (GCTGGCATCATCCACCAGTATGATCT), annealing temperature = 65°C; nested sense nex4-3′ RACE (GTGTTCCACAACGAAGCCTGGTC), annealing temperature = 63°C; nested antisense ex4R (GAGCAAGATGGCAGGGGTGGTGTG), annealing temperature = 68°C.
RH Analysis
PCR score data for the unmapped markers were obtained in our laboratory upon amplification of the Stanford G3 radiation hybrid panel (Research Genetics). The G3 panel was amplified at least twice for each marker. PCR amplification was carried out in 20 μl reactions under standard conditions. The ability of each marker to amplify DNA from an individual hybrid was scored as 0 (no amplification), 1 (clear amplification), or 2 (uncertain). PCR scores for the mapped markers were extracted from the RHdb depository. All PCR scores were analyzed using the RHMAP statistical package (Lange et al. 1995). The RH2PT program was used to calculate two-point distances and linkage groups. Size estimates between markers were calculated based on the conversion of 1 cR3000 = 270 kb for the Genebridge panel (http://www-genome.wi.mit.edu) and 1 cR10000 = 29 kb for the Stanford G3 panel (Stewart et al. 1997). The RHMAXLIK program was used to compute marker order with a maximum likelihood. The equal retention model and the stepwise ordering option were applied; the following conditions were chosen: max log10 likelihood difference to save order = 5; max log10 likelihood difference to print order = 3; max log10 likelihood support to add locus = 1.
Identification of Unmapped ESTs from RHdb
The identification of unmapped ESTs located around the target interval was done in a manner similar to that described previously (Horrigan et al. 1999). In brief, a segment on the Whitehead-MIT Genebridge4 RH map was defined, which included all positioned markers across the 8p22 chromosomal band, including MSR and D8S549. The RH mapping data were analyzed for all 24 markers in this segment, and the panel of 13 radiation hybrids with breaks within the defined interval was assembled, thus representing the breakpoint panel, which was used later at the last step of analysis. Next, 12 radiation hybrids were identified that had positive scores with all 24 core markers and presumably contained the interval in its entirety. Then, the GB4 subset of the chromosome 8-linked markers (total of 2209 markers) was extracted and all markers positive for all 12 8p22-containing hybrids were identified. These 241 markers were then analyzed using the RH2PT program to identify those within 10 cR (∼3 Mb) distance of MSR and/or D8S549, with lod scores >12. A total of 32 markers were so identified and their PCR scores were compared to the breakpoint panel. Six markers were ruled to be located outside the interval based on the fact that their PCR scores did not match PCR scores of MSR and D8S549. The remaining 26 markers had identical PCR scores and were equally likely to be close to the deletion interval, and their position could not be resolved any further, given the low resolution of the Genebridge4 RH panel. Analysis revealed 3 redundant entries; the remaining 23 markers were all ESTs and required further confirmation by PCR typing on the bacterial contig.
Acknowledgments
We thank Dr. Robert Bookstein for the invaluable help in map construction and for kindly providing PAC clone E1C and STS primers Y8 and J28. We also thank Cathrine G. Summer (Research Genetics, Inc.) for assistance with isolation of the BAC clone 112I2, which was critical to closing the gap on the map. We thank Dr. Mark Heller for the valuable discussions and help in manuscript preparation. This work has been supported by National Institutes of Health grant CA56705.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding author.
E-MAIL cwcw@uic.edu; FAX (312) 413-7963.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anbazhagan R, Fujii H, Gabrielson E. Allelic loss of chromosomal arm 8p in breast cancer progression. Am J Pathol. 1998;15:815–819. [PMC free article] [PubMed] [Google Scholar]
- Bookstein R, Levy A, MacGrogan D, Lewis TB, Weissenbach J, O'Connell P, Leach RJ. Yeast artificial chromosome and radiation hybrid map of loci in chromosome band 8p22, a common region of allelic loss in multiple human cancers. Genomics. 1994;24:317–323. doi: 10.1006/geno.1994.1622. [DOI] [PubMed] [Google Scholar]
- Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N, Jacobs SC, Robinson JC, Epstein JI, Walsh PC, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 1993;53:3869–3873. [PubMed] [Google Scholar]
- Bova GS, MacGrogan D, Levy A, Pin SS, Bookstein R, Isaacs WB. Physical mapping of chromosome 8p22 markers and their homozygous deletion in a metastatic prostate cancer. Genomics. 1996;35:46–54. doi: 10.1006/geno.1996.0321. [DOI] [PubMed] [Google Scholar]
- Boysen C, Simon MI, Hood L. Fluorescence-based sequencing directly from bacterial and P1-derived artificial chromosomes. Biotechniques. 1997;23:978–982. doi: 10.2144/97236bm01. [DOI] [PubMed] [Google Scholar]
- Buckler AJ, Chang DD, Graw SL, Brook JD, Haber DA, Sharp PA, Housman DE. Exon amplification: A strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci. 1991;88:4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino AM, Parker GJ, Boronenkov IV, Anderson RA, Chao MV. A novel interaction between the juxtamembrane region of the p55 tumor necrosis factor receptor and phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem. 1997;272:5861–5870. doi: 10.1074/jbc.272.9.5861. [DOI] [PubMed] [Google Scholar]
- Chang M, Tsuchiya K, Batchelor RH, Rabinovitch PS, Kulander BG, Haggitt RC, Burmer GC. Deletion mapping of chromosome 8p in colorectal carcinoma and dysplasia arising in ulcerative colitis, prostatic carcinoma, and malignant fibrous histiocytomas. Am J Pathol. 1994;144:1–6. [PMC free article] [PubMed] [Google Scholar]
- Chinen K, Isomura M, Izawa K, Fujiwara Y, Ohata H, Iwamasa T, Nakamura Y. Isolation of 45 exon-like fragments from 8p22 → p21.3, a region that is commonly deleted in hepatocellular, colorectal, and non-small cell lung carcinomas. Cytogenet Cell Genet. 1996;75:190–196. doi: 10.1159/000134480. [DOI] [PubMed] [Google Scholar]
- Chuaqui RF, Sanz-Ortega J, Vocke C, Linehan WM, Sanz-Esponera J, Zhuang Z, Emmert-Buck MR, Merino MJ. Loss of heterozygosity on the short arm of chromosome 8 in male breast carcinomas. Cancer Res. 1995;55:4995–4998. [PubMed] [Google Scholar]
- Church DM, Stotler CJ, Rutter JL, Murrell JR, Trofatter JA, Buckler AJ. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat Genet. 1994;6:98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- Clausen H, Bennett EP. A family of UDP-GalNAc: Polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology. 1996;6:635–646. doi: 10.1093/glycob/6.6.635. [DOI] [PubMed] [Google Scholar]
- Cunningham C, Dunlop MG, Wyllie AH, Bird CC. Deletion mapping in colorectal cancer of a putative tumour suppressor gene in 8p22-p21.3. Oncogene. 1993;8:1391–1396. [PubMed] [Google Scholar]
- Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN. Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res. 1996;56:4475–4482. [PubMed] [Google Scholar]
- Deaven LL, Van Dilla MA, Bartholdi MF, Carrano AV, Cram LS, Fuscoe JC, Gray JW, Hildebrand CE, Moyzis RK, Perlman J. Construction of human chromosome-specific DNA libraries from flow-sorted chromosomes. Cold Spring Harbor Symp Quant Biol. 1986;51:159–167. doi: 10.1101/sqb.1986.051.01.019. [DOI] [PubMed] [Google Scholar]
- Deubler DA, Williams BJ, Zhu XL, Steele MR, Rohr LR, Jensen JC, Stephenson RA, Changus JE, Miller GJ, Becich MJ, Brothman AR. Allelic loss detected on chromosomes 8, 10, and 17 by fluorescence in situ hybridization using single-copy P1 probes on isolated nuclei from paraffin-embedded prostate tumors. Am J Pathol. 1997;150:841–850. [PMC free article] [PubMed] [Google Scholar]
- El-Naggar AK, Coombes MM, Batsakis JG, Hong WK, Goepfert H, Kagan J. Localization of chromosome 8p regions involved in early tumorigenesis of oral and laryngeal squamous carcinoma. Oncogene. 1998;16:2983–2987. doi: 10.1038/sj.onc.1201808. [DOI] [PubMed] [Google Scholar]
- Emi M, Asaoka H, Matsumoto A, Itakura H, Kurihara Y, Wada Y, Kanamori H, Yazaki Y, Takahashi E, Lepert M, et al. Structure, organization, and chromosomal mapping of the human macrophage scavenger receptor gene. J Biol Chem. 1993;268:2120–2125. [PubMed] [Google Scholar]
- Fujiwara Y, Emi M, Ohata H, Kato Y, Nakajima T, Mori T, Nakamura Y. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Res. 1993;53:1172–1174. [PubMed] [Google Scholar]
- Fujiwara Y, Ohata H, Emi M, Okui K, Koyama K, Tsuchiya E, Nakajima T, Monden M, Mori T, Kurimasa A, et al. A 3-Mb physical map of the chromosome region 8p21.3-p22, including a 600-kb region commonly deleted in human hepatocellular carcinoma, colorectal cancer, and non-small cell lung cancer. Genes Chromosomes Cancer. 1994;10:7–14. doi: 10.1002/gcc.2870100103. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Ohata H, Kuroki T, Koyama K, Tsuchiya E, Monden M, Nakamura Y. Isolation of a candidate tumor suppressor gene on chromosome 8p21.3-p22 that is homologous to an extracellular domain of the PDGF receptor beta gene. Oncogene. 1995;10:891–895. [PubMed] [Google Scholar]
- Gustafson CE, Wilson PJ, Lukeis R, Baker E, Woollatt E, Annab L, Hawke L, Barrett JC, Chenevix-Trench G. Functional evidence for a colorectal cancer tumor suppressor gene at chromosome 8p22-23 by monochromosome transfer. Cancer Res. 1996;56:5238–5245. [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. The 1993-94 Genethon human genetic linkage map. Nat Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- Hennebicq-Reig S, Tetaert D, Soudan B, Kim I, Huet G, Briand G, Richet C, Demeyer D, Degand P. O-Glycosylation and cellular differentiation in a subpopulation of mucin-secreting HT-29 cell line. Exp Cell Res. 1997;235:100–107. doi: 10.1006/excr.1997.3638. [DOI] [PubMed] [Google Scholar]
- Hoon DS, Kaback MM, Lim-Steele J, Tsuchida T, Morton DL, Irie RF. Ganglioside GM2 levels in human melanoma cells: Inverse correlation with lysosomal beta-hexosaminidase A activity. Biochem Int. 1992;27:343–352. [PubMed] [Google Scholar]
- Horrigan S, Westbrook C. Construction and use of YAC contigs in disease regions. In: Boultwood J, editor. Methods in molecular biology. 68: Gene isolation and mapping protocols. Totowa, NJ: Humana Press; 1997. pp. 123–136. [DOI] [PubMed] [Google Scholar]
- Horrigan K, Bartolini L, Speer MC, Fulton N, Kravarusic J, Ramesar R, Vance JM, Yamaoka LH, Westbrook CA. Radiation hybrid breakpoint map of the acute myeloid leukemia (AML) and limb-girdl muscular distrophy 1A (LGMD1A) regions of chromosome 5q31 localizing 122 expressed sequences. Genomics. 1999;57:24–35. doi: 10.1006/geno.1999.5765. [DOI] [PubMed] [Google Scholar]
- Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, Simony-Lafontaine J, Longy M, Jacquemier J, Sobol H, Eisinger F, Birnbaum D. Genome-wide search for loss of heterozygosity shows extensive genetic diversity of human breast carcinomas. Cancer Res. 1997;57:5469–5474. [PubMed] [Google Scholar]
- Kuramochi H, Ichikawa T, Nihei N, Kawana Y, Suzuki H, Schalken JA, Takeichi M, Nagafuchi A, Ito H, Shimazaki J. Suppression of invasive ability of highly metastatic rat prostate cancer by introduction of human chromosome 8. Prostate. 1997;31:14–20. doi: 10.1002/(sici)1097-0045(19970401)31:1<14::aid-pros3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Lange K, Boehnke M, Cox DR, Lunetta KL. Statistical methods for polyploid radiation hybrid mapping. Genome Res. 1995;5:136–150. doi: 10.1101/gr.5.2.136. [DOI] [PubMed] [Google Scholar]
- Lerebours F, Olschwang S, Thuille B, Schmitz A, Fouchet P, Buecher B, Martinet N, Galateau F, Thomas G. Fine deletion mapping of chromosome 8p in non-small-cell lung carcinoma. Int J Cancer. 1999;81:854–858. doi: 10.1002/(sici)1097-0215(19990611)81:6<854::aid-ijc3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Levy A, Dang UC, Bookstein R. High-density screen of human tumor cell lines for homozygous deletions of loci on chromosome arm 8p. Genes Chromosomes Cancer. 1999;24:42–47. doi: 10.1002/(sici)1098-2264(199901)24:1<42::aid-gcc6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Lin Y, Kimmel LH, Myles DG, Primakoff P. Molecular cloning of the human and monkey sperm surface protein PH-20. Proc Natl Acad Sci. 1993;90:10071–10075. doi: 10.1073/pnas.90.21.10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrogan D, Bookstein R. Tumor suppressor genes in prostate cancer. Semin Cancer Biol. 1997;8:11–19. doi: 10.1006/scbi.1997.0048. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, Levy A, Bova GS, Isaacs WB, Bookstein R. Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics. 1996;35:55–65. doi: 10.1006/geno.1996.0322. [DOI] [PubMed] [Google Scholar]
- Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD, Pietruk T, Powell IJ. Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res. 1995;55:5390–5395. [PubMed] [Google Scholar]
- Matsumoto A, Naito M, Itakura H, Ikemoto S, Asaoka H, Hayakawa I, Kanamori H, Aburatani H, Takaku F, Suzuki H, et al. Human macrophage scavenger receptors: Primary structure, expression, and localization in atherosclerotic lesions. Proc Natl Acad Sci. 1990;87:9133–9137. doi: 10.1073/pnas.87.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Koike M, Shitara K, Kuwana Y, Kiuragi K, Igarashi S, Hasegawa M, Hanai N. Chimeric anti-ganglioside GM2 antibody with antitumor activity. Cancer Res. 1994;54:1511–1516. [PubMed] [Google Scholar]
- Nihei N, Ichikawa T, Kawana Y, Kuramochi H, Kugoh H, Oshimura M, Hayata I, Shimazaki J, Ito H. Mapping of metastasis suppressor gene(s) for rat prostate cancer on the short arm of human chromosome 8 by irradiated microcell-mediated chromosome transfer. Genes Chromosomes Cancer. 1996;17:260–268. doi: 10.1002/(SICI)1098-2264(199612)17:4<260::AID-GCC8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Ohata H, Emi M, Fujiwara Y, Higashino K, Nakagawa K, Futagami R, Tsuchiya E, Nakamura Y. Deletion mapping of the short arm of chromosome 8 in non-small cell lung carcinoma. Genes Chromosomes Cancer. 1993;7:85–88. doi: 10.1002/gcc.2870070204. [DOI] [PubMed] [Google Scholar]
- Ohata H, Fujiwara Y, Koyama K, Nakamura Y. Mapping of the human autoantigen pericentriolar material 1 (PCM1) gene to chromosome 8p21.3-p22. Genomics. 1994;24:404–406. doi: 10.1006/geno.1994.1640. [DOI] [PubMed] [Google Scholar]
- Ohgaki K, Iida A, Ogawa O, Kubota Y, Akimoto M, Emi M. Localization of tumor suppressor gene associated with distant metastasis of urinary bladder cancer to a 1-Mb interval on 8p22. Genes Chromosomes Cancer. 1999;25:1–5. doi: 10.1002/(sici)1098-2264(199905)25:1<1::aid-gcc1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Patel U, Grundfest-Broniatowski S, Gupta M, Banerjee S. Microsatellite instabilities at five chromosomes in primary breast tumors. Oncogene. 1994;9:3695–3700. [PubMed] [Google Scholar]
- Pierce JC, Sauer B, Sternberg N. A positive selection vector for cloning high molecular weight DNA by the bacteriophage P1 system: Improved cloning efficacy. Proc Natl Acad Sci. 1992;89:2056–2060. doi: 10.1073/pnas.89.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau P, Nagai H, Prigent S, Wei Y, Gyapay G, Weissenbach J, Tiollais P, Buendia MA, Dejean A. Identification of three distinct regions of allelic deletions on the short arm of chromosome 8 in hepatocellular carcinoma. Oncogene. 1999;18:3127–3134. doi: 10.1038/sj.onc.1202648. [DOI] [PubMed] [Google Scholar]
- Prasad MA, Trybus TM, Wojno KJ, Macoska JA. Homozygous and frequent deletion of proximal 8p sequences in human prostate cancers: Identification of a potential tumor suppressor gene site. Genes Chromosomes Cancer. 1998;23:255–262. [PubMed] [Google Scholar]
- Reis CA, David L, Seixas M, Burchell J, Sobrinho-Simoes M. Expression of fully and under-glycosylated forms of MUC1 mucin in gastric carcinoma. Int J Cancer. 1998;79:402–410. doi: 10.1002/(sici)1097-0215(19980821)79:4<402::aid-ijc16>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Takanishi DM, Jr, Kim SY, Kelemen PR, Yaremko ML, Kim AH, Ramesar JE, Horrigan SK, Montag A, Michelassi F, Westbrook CA. Chromosome 8 losses in colorectal carcinoma: Localization and correlation with invasive disease. Mol Diagn. 1997;2:3–10. doi: 10.1054/MODI00200003. [DOI] [PubMed] [Google Scholar]
- Takle LA, Knowles MA. Deletion mapping implicates two tumor suppressor genes on chromosome 8p in the development of bladder cancer. Oncogene. 1996;12:1083–1087. [PubMed] [Google Scholar]
- Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Konishi M, Oshimura M, Miyaki M. Suppression of tumorigenicity and invasiveness of colon carcinoma cells by introduction of normal chromosome 8p12-pter. Oncogene. 1996;12:405–410. [PubMed] [Google Scholar]
- The Third International Workshop on Human Chromosome 8 Mapping. Cytogenet Cell Genet. 1996;75:71–84. doi: 10.1159/000134460. [DOI] [PubMed] [Google Scholar]
- Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Liotta LA, Emmert-Buck MR, Linehan WM. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 1996;56:2411–2416. [PubMed] [Google Scholar]
- Wagner U, Bubendorf L, Gasser TC, Moch H, Gorog JP, Richter J, Mihatsch MJ, Waldman FM, Sauter G. Chromosome 8p deletions are associated with invasive tumor growth in urinary bladder cancer. Am J Pathol. 1997;151:753–759. [PMC free article] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, Milchgrub S, Syed S, Lam S, Mackay B, Minna JD, Gazdar AF. Allelic losses at chromosome 8p21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59:1973–1979. [PubMed] [Google Scholar]
- Wright K, Wilson PJ, Kerr J, Do K, Hurst T, Khoo SK, Ward B, Chenevix-Trench G. Frequent loss of heterozygosity and three critical regions on the short arm of chromosome 8 in ovarian adenocarcinomas. Oncogene. 1998;17:1185–1188. doi: 10.1038/sj.onc.1202028. [DOI] [PubMed] [Google Scholar]
- Wu CL, Roz L, Sloan P, Read AP, Holland S, Porter S, Scully C, Speight PM, Thakker N. Deletion mapping defines three discrete areas of allelic imbalance on chromosome arm 8p in oral and oropharyngeal squamous cell carcinomas. Genes Chromosomes Cancer. 1997;20:347–353. doi: 10.1002/(sici)1098-2264(199712)20:4<347::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]