Abstract
N-myristoylation, one of the co- or post-translational modifications of proteins, has so far been regarded as necessary for anchoring of proteins to membranes. Recently, we have revealed that Nα-myristoylation of several brain proteins unambiguously regulates certain protein–protein interactions that may affect signaling pathways in brain. Comparison of the amino acid sequences of myristoylated proteins including those in other organs suggests that this regulation is involved in signaling pathways not only in brain but also in other organs. Thus, it has been shown that myristoylated proteins in cells regulate the signal transduction between membranes and cytoplasmic fractions. An algorithm we have developed to identify myristoylated proteins in cells predicts the presence of hundreds of myristoylated proteins. Interestingly, a large portion of the myristoylated proteins thought to take part in signal transduction between membranes and cytoplasmic fractions are included in the predicted myristoylated proteins. If the proteins functionally regulated by myristoylation, a posttranslational protein modification, were understood as cross-talk points within the intracellular signal transduction system, known signaling pathways could thus be linked to each other, and a novel map of this intracellular network could be constructed. On the basis of our recent results, this review will highlight the multifunctional aspects of protein N-myristoylation in brain.
Keywords: N-myristoylation, intracellular signal transduction system, membrane lipid raft, calmodulin, CAP-23/NAP-22, MARCKS, HIV Nef
Introduction
Protein N-myristoylation was first identified in the catalytic subunit of cAMP-dependent protein kinase from bovine cardiac muscle using modern mass spectrometric techniques by K. Titani and his coworkers in 1982.1) After that, calcineurin,2) MMLV p15gag,3) NDAH cytochrome b5 reductase,4) pp 60src 5–8 were found to be myristoylated using similar techniques in succession. The N-termini of proteins are modified with myristate, a 14-carbon saturated fatty acid (Fig. 1), and the enzymology of myristoylation reaction has been well characterized.9) NMT, which exists in all eukaryotes, catalyzes the reaction. In the case of human, two enzymes are known to catalyze the reaction. The substrates are co-translationally myristoylated (Table 1). The myristoylated site is limited to N-terminal glycine, and the linkage is formed by an amide bond. Experiments using substrate peptides have revealed that the enzyme recognizes approximately only ten residues from the N-termini of substrates, and there is no consensus sequence for myristoylation besides glycine at the second position and serine at the sixth position, i.e., MGXXXSXX in the precursor proteins. For myristoylation by NMT, the removal of initiation methionine residue by methionyl aminopeptidase is needed, since exposed glycine residue at N-termini is required. The detection of myristoylation has been difficult because of low chemical reactivities of the myristoyl group, but recent advances in mass spectrometry have made the detection relatively easy. Besides myristoylation, another lipid N-modification was also identified.10) In this article, recent studies on myristoylated proteins including post-genome research studies are described.
Figure 1.
Chemical structure of the myristoyl moiety. A myristoyl group binds to an N-terminal glycine residue covalently through an amide linkage.
Table 1.
Comparison between myristoylation and palmitoylation
| N-myristoylation | S-palmitoylation | |
|---|---|---|
| Modifying group; | myristate | palmitate |
| Chemical structure; | 14-carbon saturated fatty acid | 16-carbon saturated fatty acid |
| Modification enzyme; | N-myristoyl transferase | dependent on each case |
| Timing; | co-translational | post-translational |
| Linkage; | Gly | Cys |
| Chemical bond; | amide | thio-ester |
| Modified proteins; | Src family members | G protein coupled receptors |
| α subunits of G proteins | HLA | |
| HIV Nef | caveolin | |
| Calcineurin B | CD4 | |
| recoverin | influenza HA | |
| catalytic subunit of A kinase | GAP43 | |
| cytochrome b5 reductase | H-Ras, N-Ras | |
| NAP22 | α subunits of G proteins | |
| MARCKS | Src family members |
Function of myristoyl moiety
Anchoring to membranes.
In consideration of its strong hydrophobicity, myristoylation has been thought to act as an anchor for modified proteins to biomembranes.11,12) However, it is obvious that the myristoyl moiety alone is not sufficient to capture large molecules, such as proteins, at membrane fractions. Other protein fatty acylations such as S-palmitoylation and the presence of other basic regions in the molecule in addition to myristoylation might function to strengthen affinities of the myristoylated proteins to membranes.11,13) Unlike membrane proteins with trans-membrane domains, myristoylated proteins can leave membranes under their regulation using certain signaling systems.
Interaction with CaM.
We have found that CAP-23/NAP-22, a neuron specific protein isolated from rat brain, is N-myristoylated, and that the myristoylation is essential for its interaction with CaM in the presence of Ca2+. In addition, the CaM-binding site has been narrowed down to the myristoyl moiety together with the N-terminal basic domain of 9 amino acid residues, GGKLSKKKK.14,15)
CaM is a small calcium-binding protein (16.7 kDa) involved in a wide range of cellular Ca2+-dependent signaling pathways through various enzymes, including protein kinases, protein phosphatases, nitric oxide synthase, inositol triphosphate kinase, nicotinamide adenine dinucleotide kinase, and cyclic nucleotide phosphodiesterase.16–19) CAP-23/NAP-22, a neuron-specific protein, was first isolated from chicken brain and characterized as a 23 kDa cortical cytoskeleton-associated protein (CAP-23),20) and the rat homologue was later isolated as NAP-22.21)
To examine the effects of myristoylation on the interaction of CAP-23/NAP-22 with CaM, two recombinant proteins, i.e., non-myristoylated and myristoylated CAP-23/NAP-22 proteins, were produced in E. coli. For the myristoylated protein, a pBB131 vector (a gift from Dr. J. I. Gordon) containing yeast N-myristoyl transferase cDNA was co-transformed.22,23) Both of its proteins were purified by successive column chromatography on Phenyl-Sepharose and Resource RPC (Amersham Pharmacia Biotech), and the authenticity of the two proteins was established by electrospray mass spectrometry (Fig. 2). The mass of the non-myristoylated protein was determined to be 21,629.2 ± 2.9 Da (the theoretical mass; 21,629.1 Da), while that of the myristoylated protein was 21,839.5 ± 2.0 Da (the theoretical mass; 21,839.5 Da). These results indicated that the two proteins differed only in their N-terminal myristoylation.
Figure 2.
Liquid chromatography (left panel)/electrospray mass spectrometry analyses (right panel) of recombinant non-myristoylated (A) and myristoylated (B) CAP-23/NAP-22. The retention time of myristoylated CAP-23/NAP-22 on the reversed-phase liquid chromatography (left panel of B) was longer than that of non-myristoylated CAP-23/NAP-22 (left panel of A). Each fraction was directly injected into the electrospray mass spectrometry apparatus (right panels). The difference between the observed molecular weights (213.7 Da) corresponded to that of the myristoyl group (210.0 Da).
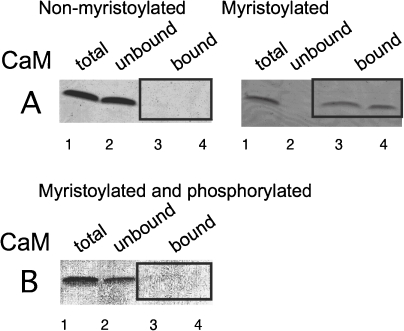
Interaction of the two recombinant proteins with CaM was analyzed by the binding to CaM-agarose beads (Fig. 3). Clearly, only the myristoylated protein bound to the CaM-beads, and most of the bound protein was eluted with a Ca2+-free buffer. The non-myristoylated protein did not bind to the CaM beads to any significant extent. Therefore, CAP-23/NAP-22 bound to CaM in a Ca2+ and myristoylation-dependent manner. It was also shown that the binding of mC/N9, N-myristoylated 9 residue peptide corresponding to the N-terminal CaM binding site of CAP-23/NAP-22, to CaM was dependent on the existence of the myristoyl moiety (Fig. 4). Furthermore, phosphorylation of Ser5 in the N-terminal region of CAP-23/NAP-22 by PKC abolished the binding of CAP-23/NAP-22 to Ca2+/CaM (Fig. 4).14) This was assumed to be caused by the introduction of an oppositely charged group into the middle of the basic residues that were essential in making the ionic contact with negatively charged CaM.
Figure 3.
Effects of myristoylation of recombinant CAP-23/NAP-22 on the interaction with CaM. Binding of the non-myristoylated (A) and myristoylated (B) recombinant CAP-23/NAP-22 to CaM was determined using CaM-agarose. The proteins (Lane 1) were mixed with calmodulin-agarose in 50 mM Tris-HCl buffer (pH 6.8) containing 1 mM CaCl2 and 0.2 M NaCl. After a short period of centrifugation in a tabletop centrifuge, the supernatants were removed (Lane 2). The calmodulin-agarose was washed twice with the same buffer. To the sedimented gels, 50 mM Tris-HCl buffer (pH 6.8) containing 0.2 M NaCl and 5 mM EGTA was added. After centrifugation, the supernatants were removed (Lane 3), and the remaining proteins were eluted with the sample buffer containing 1% SDS (Lane 4). The fractions obtained were analyzed by SDS gel electrophoresis.
Figure 4.
Effects of myristoylation of NAP-22 N-terminal peptide on the interaction with CaM (A), and the effect of phosphorylation by protein kinase C (B). Bindings of the non-myristoylated (left panel of A), myristoylated (right panel of A), and myristoylated-phosphorylated (B) NAP-22 N-terminal peptide (Gly1-Lys9) to CaM were determined using CaM-agarose as described in Fig. 3. The fractions obtained were analyzed by SDS gel electrophoresis. The synthetic peptide was phosphorylated by PKC purified from bovine brain as described previously.14)
Gel shift assay for stoichiometric analyses of the interaction between CAP-43/NAP-22 and Ca2+/CaM clearly indicated that two molecules of CAP-43/NAP-22 bound to one molecule of CaM (Fig. 5). Ca2+/CaM molecule adopted an ‘elongated’ structure that comprised two globular domains connected by a highly flexible linker.24–30) The binding of Ca2+/CaM to the target peptide induced a compact globular structure caused by the bending of the domain linker.31–33) The target peptides formed an α-helix in the complexes in a basic amphiphilic nature. Besides the traditional mechanism for the target recognition of CaM described above, other novel mechanisms were identified. It was shown that a single unique complex of Ca2+/CaM was formed with two peptides that corresponded to the C-terminal region of petunia glutamate decarboxylase (PGD). The formation of a 1:2 protein–protein complex was unusual; normally, Ca2+/CaM forms 1:1 complexes with the majority of its target proteins.34) It has previously been shown that a peptide corresponding to the N-terminal portion of the CaM-binding domain in plasma membrane calcium pump bound only to the C-terminal half of CaM, and that, in binding to the peptide, CaM did not form any of the collapsed structures observed in the previous studies.35)
Figure 5.
Stoichiometric analysis using band shift assay of non-denatured gel electrophoresis revealed the formation of Ca2+/CaM-NAP22 complex with the molar ratio of 1:2. Lanes 1 and 6 are results of Ca2+/CaM and NAP22. Lanes 2, 3, 4 and 5 are results of Ca2+/CaM-NAP22 mixture with the molar ratio of 2:1, 1:1, 1:2 and 1:3, respectively. Bands 1, 2, and 3 correspond to Ca2+/CaM-NAP22 complex, NAP22 and Ca2+/CaM, respectively. In the presence of the calcium ion (A), upon the formation of the Ca2+/CaM-NAP22 complex, the isolated band of CaM disappeared, and, when the molar ratio of Ca2+/CaM and NAP22 was over 1:3, the isolated band of NAP22 appeared. In the absence of calcium ion (B), no shift was observed.
SAXS can capture structural transformations of proteins in solution in terms of changes in the radius of gyration. The SAXS analysis indicated that the binding of two mC/N9 molecules induced a drastic structural change in Ca2+/CaM (Fig. 6). The radius of gyration for the Ca2+/CaM-mC/N9 complex was 19.8 ± 0.3 Å (Table 2). This value was significantly smaller than that of Ca2+/CaM (21.9 ± 0.3 Å), which adopted a dumbbell structure and was 2–3 Å larger than those of the complexes of Ca2+/CaM with the non-myristoylated target peptides of MLCK or CaM kinase II, which adopted a compact globular structure.36) The pair distance distribution function had no shoulder peak at around 40 Å which was mainly due to the dumbbell structure. These results suggested that Ca2+/CaM interacted with Nα-myristoylated CAP-23/NAP-22 differently than it did with other non-myristoylated target proteins.
Figure 6.
The radius of gyration as a function of the molar ratio of mC/N9 to Ca2+/CaM at a CaM concentration of 9.0 mg/mL.
Table 2.
Radius of gyration Rg and maximum dimension dmax for Ca2+/CaM and its complexes
| Rg [Å] | dmax [Å] | reference | |
|---|---|---|---|
| Ca2+/CaMa | 21.9 ± 0.3 | 62 | 36 |
| Ca2+/CaM - myristoylated NAP22 peptidea |
19.8 ± 0.3 | 50 | 36 |
| Ca2+/CaMa | 21.5 ± 0.3 | 69 | 61 |
| Ca2+/CaM - M13a,b | 16.4 ± 0.2 | 49 | 27 |
| Ca2+/CaM - W-7a | 17.6 ± 0.3 | 47 | 62 |
aValues at zero protein concentration obtained by SAXS experiment.
bM13: a peptide based on the CaM-binding domain of MLCK.
We analyzed the interaction between mC/N9 and Ca2+/CaM using 15N labeled CaM and two-dimensional 1H-15N HSQC NMR spectroscopy. Portions of the NMR spectra for Ca2+/CaM in the absence or presence of mC/N9 are shown in Fig. 7. When mC/N9 was added, shifts of certain peaks were observed in the 1H-15N HSQC NMR spectra of Ca2+/CaM. Some drastic shifts of the peaks were also observed by addition of 2 molar equivalents of mC/N9.
Figure 7.
Titration of Ca2+/CaM with mC/N9 studied by CaM uniformly labeled with 15N and 1H-15N HSQC NMR spectroscopy. The sample contained 0.5 mM CaM, 120 mM NaCl, 2.5 mM CaCl2, and 50 mM deuterated TrisHCl (pH 7.5) in 90% H2O and 10% D2O. The resonance assignments were made with reference to Ikura et al.83) The three well isolated regions are indicated. The tentative assignments (Lys21, Ile27, Ala57) are shown. The spectra of Ca2+/CaM in the presence of 0, 1, 2, 3, 4 and 5 molar equivalents of mC/N9 are shown by the number, respectively.
Unlike other CaM target proteins, CAP-23/NAP-22 lacked any canonical CaM-binding motif of a basic amphiphilic nature, suggesting that the myristoyl moiety of the protein plays a direct role in the protein–protein interaction.
In the case of M13 (the CaM-binding domain of MLCK), the amphiphilic nature of the peptide required for its binding to CaM was induced by the α-helical conformation. The CaM-binding domain of CAP-23/NAP-22 adopted a non-helical conformation in the Ca2+/CaM-complex.14) The N-terminal domain of CAP-23/NAP-22 contained one hydrophobic residue (Leu4) in addition to five basic residues (Lys3, Lys6, Lys7, Lys8 and Lys9). In this domain, one hydrophobic acyl group (N-terminal myristoyl moiety) was followed by one basic residue (Lys3) and then one hydrophobic residue (Leu4). This result resembled the canonical CaM-binding motif, in which positively charged hydrophilic and hydrophobic residues alternated.37,38) If the acyl group had been substituted for a large hydrophobic residue, such as Trp or Leu found in the canonical CaM-binding motif, the overall structural characteristics would have appeared to be very similar to each other. The distance between the myristoyl moiety and Leu4 was comparable to that between the two critical hydrophobic residues found in M13 (Fig. 8 ; shown in red). TFP and N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7) are small compounds known to bind to CaM.39) Although the chemical structure of TFP/W-7 clearly differed from that of mC/N9 or M13, it also contained hydrophobic groups (Fig. 8; shown in red) as well as positively charged groups (Fig. 8; shown in blue), and these groups were the cause of the amphiphilic nature of the molecule. All these observations together emphasized the importance of the amphiphilic nature required for the binding of CaM-binding molecules, not only proteins but also bioactive small molecules, to Ca2+/CaM.
Figure 8.
A comparison between the canonical CaM-binding peptide, TFP, and the myristoylated mC/N9. A space-filling model of the M13 peptide derived from skeletal muscle MLCK in a helical conformation (top); the hydrophobic amino acid residues that play important roles in the CaM interaction are shown in red, and the positively charged amino acid residues are shown in blue. TFP (middle); the hydrophobic aromatic group is shown in red, and the positively charged group is shown in blue. The myristoylated N-terminal peptide of CAP-23/NAP-22 (myr-GGKLSKKKK) in an elongated structure (bottom); the myristoyl moiety and Leu4 are shown in red; the positively charged amino acid residues are shown in blue, and one phosphorylatable amino acid residue, Ser5, is shown in yellow. All of these molecules include the basic amphiphilic natures (basic group—blue, hydrophobic group—red) in them.
Physiological function of myristoylation
Myristome; comprehensive analysis of Nαmyristoylated proteins.
Using the complete genome data, a comprehensive prediction of myristoylated proteins was performed. A substrate of a known complex of myristoylation enzyme was replaced by peptides with the N-terminal sequence of all the gene products anticipated from the genome data, and the affinities were evaluated (http://mendel.imp.ac.at/myristate/myrbase/).40) Then, the prediction was experimentally verified by mass spectrometry and a peptide array method. Peptides corresponding to the N-terminal amino acid sequences of the candidates were arranged. Incorporations of the myristoyl group were detected by radioisotope. Apart from that, the prediction was also assayed by mass spectrometric analyses (http://mendel.imp.ac.at/myristate/myrbase/MYRBASE_additional_data_file_1.pdf). Surprisingly, the results indicated that 0.5% of proteins in eukaryotic cells are myristoylated.
In brain.
The physiological function of CAP-23/NAP-22 has yet to be determined, but its involvement in synaptogenesis and neuronal plasticity has been suggested.41,42) CAP-23/NAP-22 is related to other neuron-specific acidic proteins, such as GAP-43 and MARCKS20,21,43) because it is also a prominent substrate of PKC. MARCKS family proteins have their own common properties; they are natively unfolded proteins,44) heat stable, major PKC substrates in neuronal cells, fatty-acylated, and they interact with CaM.44) The phosphorylation site, the myristoylation site, and the CaM binding site of CAP-23/NAP-22 are located in the same region. Phosphorylation of a single serine residue in the N-terminal domain by PKC abolished the binding of CAP-23/NAP-22 to Ca2+/CaM. These results strongly suggested that crosstalks among several distinct intracellular signal transduction systems in brain are being carried out in the N-terminal region of CAP-23/NAP-22. Besides CAP-23/NAP-22, myristoylation of Cdk5 has been found to regulate the localization in the cells,45) and many other myristoylated proteins are thought to play key roles in brain where rapid and flexible responses are required.
Oncogene products.
Table 3 shows an alignment of the N-termini of some of the myristoylated proteins. Lysine (K) is required to form the amphiphilicity necessary for association with CaM. Serine (S) can be used for a phosphorylation site to regulate the proteins. Myristoylated proteins having both lysine and serine in the N-terminal myristoylation domains might be under the regulatory control described above. The myristoylated N-terminal region of Src kinase meets the requirements for CaM binding, and it also has serine residues phosphorylated by other protein kinases. In fact, it has been suggested that Src kinase may be regulated by the same mechanism found in the case of CAP-23/NAP-22.46) To stably anchor src kinase on the membranes, certain coupling factors are likely to be needed in addition to myristoylation of the protein.
Table 3.
Amino acid sequence alignment of human myristoylated proteins predicted by the genome analysisa
| Nameb | Sequencec | Ref.d |
|---|---|---|
| CAP-23/NAP-22 | GGKLSKKKKG | 14 |
| tyrosine protein kinase transforming protein src (p60-Src) | GSSKSKPKDP | 63 |
| proto-oncogene tyrosine protein kinase src (p60-Src) | GSNKSKPKDA | 63 |
| HIV Nef | GGKWSKSSVV | 64 |
| (2′-5′)oligoadenylate synthetase | GNGESQLSSV | 65 |
| annexin XIII | GNRHAKASSP | 66 |
| guanylate cyclase activating protein 1 | GNVMEGKSVE | 67 |
| NADH-ubiquinone oxidoreductase B18 subunit | GAHLVRRYLG | * |
| NADH-cytochrome b5 reductase | GAQLSTLGHM | 68 |
| endothelial nitric oxide synthase (eNOS) | GNLKSVAQEP | 69 |
| acetylcholine receptor-associated 43 kD protein | GQDQTKQQIE | 70 |
| T-lymphoma invasion and metastasis inducing protein 1 | GNAESQHVEH | * |
| visinin-like protein 1 | GKQNSKLAPE | 71 |
| recoverin | GNSKSGALSK | 72 |
| calcineurin B | GNEASYPLEM | 73 |
| neuron-specific calcium-binding protein hippocalcin | GKQNSKLRPE | 74 |
| neurocalcin d | GKQNSKLRPE | 75 |
| calcium-binding protein P22/calcineurin homologous protein | GSRASTLLRD | * |
| a subunit of cAMP-dependent protein kinase | GNAAAAKKGS | 1 |
| b subunit of cAMP-dependent protein kinase | GNAATAKKGS | 1 |
| g subunit of cAMP-dependent protein kinase | GNAPAKKDTE | 1 |
| b lymphocyte tyrosine protein kinase | GLVSSKKPDK | 76 |
| proto-oncogene tyrosine protein kinase Fyn (p59-Fyn) | GCVQCKDKEA | 77 |
| tyrosine protein kinase Hck (hemopoietic cell kinase) | GGRSSCEDPG | 78 |
| proto-oncogene tyrosine protein kinase Lck | GCGCSSHPED | 79 |
| tyrosine protein kinase Lyn | GCIKSKGKDS | * |
| proto-oncogene tyrosine protein kinase Yes (p61-Yes) | GCIKSKENKS | 76 |
| ADP-ribosylation factor 1 | GNIFANLFKG | 80 |
| ADP-ribosylation factor 3 | GNIFGNLLKS | 80 |
| ADP-ribosylation factor 4 | GLTISSLFSR | 80 |
| ADP-ribosylation factor 5 | GLTVSALFSR | 80 |
| ADP-ribosylation factor 6 | GKVLSKIFGN | 80 |
| HIV Gag | GARASVLSGG | 81 |
| guanine nucleotide-binding protein Go, α subunit 1 | GCTLSAEERA | 82 |
| guanine nucleotide-binding protein Go, α subunit 2 | GCTLSAEERA | 82 |
| guanine nucleotide-binding protein Gi, α subunit 1 | GCTLSAEDKA | 82 |
| guanine nucleotide-binding protein Gi, α subunit 2 | GCTVSAEDKA | 82 |
| guanine nucleotide-binding protein Gt, α subunit 1 | GAGASAEEKH | 82 |
| guanine nucleotide-binding protein Gt, α subunit 2 | GSGASAEDKE | 82 |
| Residues required for the CaM bindinge | G-KLS- - - - - | |
aWhen the modification has been identified in the protein from other species, the references are shown.
bNames of myristoylated proteins including the abbreviations. Refer to the superscripts for the formal name (see below).
cN-terminal ten-residue sequences.
dReferences for the myristoylation. Asterisks (*) show that the myristoylation of the protein has not yet been directly identified, but the possibility of its eventual identification is suggested from the sequence similarity.
eResidues in CAP-23/NAP-22 required for the CaM binding (ref. 14) are shown with the phosphorylatable serin residue in the domain.
HIV Nef.
HIV Nef (one of the human immunodeficiency virus gene products) is another example. Using an algorithm for the prediction of myristoylation,40) it has been found that all of the several hundred Nef isoforms are perhaps myristoylated. This result suggests that Nef myristoylation is necessary for its function. The myristoylated N-terminal region of Nef also meets the requirements for CaM binding, and it also has serine residues phosphorylated by other protein kinases. Therefore, Nef is considered to use the same mechanism to intervene in the signaling systems of the host cells.15,47)
Signal transduction between membranes and cytoplasmic fractions through myristoylated proteins
Functional implication of myristoyl moiety.
Because of the reversibility of these transitions, they are considered to play a role in communications between membranes and cytoplasmic fractions. Recently, some reports have shown that myristoylated proteins exist in membrane micro domains, called rafts,48) and they are thought to function for processing the signals rapidly and flexibly. Therefore, myristoylated proteins may be designed as multi-functional molecules. Figure 9 shows a summary of reversible translocations of myristoylated proteins between membranes and cytoplasmic fractions under the regulation of signaling system crosstalks. Phosphorylation of myristoylated proteins abolishes their interactions with CaM, and it might also reduce their affinities to membranes because of the introduction of negative charges. Their interactions with CaM might also inhibit their localization to membranes because CaM which binds directly to the myristoyl moiety is essential for the membrane binding by serving as a membrane anchor. The state of membranes may also have effects on their interactions. Some myristoylated proteins have been reported to localize transiently to membrane micro-domains according to the state of cells.49)
Figure 9.
Scheme of reversible translocation of myristoylated proteins between the membrane and cytoplasmic fractions.
The involvement of protein myristoylation in protein–protein interactions has been implied in various studies,50–52) but it has never been clearly demonstrated. To the best of our knowledge, our result is the first report directly demonstrating the involvement of myristoylation in protein–protein interactions. Protein myristoylation has been implicated in the regulation of various signal transduction proteins,11,53) and in addition, there are many other potential myristoylated proteins whose myristoylation can be predicted from their amino acid sequences. Among these proteins, some have functionally important features besides myristoylation, such as the possession of basic residues (lysine), and the target residue of phosphorylation by protein kinase C (serine) (Table 3). Therefore, there might be a strong possibility that myristoylation-dependent protein–protein interactions play important roles in at least some of these cases.36)
Subcellular localization regulated by myristoylated domains.
A simulation of the membrane binding of a myristoylated domain shows that not only the myristoyl group, but also the N-terminal region, might contribute to the membrane binding.12) Myristoylated proteins have their own myristoyl groups in common. However, their N-terminal amino acid sequences are considerably divergent. The amino acid sequences of the first 10 members in the database of predicted myristoylated proteins exhibit very different pI values and hydrophobicity profiles, as shown in Fig. 10. It can be speculated that a variety of the properties in the N-terminal regions of myristoylated proteins could result from differences in their membrane targeting regions. Both the N-terminal regions of the myristoylated proteins and the contents of their target membranes may exert great influence on the affinities between the proteins and the membranes. In fact, different myristoylated proteins have been isolated from different membrane fractions.54) Furthermore, several groups have reported that different modifications of the amino acid sequences at myristoylated domains altered their localizations in cells.55–57)
Figure 10.
pI values and hydrophobicity profiles of the N-terminal amino acid sequences of some members of the predicted myristoylated protein database. pI values were calculated using Protein Identification and Analysis Tools on the ExPASy Server.84) Hydrophobicity profiles were calculated by the Kyte–Doolittle method85) using the Molecular Toolkit of Colorado State University. The attached numbers of each panel correspond to numbers in the list of the predicted myristoylated proteins shown below.
Future perspectives
New techniques for analyses of delicate interactions.
The biological significances of myristoylated proteins could be elucidated by observing the behavior of each myristoylated protein in living cells. One particular molecule tracking technique seems to be a powerful tool for this purpose, but the target molecule is required to be labeled with a fluorescent probe for this analysis. However, the anchoring affinity of myristoyl proteins to membranes is generally not so strong, and the proteins of more than 20 kDa can remain at membrane fractions unstably only by the myristoyl group,58) and the heavy probes generally used, like those for green fluorescent protein (GFP), are predicted to change the dynamics of the target proteins and result in some artifacts. Thus, a new methodology for site-selective post-translational modification of proteins has been developed.59) Using this technique, any probe could be introduced to any site of the protein.
Also, in order to observe the motion of target proteins in living cells, an in-cell NMR technique has been developed.60) Combination of existing methodologies and these new technologies would be indispensable to elucidate the regulatory mechanisms of delicate and complicated signal processing involving myristoylated proteins.
New insight into rapid and flexible signal processing on the cell surface.
As is shown herein, a wide variety of myristoylated proteins are suggested to be involved in various intracellular signaling pathways between membranes and cytoplasm fractions. Although these myristoylated proteins are produced by the same mechanism, their biological significances must be different with one another. Considering the proteins whose functions are regulated by myristoylation as cross-talk points in the intracellular signal transduction systems, the known signaling pathways could be linked to one another. Accordingly, a novel map of their intracellular signal transduction network could be constructed.
Acknowledgments
We thank all our collaborators who have contributed our research projects described herein. We also thank Dr. T. Yamakawa, M.J.A., for inviting us to submit this review to the journal, Dr. S. Nakanishi, M.J.A., for communicating it to the jounal and Mr. Ron Belisle for editing our manuscript.
Abbreviations
- CaM
calmodulin
- NAP-22
22kDa neuron-specific acidic protein of rat
- NMT
N-myristoyl transferase
- MLCK
myosin light chain kinase
- GAP-43
growth associated protein-43
- MARCKS
myristoylated alanine-rich protein kinase C substrate
- HIV Nef
negative factor, a human immunodeficiency virus gene products
- SAXS
small angle X-ray scattering
- NMR
nuclear magnetic resonance
- HSQC
hetero nuclear single quantum coherence spectroscopy
- TFP
trifluoperazine
- PKC
protein kinase C
Biographies
Profile
Nobuhiro Hayashi was born in Kyoto, Japan, in 1964. He is interested in understanding life at molecular level, and uses freely physicochemical techniques such as NMR, mass spectroscopy, solution small angle X-ray/neutron scattering, fluorescence spectroscopy, circular dichroism spectroscopy, surface plasmon resonance spectroscopy, and so on. He graduated Department of Physics, Faculty of Science, University of Tokyo in 1989. He started his research from 1989 and received Ph. D. with the thesis entitled “Functional and structural analyses of specific conformation of E. coli lysyl-tRNA anti-codon using NMR” from Tokyo Institute of Technology in 1994 with Prof. Kimitsuna Watanabe. From 1994 to 2000, he studied function of N-myristoylation of protein, and found novel regulatory mechanism of intracellular signal transduction system through reversible translocation of myristoylated proteins as Research Associate and Assistant Professor with Prof. Koiti Titani at Division of Biomedical Polymer Science, Institute of Comprehensive Medical Science, Fujita Health University. The results are described in the paper this time. From 2000 to 2008, he expanded the research, and revealed that there are many myristoylated proteins in the cells and some of them participate in functional regulation of micro-domain on the membrane, membrane lipid raft, as Associate Professor with Prof. Keiichiro Hashimoto at Division of Biomedical Polymer Science, Institute of Comprehensive Medical Science, Fujita Health University. Parts of the results are also mentioned in the paper. In those days, he has developed high performance proteomics technique independently, and site-selective labeling methodology by joint researches. From 2008, he started structural and functional analysis of membrane lipid raft using these techniques as Associate Professor at Department of Life Science, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology. Besides the raft researches, he has applied these new techniques to various objects to open up a new field of life science by high throughput and comprehensive analyses of gene products of the cells. So far, he had contributed to publications of 50 original papers in English in the first-rate journals including Nature, Proc. Natl. Acad. Sci. USA, J. Biol. Chem., J. Mol. Biol., Biochemistry, Biochem. J., J. Neurocham., and Protein Sci.
Koiti Titani was born in Tokyo, Japan, in 1930. He had devoted himself to the development of “Protein Science” consistently until his retirement in 2000. He graduated Department of Chemistry, Faculty of Science, University of Tokyo in 1955. He started his research from 1954 and received Ph. D. with the thesis entitled “Relationship between Structure and Function of Cytochrome c” from University of Tokyo Graduate School of Chemistry in 1960. He determined the complete amino acid sequence of cytochrome c from baker’s yeast in 1963, the first complete chemical structure of a protein determined in Japan, as Instructor and Assistant Professor with Prof. K. Narita at the Division of Chemical Structure, Institute for Protein Research, Osaka University (1960–1969). He worked with Prof. F. Putnam at Department of Biochemistry, School of Medicine, University of Florida, and at Division of Biological Science, Indiana University (1964–1967) to determine the complete amino acid sequence of the first several κ- and λ-type Bence-Jones Proteins secreted in urine of myeloma patients, which are equivalent to the light chain of immunoglobulins, and contributed to proposal for the structural model and evolution of antibody. He was appointed to Research Associate Professor (1969–1976) and Research Professor (1976–1986) at Department of Biochemistry, School of Medicine, University of Washington (also Investigator of Howard Hughes Medical Institute, 1977–1983) and determined the amino acid sequence of many proteases in a research group on proteolytic enzymes of Prof’s K. Walsh and H. Neurath, including thermolysin (the first characterized thermostable protease), bovine blood coagulation factor X and IX (the first characterized non-digestive proteases) in collaboration with Prof’s. K. Fujikawa and E. Davie et al. and rat cathepsin B and H (the first characterized SH proteases in animals) with Prof. N. Katsunuma et al. at Tokushima University. He also first elucidated the amino acid sequence and its function relationship of rabbit glycogen phosphorylase b, which is activated by phosphorylation, and various bovine protein kinases including cAMP-dependent, cGMP-dependent, glycogen phosphorylase b and Ca-dependent myosin light chain kinases in collaboration with Prof’s E. Krebs and E. Fischer who were Nobel Prize laureates in Physiology and Medicine in 1992. These studies have been highly evaluated in the world as one of pioneer works on signal transduction in cells. During his structural studies on protein kinases, he and his collaborators first discovered that the N-terminus of the catalytic subunit of bovine cAMP-dependent kinase is myristoylated (a new post- or co-translational modification of proteins reviewed in the present article). He came back to Japan in 1985 as Professor of Division of Biomedical Polymer Science, Institute of Comprehensive Medical Science, Fujita Health University and also as Head of Laboratory for Aging Process Research, Frontier Research Program, Riken (1986–1992). At Riken, he mainly studied proteins related to Alzheimer’s disease in collaboration with Prof. Y. Ihara et al. at University of Tokyo. At the former Institute, he continued the studies on the functional domains and the sugar chains including those of ABO blood type of human von Willebrand factor (VWF), an ultra-large protein which exists in plasma as heteropolymers of the subunit composed of 2,050 amino acid residues and essential for platelet aggregation, of which he had already completed the amino acid sequence in collaboration with Prof. E. Davie et al. in USA, and other cell adhesion proteins such as fibronectin and N-myristoylated brain proteins such as CAP-22/NAP-23. In addition, he also started the studied on snake venom proteins which regulate the interaction of VWF with platelets including “botrocetin” in collaboration with Prof. Y. Fujimura et al. at Nara Medical University and with staff members in his own laboratory until his retirement in 2000. By that time, he had contributed to publications of 314 original papers in English in the first-rate journals including Science, Nature, Neuron, Proc. Natl. Acad. Sci. USA, J. Biol. Chem., Biochemistry, Biochem. J., Protein Sci., Blood, J. Clin. Invest., Cancer Res., and Brain Res., and additional 28 review articles in English in proceedings, journals and books. He is now a professor emeritus at Fujita Health University. He was the president of Protein Engineering Society of Japan (now Protein Science Society of Japan) in 1987–1988. He received Tokai Yomiuri Award in Medicine for his contribution to medicine by structural studies on many functional proteins in 1997.
References
- 1).Carr S.A., Biemann K., Shoji S., Parmelee D.C., Titani K. (1982) n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc. Natl. Acad. Sci. USA 79, 6128–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Aitken A., Cohen P., Santikarn S., Williams D.H., Calder A.G., Smith A., et al. (1982) Identification of the NH2-terminal blocking group of calcineurin B as myristic acid. FEBS Lett. 150, 314–318 [DOI] [PubMed] [Google Scholar]
- 3).Bryant M., Ratner L. (1990) Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Ozols J., Korza G., Heinemann F.S., Hediger M.A., Strittmatter P. (1985) Complete amino acid sequence of steer liver microsomal NADH-cytochrome b5 reductase. J. Biol. Chem. 260, 11953–11961 [PubMed] [Google Scholar]
- 5).Cross F.R., Garber E.A., Pellman D., Hanafusa H. (1984) A short sequence in the p60src N terminus is required for p60src myristylation and membrane association and for cell transformation. Mol. Cell. Biol. 4, 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Kamps M.P., Buss J.E., Sefton B.M. (1985) Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc. Natl. Acad. Sci. USA 82, 4625–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Schultz A.M., Henderson L.E., Oroszlan S., Garber E.A., Hanafusa H. (1985) Amino terminal myristylation of the protein kinase p60src, a retroviral transforming protein. Science 227, 427–429 [DOI] [PubMed] [Google Scholar]
- 8).Song K.S., Sargiacomo M., Galbiati F., Parenti M., Lisanti M.P. (1997) Targeting of a G alpha subunit (Gi1 alpha) and c-Src tyrosine kinase to caveolae membranes: clarifying the role of N-myristoylation. Cell. Mol. Biol. 43, 293–303 [PubMed] [Google Scholar]
- 9).Resh M.D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 10).Kokame K., Fukada Y., Yoshizawa T., Takao T., Shimonishi Y. (1992) Lipid modification at the N terminus of photoreceptor G-protein α-subunit. Nature 359, 749–752 [DOI] [PubMed] [Google Scholar]
- 11).Resh M.D. (1996) Regulation of cellular signalling by fatty acid acylation and prenylation of signal transduction proteins. Cell. Signal. 8, 403–412 [DOI] [PubMed] [Google Scholar]
- 12).Arbuzova A., Wang J., Murray D., Jacob J., Cafiso D.S., McLaughlin S. (1997) Kinetics of interaction of the myristoylated alanine-rich C kinase substrate, membranes, and calmodulin. J. Biol. Chem. 272, 27167–27177 [DOI] [PubMed] [Google Scholar]
- 13).Matsubara M., Titani K., Taniguchi H., Hayashi N. (2003) Direct involvement of protein myristoylation in myristoylated alanine-rich C kinase substrate (MARCKS)-calmodulin interaction. J. Biol. Chem. 278, 48898–48902 [DOI] [PubMed] [Google Scholar]
- 14).Takasaki A., Hayashi N., Matsubara M., Yamauchi E., Taniguchi H. (1999) Identification of the calmodulin-binding domain of neuron-specific protein kinase C substrate protein CAP-22/NAP-22. Direct involvement of protein myristoylation in calmodulin-target protein interaction. J. Biol. Chem. 274, 11848–11853 [DOI] [PubMed] [Google Scholar]
- 15).Hayashi N., Matsubara M., Jinbo Y., Titani K., Izumi Y., Matsushima N. (2002) Nef of HIV-1 interacts directly with calcium-bound calmodulin. Protein Sci. 11, 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Crivici A., Ikura M. (1995) Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24, 85–116 [DOI] [PubMed] [Google Scholar]
- 17).Hayashi N., Matsubara M., Titani K., Taniguchi H. (1997) Circular dichroism and 1H nuclear magnetic resonance studies on the solution and membrane structures of GAP-43 calmodulin-binding domain. J. Biol. Chem. 272, 7639–7645 [DOI] [PubMed] [Google Scholar]
- 18).Matsubara M., Hayashi N., Jing T., Titani K. (2003) Regulation of endothelial nitric oxide synthase by protein kinase C. J. Biochem. 133, 773–781 [DOI] [PubMed] [Google Scholar]
- 19).Matsubara M., Hayashi N., Titani K., Taniguchi H. (1997) Circular dichroism and 1H NMR studies on the structures of peptides derived from the calmodulin-binding domains of inducible and endothelial nitric-oxide synthase in solution and in complex with calmodulin. Nascent α-helical structures are stabilized by calmodulin both in the presence and absence of Ca2+. J. Biol. Chem. 272, 23050–23056 [DOI] [PubMed] [Google Scholar]
- 20).Widmer F., Caroni P. (1990) Identification, localization, and primary structure of CAP-23, a particle-bound cytosolic protein of early development. J. Cell Biol. 111, 3035–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Maekawa S., Maekawa M., Hattori S., Nakamura S. (1993) Purification and molecular cloning of a novel acidic calmodulin binding protein from rat brain. J. Biol. Chem. 268, 13703–13709 [PubMed] [Google Scholar]
- 22).Duronio R.J., Jackson-Machelski E., Heuckeroth R.O., Olins P.O., Devine C.S., Yonemoto W., et al. (1990) Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc. Natl. Acad. Sci. USA 87, 1506–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Knoll L.J., Johnson D.R., Bryant M.L., Gordon J.I. (1995) Functional significance of myristoyl moiety in N-myristoyl proteins. Methods Enzymol. 250, 405–435 [DOI] [PubMed] [Google Scholar]
- 24).Seaton B.A., Head J.F., Engelman D.M., Richards F.M. (1985) Calcium-induced increase in the radius of gyration and maximum dimension of calmodulin measured by small-angle X-ray scattering. Biochemistry 24, 6740–6743 [DOI] [PubMed] [Google Scholar]
- 25).Kretsinger R.H., Rudnick S.E., Weissman L.J. (1986) Crystal structure of calmodulin. J. Inorg. Biochem. 28, 289–302 [DOI] [PubMed] [Google Scholar]
- 26).Persechini A., Kretsinger R.H. (1988) The central helix of calmodulin functions as a flexible tether. J. Biol. Chem. 263, 12175–12178 [PubMed] [Google Scholar]
- 27).Heidorn D.B., Seeger P.A., Rokop S.E., Blumenthal D.K., Means A.R., Crespi H., et al. (1989) Changes in the structure of calmodulin induced by a peptide based on the calmodulin-binding domain of myosin light chain kinase. Biochemistry 28, 6757–6764 [DOI] [PubMed] [Google Scholar]
- 28).Barbato G., Ikura M., Kay L.E., Pastor R.W., Bax A. (1992) Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 29).Finn B.E., Evenas J., Drakenberg T., Waltho J.P., Thulin E., Forsen S. (1995) Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 2, 777–783 [DOI] [PubMed] [Google Scholar]
- 30).van der Spoel D., de Groot B.L., Hayward S., Berendsen H.J., Vogel H.J. (1996) Bending of the calmodulin central helix: a theoretical study. Protein Sci. 5, 2044–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Ikura M., Clore G.M., Gronenborn A.M., Zhu G., Klee C.B., Bax A. (1992) Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 32).Meador W.E., Means A.R., Quiocho F.A. (1992) Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 33).Meador W.E., Means A.R., Quiocho F.A. (1993) Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science 262, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 34).Yuan T., Vogel H.J. (1998) Calcium-calmodulin-induced dimerization of the carboxyl-terminal domain from petunia glutamate decarboxylase. A novel calmodulin-peptide interaction motif. J. Biol. Chem. 273, 30328–30335 [DOI] [PubMed] [Google Scholar]
- 35).Elshorst B., Hennig M., Forsterling H., Diener A., Maurer M., Schulte P., et al. (1999) NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38, 12320–12332 [DOI] [PubMed] [Google Scholar]
- 36).Hayashi N., Izumi Y., Titani K., Matsushima N. (2000) The binding of myristoylated N-terminal nonapeptide from neuron-specific protein CAP-43/NAP-22 to calmodulin does not induce the globular structure observed for the calmodulin-nonmyristoylated peptide complex. Protein Sci. 9, 1905–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Blumenthal D.K., Takio K., Edelman A.M., Charbonneau H., Titani K., Walsh K.A., et al. (1985) Identification of the calmodulin-binding domain of skeletal muscle myosin light chain kinase. Proc. Natl. Acad. Sci. USA 82, 3187–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).O’Neil K.T., DeGrado W.F. (1990) How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64 [DOI] [PubMed] [Google Scholar]
- 39).Matsushima N., Hayashi N., Jinbo Y., Izumi Y. (2000) Ca2+-bound calmodulin forms a compact globular structure on binding four trifluoperazine molecules in solution. Biochem. J. 347, Pt. 1, 211–215 [PMC free article] [PubMed] [Google Scholar]
- 40).Maurer-Stroh S., Gouda M., Leite F., Novatchkova M., Schleiffer A., Schneider G., et al. (2004) MYRbase: Evaluation of genome-wide glycine myristoylation enlarges functional spectrum of eukaryotic myristoylated proteins. Genome Biol. 5, R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Caroni P., Aigner L., Schneider C. (1997) Intrinsic neuronal determinants locally regulate extrasynaptic and synaptic growth at the adult neuromuscular junction. J. Cell Biol. 136, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Frey D., Laux T., Xu L., Schneider C., Caroni P. (2000) Shared and unique roles of CAP23 and GAP43 in actin regulation, neurite outgrowth, and anatomical plasticity. J. Cell Biol. 149, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Blackshear P.J. (1993) The MARCKS family of cellular protein kinase C substrates. J. Biol. Chem. 268, 1501–1504 [PubMed] [Google Scholar]
- 44).Tapp H., Al-Naggar I.M., Yarmola E.G., Harrison A., Shaw G., Edison A.S., et al. (2005) MARCKS is a natively unfolded protein with an inaccessible actin-binding site: evidence for long-range intramolecular interactions. J. Biol. Chem. 280, 9946–9956 [DOI] [PubMed] [Google Scholar]
- 45).Asada A., Yamamoto N., Gohda M., Saito T., Hayashi N., Hisanaga S. (2008) Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cyclin-dependent kinase 5 complexes. J. Neurochem. 106, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 46).Hayashi N., Nakagawa C., Ito Y., Takasaki A., Jinbo Y., Yamakawa Y., et al. (2004) Myristoylation-regulated direct interaction between calcium-bound calmodulin and N-terminal region of pp60v-src. J. Mol. Biol. 338, 169–180 [DOI] [PubMed] [Google Scholar]
- 47).Matsubara M., Jing T., Kawamura K., Shimojo N., Titani K., Hashimoto K., et al. (2005) Myristoyl moiety of HIV Nef is involved in regulation of the interaction with calmodulin in vivo. Protein Sci. 14, 494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Lucero H.A., Robbins P.W. (2004) Lipid rafts-protein association and the regulation of protein activity. Arch. Biochem. Biophys. 426, 208–224 [DOI] [PubMed] [Google Scholar]
- 49).Epand R.M., Vuong P., Yip C.M., Maekawa S., Epand R.F. (2004) Cholesterol-dependent partitioning of PtdIns(4,5)P2 into membrane domains by the N-terminal fragment of NAP-22 (neuronal axonal myristoylated membrane protein of 22 kDa). Biochem. J. 379, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Chow M., Newman J.F., Filman D., Hogle J.M., Rowlands D.J., Brown F. (1987) Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327, 482–486 [DOI] [PubMed] [Google Scholar]
- 51).Kawamura S., Cox J.A., Nef P. (1994) Inhibition of rhodopsin phosphorylation by non-myristoylated recombinant recoverin. Biochem. Biophys. Res. Commun. 203, 121–127 [DOI] [PubMed] [Google Scholar]
- 52).Senin I.I., Zargarov A.A., Alekseev A.M., Gorodovikova E.N., Lipkin V.M., Philippov P.P. (1995) N-myristoylation of recoverin enhances its efficiency as an inhibitor of rhodopsin kinase. FEBS Lett. 376, 87–90 [DOI] [PubMed] [Google Scholar]
- 53).Towler D.A., Gordon J.I., Adams S.P., Glaser L. (1988) The biology and enzymology of eukaryotic protein acylation. Annu. Rev. Biochem. 57, 69–99 [DOI] [PubMed] [Google Scholar]
- 54).Pike L.J., Han X., Gross R.W. (2005) Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: a shotgun lipidomics study. J. Biol. Chem. 280, 26796–26804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Wingard J.N., Ladner J., Vanarotti M., Fisher A.J., Robinson H., Buchanan K.T., et al. (2008) Structural insights into membrane targeting by the flagellar calcium-binding protein (FCaBP), a myristoylated and palmitoylated calcium sensor in Trypanosoma cruzi. J. Biol. Chem. 283, 23388–23396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Nelson A.R., Borland L., Allbritton N.L., Sims C.E. (2007) Myristoyl-based transport of peptides into living cells. Biochemistry 46, 14771–14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Colombo S., Longhi R., Alcaro S., Ortuso F., Sprocati T., Flora A., et al. (2005) N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 168, 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Peitzsch R.M., McLaughlin S. (1993) Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 32, 10436–10443 [DOI] [PubMed] [Google Scholar]
- 59).Ohno S., Matsui M., Yokogawa T., Nakamura M., Hosoya T., Hiramatsu T., et al. (2007) Site-selective post-translational modification of proteins using an unnatural amino acid, 3-azidotyrosine. J. Biochem. 141, 335–343 [DOI] [PubMed] [Google Scholar]
- 60).Sakakibara D., Sasaki A., Ikeya T., Hamatsu J., Hanashima T., Mishima M., et al. (2009) Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 458, 102–105 [DOI] [PubMed] [Google Scholar]
- 61).Matsushima N., Izumi Y., Matsuo T., Yoshino H., Ueki T., Miyake Y. (1989) Binding of both Ca2+ and mastoparan to calmodulin induces a large change in the tertiary structure. J. Biochem. 105, 883–887 [DOI] [PubMed] [Google Scholar]
- 62).Osawa M., Kuwamoto S., Izumi Y., Yap K.L., Ikura M., Shibanuma T., et al. (1999) Evidence for calmodulin inter-domain compaction in solution induced by W-7 binding. FEBS Lett. 442, 173–177 [DOI] [PubMed] [Google Scholar]
- 63).Glover C.J., Goddard C., Felsted R.L. (1988) N-myristoylation of p60src. Identification of a myristoyl- CoA:glycylpeptide N-myristoyltransferase in rat tissues. Biochem. J. 250, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Guy B., Kieny M.P., Riviere Y., Le Peuch C., Dott K., Girard M., et al. (1987) HIV F/3′ orf encodes a phosphorylated GTP-binding protein resembling an oncogene product. Nature 330, 266–269 [DOI] [PubMed] [Google Scholar]
- 65).Sarkar S.N., Bandyopadhyay S., Ghosh A., Sen G.C. (1999) Enzymatic characteristics of recombinant medium isozyme of 2′-5′ oligoadenylate synthetase. J. Biol. Chem. 274, 1848–1855 [DOI] [PubMed] [Google Scholar]
- 66).Wice B.M., Gordon J.I. (1992) A strategy for isolation of cDNAs encoding proteins affecting human intestinal epithelial cell growth and differentiation: characterization of a novel gut-specific N-myristoylated annexin. J. Cell Biol. 116, 405–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Olshevskaya E.V., Hughes R.E., Hurley J.B., Dizhoor A.M. (1997) Calcium binding, but not a calcium-myristoyl switch, controls the ability of guanylyl cyclase-activating protein GCAP-2 to regulate photoreceptor guanylyl cyclase. J. Biol. Chem. 272, 14327–14333 [DOI] [PubMed] [Google Scholar]
- 68).Borgese N., Aggujaro D., Carrera P., Pietrini G., Bassetti M. (1996) A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J. Cell Biol. 135, 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Liu J., Sessa W.C. (1994) Identification of covalently bound amino-terminal myristic acid in endothelial nitric oxide synthase. J. Biol. Chem. 269, 11691–11694 [PubMed] [Google Scholar]
- 70).Musil L.S., Carr C., Cohen J.B., Merlie J.P. (1988) Acetylcholine receptor-associated 43K protein contains covalently bound myristate. J. Cell Biol. 107, 1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Spilker C., Gundelfinger E.D., Braunewell K.H. (1997) Calcium- and myristoyl-dependent subcellular localization of the neuronal calcium-binding protein VILIP in transfected PC12 cells. Neurosci. Lett. 225, 126–128 [DOI] [PubMed] [Google Scholar]
- 72).Ames J.B., Porumb T., Tanaka T., Ikura M., Stryer L. (1995) Amino-terminal myristoylation induces cooperative calcium binding to recoverin. J. Biol. Chem. 270, 4526–4533 [DOI] [PubMed] [Google Scholar]
- 73).Zhu D., Cardenas M.E., Heitman J. (1995) Myristoylation of calcineurin B is not required for function or interaction with immunophilin-immunosuppressant complexes in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 24831–24838 [DOI] [PubMed] [Google Scholar]
- 74).Kobayashi M., Takamatsu K., Saitoh S., Miura M., Noguchi T. (1993) Molecular cloning of hippocalcin, a novel calcium-binding protein of the recoverin family exclusively expressed in hippocampus. Biochem. Biophys. Res. Commun. 196, 1017. [DOI] [PubMed] [Google Scholar]
- 75).Faurobert E., Chen C.K., Hurley J.B., Teng D.H. (1996) Drosophila neurocalcin, a fatty acylated, Ca2+-binding protein that associates with membranes and inhibits in vitro phosphorylation of bovine rhodopsin. J. Biol. Chem. 271, 10256–10262 [DOI] [PubMed] [Google Scholar]
- 76).Koegl M., Zlatkine P., Ley S.C., Courtneidge S.A., Magee A.I. (1994) Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif. Biochem. J. 303, 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).van’t Hof W., Resh M.D. (1999) Dual fatty acylation of p59Fyn is required for association with the T cell receptor ζ chain through phosphotyrosine-Src homology domain-2 interactions. J. Cell Biol. 145, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Robbins S.M., Quintrell N.A., Bishop J.M. (1995) Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol. Cell Biol. 15, 3507–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Zlatkine P., Mehul B., Magee A.I. (1997) Retargeting of cytosolic proteins to the plasma membrane by the Lck protein tyrosine kinase dual acylation motif. J. Cell Sci. 110, 673–679 [DOI] [PubMed] [Google Scholar]
- 80).Haun R.S., Tsai S.C., Adamik R., Moss J., Vaughan M. (1993) Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J. Biol. Chem. 268, 7064–7068 [PubMed] [Google Scholar]
- 81).Shoji S., Tashiro A., Kubota Y. (1988) Antimyristoylation of gag proteins in human T-cell leukemia and human immunodeficiency viruses with N-myristoyl glycinal diethylacetal. J. Biochem. 103, 747–749 [DOI] [PubMed] [Google Scholar]
- 82).Mumby S.M., Heukeroth R.O., Gordon J.I., Gilman A.G. (1990) G-protein α-subunit expression, myristoylation, and membrane association in COS cells. Proc. Natl. Acad. Sci. USA 87, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Ikura M., Kay L.E., Bax A. (1990) A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 84).Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M.R., Appel, R.D. etal. (2005) Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (ed. Walker, J.M.). Humana Press, Totowa, pp. 571–607. [Google Scholar]
- 85).Kyte J., Doolittle R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]