Abstract
Natural killer (NK) cells can recognize and kill tumor cells lacking “self” markers, such as class I MHC, but the basis for this recognition is not completely understood. NKR-P1 receptors are members of the C-type lectin-related NK receptor superfamily that are conserved from rodents to humans. Identification of Clr ligands for the NKR-P1 receptors has facilitated functional analysis of MHC-independent target cell recognition by NK cells. One receptor-ligand pair, NKR-P1B:Clr-b, can mediate “missing-self” recognition of tumor and infected cells, but the role of this axis in sensing stressed cells remains unknown. Here, we show that Clr-b is rapidly downregulated in cells undergoing genotoxic and cellular stress at the level of both RNA and surface protein. Stress-mediated loss of Clr-b on leukemia cells enhanced cytotoxicity mediated by NKR-P1B+ NK cells. Notably, Clr-b downregulation was coordinated functionally with stress-mediated upregulation of NKG2D ligands (but not class I MHC). Our findings highlight a unique role for the MHC-independent NKR-P1B:Clr-b missing-self axis in recognition of stressed cells, and provide evidence of two independent levels of Clr-b regulation in stressed cells.
Introduction
Innate immunity constitutes an important front-line defense and barrier to infectious disease and malignancies (1). Natural killer (NK) cells are innate lymphocytes capable of recognizing and eliminating a wide variety of target cells, including transformed, infected, transplanted, antibody-coated, and stressed cells (2, 3). NK effector mechanisms include cellular cytotoxicity mediated by perforin, granzymes, and cell surface molecules (e.g., FasL, tumor necrosis factor–related apoptosis-inducing ligand), and rapid secretion of cytokines (e.g., IFN-γ, tumor necrosis factor-α, chemokines; ref. 2). Although NK cells are tolerant to normal “self” cells, they frequently respond to abnormal cells undergoing pathologic alterations. How NK cells mediate self–nonself discrimination at the molecular level remains the focus of intense research.
The frequent correlation between heightened NK cytotoxicity and a lack of MHC-I expression on tumor cell lines led to the hypothesis that NK cells sense the absence of self MHC-I markers on target cells. Termed “missing-self” recognition (4), this pathway is governed by inhibitory receptors specific for self MHC-I molecules that normally override stimulatory NK-target interactions. In turn, the loss of MHC-I molecules upon malignant transformation or infection becomes sufficient to trigger NK cytotoxicity via disinhibition of effector function (5). However, this mechanism alone is insufficient to explain the complex outcomes that regulate NK-target interplay.
Indeed, other potent stimulatory NK receptors such as NKG2D recognize “induced-self” ligands dynamically upregu-lated during stress responses. Thus, a contemporary dual-recognition model proposes that NK cell function is regulated by a balance of stimulatory and inhibitory receptor signals, which in turn are determined by target cell modulation of numerous cognate ligands (3, 6). This more target-centric model highlights the need to understand how both NK cell receptors and their cognate ligands are modulated in real time under pathologic conditions to fully appreciate complex NK recognition events in vivo.
The underlying molecular basis behind the ability of NK cells to recognize “stressed” cells remains incompletely understood. Recent evidence indicates that cells exposed to chemotherapeutic agents, genotoxic stimuli, or stalled DNA replication cycles dynamically upregulate stimulatory NKG2DL (7). This induced-self ligand modulation is mediated in part through the DNA damage response (DDR) pathway, in particular involving ATR, ATM, and Chk1 (8). NKG2DL upregulation has been observed in response to diverse genotoxic stimuli (including aphidicolin, cisplatin, 5-fluorouracil, γ-irradiation, and UV radiation; ref. 9), as well as heat shock responses (10), antigen-mediated activation of T cells (11–13), and dysregulation of Dicer expression (14). In addition, ligands for other stimulatory NK receptors, such as DNAM-1 (CD226), seem to be similarly upregulated (15). However, the modulation of inhibitory NK ligands during stress responses remains undocumented to date.
In addition to MHC-I molecules, several MHC-independent inhibitory ligands have recently been characterized, including Clr-b (Clec2d), the ligand for the NKR-P1B (Klrb2) receptor (3, 16–20). Importantly, the expression pattern of Clr-b closely resembles that of classic MHC-I antigens. Clr-b is widely expressed on normal hematopoietic and non-hematopoietic cells and is frequently lost on tumor cells (16). Moreover, like MHC-I, Clr-b expression is rapidly downregulated following cytomegalovirus (CMV) infection (21). Thus, Clr-b seems to be lost under numerous pathologic circumstances, and might function as an inhibitory rheostat in the routine detection of abnormal cells. Although the mechanism(s) governing Clr-b regulation remain to be elucidated, events leading to transformation or infection seem to initiate a programmed cellular response pathway culminating in the loss of Clr-b at the cell surface (17, 21). This intrinsic host response pathway differs from the extrinsic CTL-selected or immunoevasin-induced loss of MHC-I on tumor or infected cells, and thus, might share common elements known to be involved in the DDR pathway responsible for NKG2DL upregulation.
In this report, we investigated the influence of cell stress on Clr-b expression. Various chemotherapeutic agents and inducers of genotoxic or physiologic stress were found to promote a rapid functional downregulation of Clr-b transcripts and cell surface protein. Notably, MHC-I cell surface molecules were not substantially altered by similar treatments (or actually increased). Extending previous work, NKG2DL upregulation was found to be blocked by both caffeine and short hairpin RNA (shRNA)-mediated silencing of the ATR and Chk1 pathway (9). However, Clr-b downregulation was differentially affected by caffeine treatment and remained largely unaffected by shRNA-mediated knockdown of the ATR/Chk1 and ATM/Chk2 pathways. This suggests that loss of Clr-b following genotoxic stress may occur independently of the classic DDR pathway. Interestingly, ectopic expression of Clr-b transcripts prevented stress-mediated loss of Clr-b surface protein. Moreover, pharmacologic inhibition of the ubiquitin-proteasome degradation pathway uncoupled the downregulation of Clr-b surface protein from the stress-mediated loss of endogenous Clr-b transcripts. Collectively, these results show that genotoxic stress promotes missing-self regulation of the NK inhibitory ligand, Clr-b, at both the RNA and surface protein levels.
Materials and Methods
Chemicals and chemotherapeutic agents
Aphidicolin, bleomycin, cisplatin, camptothecin, etoposide, 5-fluorouracil, phleomycin, roscovitine, Scriptaid, trichostatin-A, tunicamycin, 5-aza-2′-deoxycytidine, caffeine, H2O2, α-amanitin, actinomycin-D, and cycloheximide were purchased from Sigma-Aldrich. MG132 and lactacystin were purchased from Calbiochem. γ-Irradiation was performed using a Gammacell-1000 Cs-source irradiator. UV-C irradiation was performed using a XL-1000 Spectrolinker. Chemicals were dissolved in DMSO, water, or PBS, according to the instructions of the manufacturer.
Mice, cell lines, and tissue culture
C1498 and NIH3T3 cells were purchased directly from American Type Culture Collection and validated independently using our own in-house stocks. MNK-1 was generated in our laboratory from primary mouse fetal thymic NK cells, as described previously (22). Primary mouse embryonic fibroblasts (MEF) and interleukin (IL)-2 lymphokine-activated killer (LAK) cells were generated from B6 and CD-1 mice bred and maintained in our own facilities. Cells were maintained in complete medium (DMEM-HG plus 10% FCS and supplements); MNK-1 cells were maintained in rhIL-2 (300 units/mL of Proleukin; Novartis) and routinely tested for IL-2 dependence (22). All cell lines (C1498, NIH3T3, MNK-1, and MEF) were authenticated by flow cytometry for expression of MHC-I and CD45 alleles, Clr-b, and various lineage markers (including NK1.1, CD3, CD4, CD8, CD19, CD11b, CD11c, and Gr-1). Cell lines were maintained in culture for less than 6 months (frequently 1–2 months) following resuscitation from stocks within one passage of expansion prior to freezing. Fibroblasts were maintained in a subconfluent state at all times to prevent the formation of foci, and all cells were maintained in logarithmic growth phase prior to treatment. Cell responses were independently validated following 1 week of culture in Normocin (Invivogen) to ensure the absence of Mycoplasma. For most treatments, 2 × 105 to 5 × 105 cells were seeded in 2 mL of complete medium in six-well plates and dosed for 24 hours, with or without 1-hour pretreatments with caffeine, MG132, or lactacystin.
Flow cytometric analysis
All commercial monoclonal antibodies (mAb) were purchased from BD PharMingen or eBioscience. Biotinylated antimouse Clr-b mAb (4A6 mAb; ref. 16) was generated previously. NKG2DL was visualized using NKG2D/hIgG fusion protein (23) plus goat anti-hIgG-PE (Jackson Immuno-Research). MHC-I levels were analyzed using anti-KbDb mAb. Streptavidin-R-phycoerythrin or streptavidin-allophycocyanin were used as secondary reagents (Invitrogen). Cells were stained as described (22), then analyzed using a BD FACSCa-libur flow cytometer and FlowJo software (TreeStar). All plots show cells gated for viable cells, as determined by live-cell gating by forward scatter, side scatter, and lack of propidium iodide uptake; viability for all experiments was >80% to 90%. Untreated control stains show analysis of cells treated with solvent alone.
BWZ reporter assay
BWZ.36 reporter cells (24) expressing CD3ζ-fusion receptors were described previously (16). For cell-based assays, 105 stimulator cells were treated for 24 hours, washed, and then cocultured overnight with 5 × 104 BWZ reporter cells prior to analysis of β-galactosidase activities (16).
Reverse transcription-PCR/quantitative reverse transcription-PCR
Total RNA was prepared using either Trizol or the PureLink RNA Mini Kit (Invitrogen). RNA was reverse-transcribed using the Super-Script III Kit and oligo-dT primer (Invitrogen). Reverse transcription-PCR (RT-PCR) was performed using a PTC-240G Tetrad (Bio-Rad) for 35 cycles (Clr-b) or 28 cycles (G3PDH), as follows: 94°C for 30 seconds, 53°C for 30 seconds, and 72°C for 60 seconds. Primers used were Clrb (624 bp), F-5′-ATGTGTGTCA-CAAAGGCTTCC-3′, R-5′-CTAGGAAGGAAAAAAAGGAGTTTG-3′; G3PDH (452 bp), F-5′-ACCACAGTCCATGCCATCAC-3′, R-5′-TCCACCACCCTGTTGCTGTA-3′.
qRT-PCR was performed using iTaq SYBR Green with ROX (Bio-Rad) on an ABI Prism 7000 (Applied Biosystems) for 37 cycles, as follows: 95°C for 30 seconds and 61°C for 60 seconds. Products were confirmed by dissociation curve analysis. Primers used were mClrb (120 bp), F-5′-CATCTCCCCTGAGTCTTGTG-3′, R-5′-ACTGGGATCTGTTCTGTCTTTG-3′; G3PDH (120 bp), F-5′-GACTTCAACAGCAACTCCCACTCTT-3′, R-5′-CACCCTGTTGCTGTAGCCGTATTC-3′.
51Cr-release cytotoxicity assays
LAK effector cells were prepared from bone marrow cells and splenocytes grown in complete medium containing 1,000 to 2,000 units/mL of rhIL-2 plus 15 ng/mL of mIL-15 (Peprotech) for 5 to 6 days. Lysis assays were performed as described (16).
Retroviral transduction
The full-length Clr-b coding sequence was subcloned into pMSCV2.2-CMV-IRES-GFP. Retroviral shRNA constructs based on the pMND-Banshee vector (which contains an LTR-driven GFP reporter gene) were previously characterized and validated (9). Stable transductants were generated as described (16), sorted for GFP, and expanded for 5 days before analysis.
Results
Rapid and dose-dependent loss of cell surface Clr-b in response to diverse genotoxic agents
We have previously shown that Clr-b is broadly expressed on most normal cells, yet frequently lost on tumor cells; this renders transformed target cells more susceptible to NK cytotoxicity via NKR-P1B–mediated missing-self recognition (16). However, the underlying basis for the loss of Clr-b during transformation remains unclear (3). To gain better insight into the regulation of Clr-b expression, we investigated whether Clr-b levels are influenced by genotoxic stress, an initiating event in transformation (9). To this end, several Clr-b+ mouse cell lines were screened for their responses to various geno-toxic agents, then evaluated for surface expression of Clr-b (16). For comparison, we also monitored the expression of classic MHC-I molecules and NKG2DL (23).
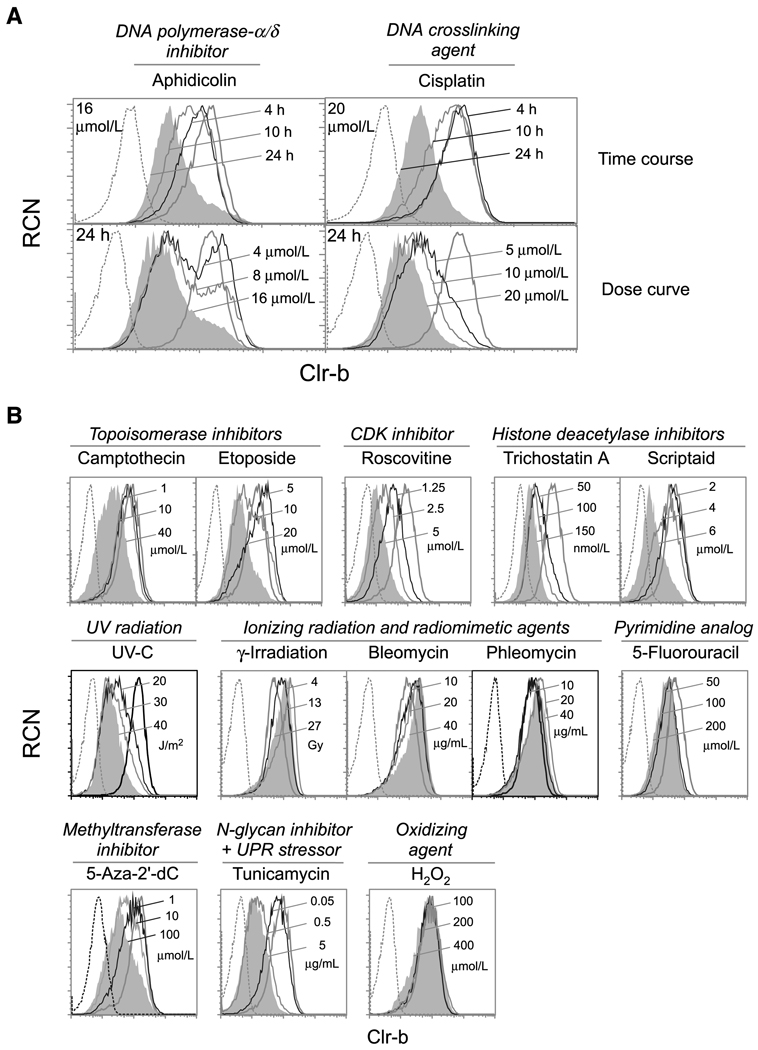
As previously shown (16), C1498 cells (NKT-like acute leukemia; ref. 25), MNK-1 cells (IL-2–dependent thymic pre-NK line; ref. 22), NIH3T3 fibroblasts, and MEF cells express high levels of cell surface Clr-b. Strikingly, treatment of these cells with chemotherapeutic agents and chemicals known to induce genotoxic stress promoted a rapid downregulation of Clr-b surface protein (Fig. 1; Supplementary Fig. S1A). Loss of Clr-b expression was a rapid and dynamic event, and the magnitude of downregulation was controlled in a dose-and time-dependent manner (Fig. 1A). To investigate distinct modes of cellular stress, Clr-b modulation was evaluated in response to a variety of chemotherapeutic and genotoxic agents, as well as agents that induce alternative stress responses. Several but not all genotoxic agents efficiently promoted Clr-b downregulation in four cell lines at pharmacologic doses within 24 hours (Fig. 1B; Supplementary Fig. S1A). Interestingly, analogous treatments had similar effects on Clr-b expression, including (a) camptothecin and etoposide (topoisomerase-I/II inhibitors); (b) trichostatin-A and Scriptaid (histone deacetylase inhibitors); and (c) γ-irradiation, bleomycin, and phleomycin (ionizing radiation and radiomimetic agents). This highlights a correlation between similar stress response pathways and the extent of Clr-b downregulation.
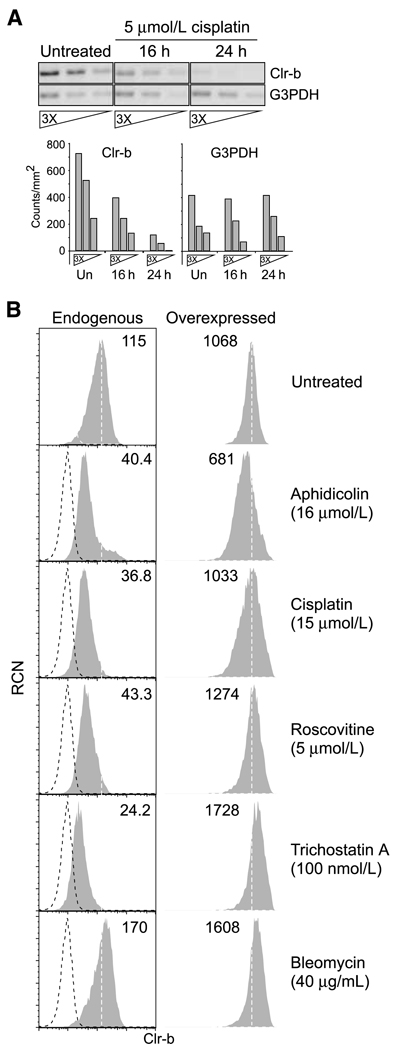
Figure 1.
Rapid and dose-dependent downregulation of Clr-b on C1498 leukemia cells in response to genotoxic and cellular stress. A, C1498 cells were treated as indicated and analyzed by flow cytometry. Histograms show untreated cells (thick black line), treated cells (thin solid lines, with maximal treatment indicated by gray shading), or secondary reagent alone (dotted line). B, C1498 cells were treated as indicated for 24 h, then analyzed as in A. RCN, relative cell number. Representative of three independent experiments.
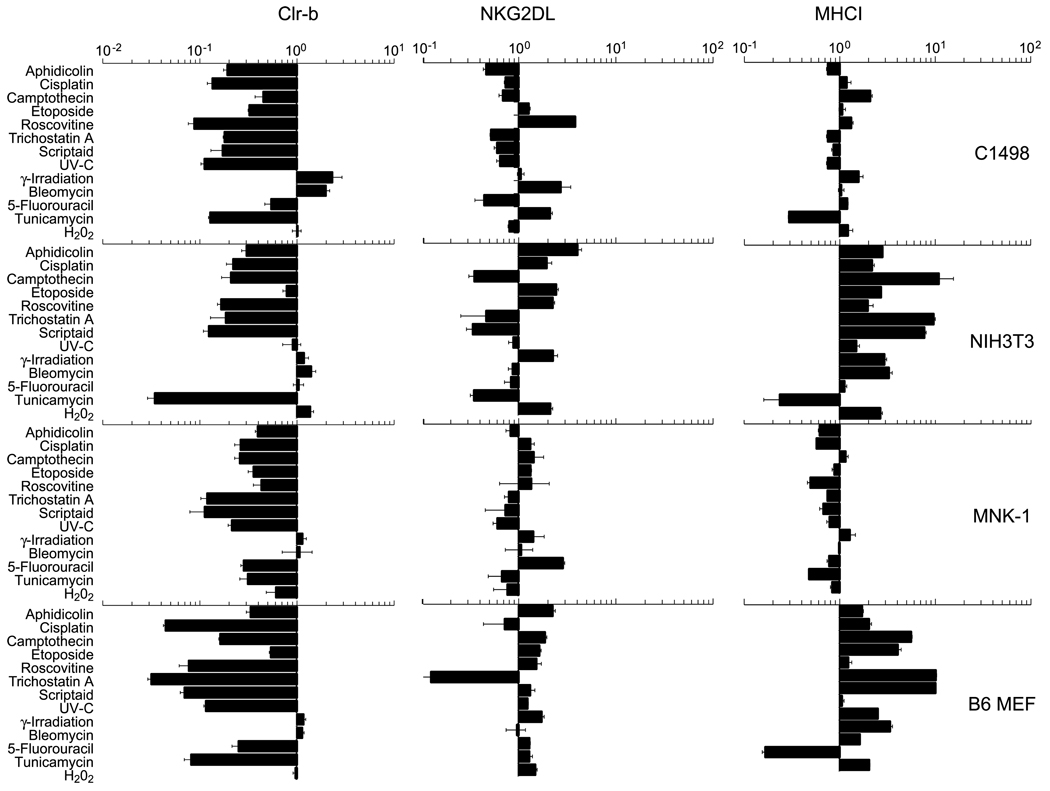
In contrast, NKG2DL was frequently upregulated under these conditions (Supplementary Fig. S1B; ref. 9), whereas MHC-I surface levels remained largely unaffected (or increased somewhat; Supplementary Fig. S1C). Collectively, these combined responses are expected to enhance the susceptibility of target cells to NK cytotoxicity. Because heterogeneity in responses existed among the cells tested, factors such as transformation state, cell type, or other cell-intrinsic properties might influence the degree or magnitude of Clr-b or NKG2DL modulation. Notably, Clr-b was strongly and consistently downregulated on several cell lines in response to treatment with aphidicolin (DNA replication inhibitor), cisplatin (DNA crosslinking agent), and roscovitine (CDK inhibitor), but not bleomycin (DNA-cleaving radiomimetic; Figs. 1 and 2; Supplementary Fig. S1A). Therefore, we tested whether the responses to these agents had functional consequences on NK recognition.
Figure 2.
Clr-b downregulation on four different cell lines in response to genotoxic and cellular stress. C1498, NIH3T3, MNK-1, and B6 strain MEF cells were treated as indicated in Fig. 1, and analyzed by flow cytometry. Bar graphs indicate the fold up/downregulation in median fluorescence intensity (MFI) levels of Clr-b, NKG2DL, and MHC-I, relative to untreated control cells. Error bars indicate SDs of three experiments.
Genotoxic stress enhances susceptibility to NK cytotoxicity and diminishes NKR-P1B–mediated recognition of Clr-b ligand
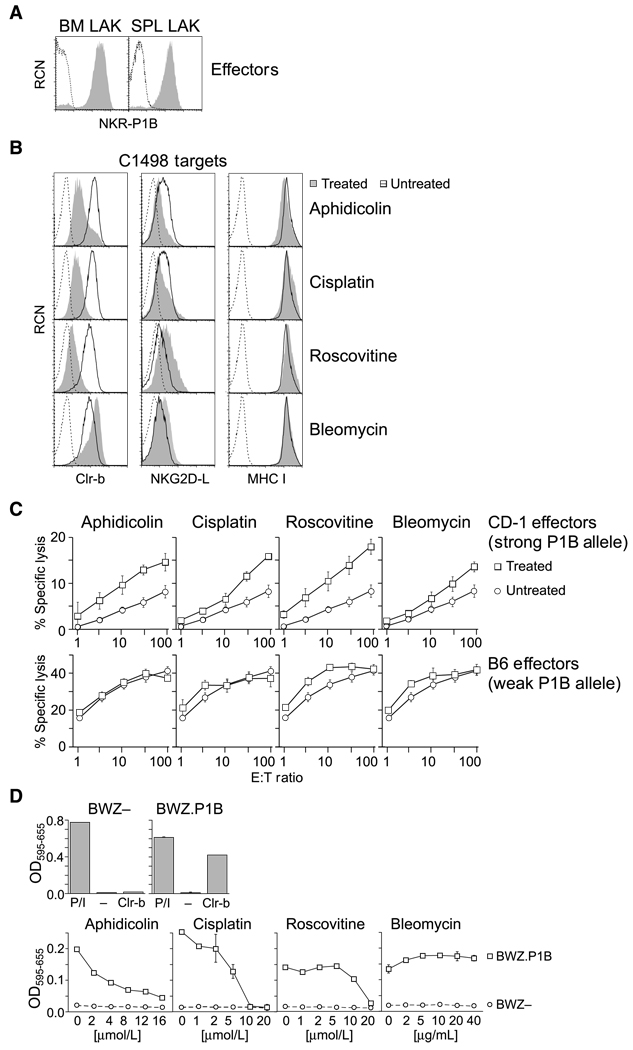
Loss of surface Clr-b on cells undergoing genotoxic stress would be expected to enhance NK cytotoxicity, as previously observed for tumor cells (16). To address this, treated target cells were analyzed in standard 51Cr-release cytotoxicity assays. C1498 targets were used because they represent a model acute leukemia cell line used broadly in cytotoxicity assays (26, 27), they are one of few tumor lines that express high levels of Clr-b (16), and they modulate Clr-b but not other known NK ligands in response to several genotoxic agents. To further control for receptor specificity, we took advantage of known NKR-P1B allelic polymorphisms between two mouse strains, CD-1 and B6, which have been previously shown to influence the magnitude of Clr-b–dependent inhibition in cytotoxicity assays: CD-1 strain LAK cells are strongly inhibited via the NKR-P1BCD-1 allele, whereas B6-strain LAK cells are only weakly inhibited via the NKR-P1BB6 allele (a.k.a., NKR-P1D; ref. 28), in response to target cells expressing Clr-b (16, 29). Because the NKR-P1B receptor is known to exhibit variegated expression on NK subsets (3, 29), the use of CD-1 strain LAK allowed us to monitor surface expression of the NKR-P1BCD-1 allele (via PK136 mAb; refs. 3, 16, 22, 28), whereas mAbs specific for the NKR-P1BB6 allele are not commercially available (18, 29).
As shown in Fig. 3A, both bone marrow–derived and splenic LAK cells from CD-1 mice express high levels of NKR-P1B. Furthermore, C1498 targets treated with aphidicolin or cisplatin possess strongly reduced levels of surface Clr-b, whereas NKG2DL and MHC-I surface levels remain largely unaltered (Figs. 1, 2, and 3B). In contrast, C1498 cells treated with roscovitine lose Clr-b and induce NKG2DL, whereas cells treated with bleomycin maintain Clr-b levels and moderately upregulate NKG2DL. As expected, treatment with genotoxic agents rendered C1498 targets more sensitive to cytotoxicity mediated by CD-1 strain LAK (Fig. 3C). Moreover, target cytotoxicity was increased to a greater extent in response to aphidicolin, cisplatin, or roscovitine (which strongly down-regulate Clr-b), compared with bleomycin (which does not promote loss of Clr-b). On the other hand, cytotoxicity mediated by B6-strain LAK was minimally affected by aphidicolin or cisplatin whereas cytotoxicity was increased by roscovitine or bleomycin, consistent with the moderate increase in NKG2DL expression observed in the latter treatments. Importantly, known receptor polymorphisms affecting CD-1 versus B6 LAK function are limited to the NKR-P1B and Ly49 receptors, yet MHC-I levels do not change upon aphidicolin or cisplatin treatment. This rules out a role for loss of MHC-I ligands or induction of stimulatory ligands recognized by nonpolymorphic NK receptors in contributing to the augmented cytotoxicity observed for CD-1 strain LAK. This strongly argues that enhanced cytotoxicity mediated by NKR-P1B+ LAK is due to loss of Clr-b expression on C1498 targets exposed to the genotoxic agents, aphidicolin and cisplatin.
Figure 3.
Loss of NKR-P1B–mediated inhibition of NK cytotoxicity and Clr-b ligand function in response to genotoxic stress. A, CD-1 strain bone marrow and splenic LAK cultures were analyzed by flow cytometry for NKR-P1B expression (shaded histogram) versus secondary reagent alone (dotted line). B, flow cytometric analysis of Clr-b, NKG2DL, and MHC-I (KbDb) expression on treated C1498 cells (shaded area), untreated cells (DMSO alone; solid line), or secondary reagent alone (dotted line). C1498 cells were treated overnight (24 h) with aphidicolin (20 µmol/L), cisplatin (10 µmol/L), roscovitine (10 µmol/L), or bleomycin (40 µg/mL). C, standard 51Cr-release cytotoxicity assay of CD-1 or B6 LAK cells versus C1498 target cells treated as in B. Plots indicate mean of triplicate percentage of specific lysis values ± SEM for the indicated effector/ target (E:T) ratios. D, top, BWZ–parental cells or BWZ.CD3ζ/ NKR-P1B reporter cells were plated overnight in medium alone (−), stimulated using PMA + ionomycin (P/I), or mixed with BWZ.Clr-b stimulator cells (Clr-b). Normalized OD595–655 values are shown. Bottom, C1498 stimulator cells were treated with the indicated concentrations of agents in situ, then incubated with BWZ.P1B reporter cells overnight. Representative of three independent experiments.
To specifically address the loss of surface Clr-b ligand function in isolation, BWZ reporter assays were performed. Here, BWZ.36 reporter cells (24) expressing a chimeric CD3ζ/NKR-P1B fusion-receptor (BWZ.CD3ζ/P1B cells; ref. 16) were used to assess NKR-P1B–dependent recognition of treated stimulator cells. As shown in Fig. 3D, BWZ.CD3ζ/P1B reporter cells respond specifically to stimulator cells expressing Clr-b ligand, whereas BWZ– parental cells fail to respond. Next, C1498 stimulator cells were incubated in situ for 24 hours with various genotoxic agents in 96-well plates, followed by washing and overnight incubation with BWZ- or BWZ.CD3ζ/P1B reporter cells. Importantly, these results confirm that Clr-b ligand function is lost in a dose-dependent manner, yet only in response to genotoxic agents that downregulate Clr-b cell surface expression (i.e., aphidicolin, cisplatin, roscovitine, but not bleomycin; Fig. 3D; Figs. 1 and 2). This loss of cognate ligand function was independently confirmed with three other cell lines treated with the same four genotoxic agents (data not shown).
Potential involvement of the ATM/ATR pathways in stress-mediated Clr-b downregulation
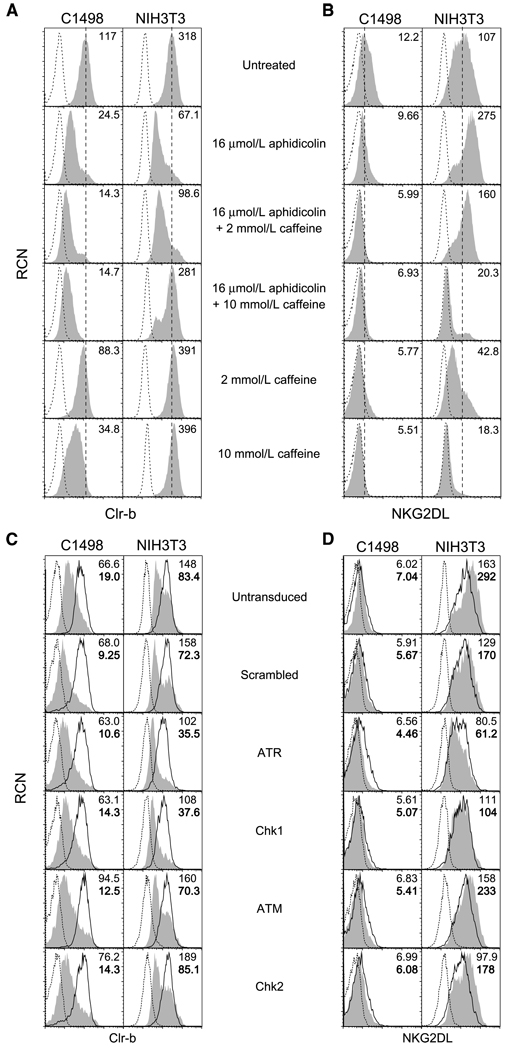
Previous studies showed a role for the DDR pathway in aphidicolin-induced NKG2DL upregulation on fibroblasts (via ATR/Chk1), as well as constitutive NKG2DL expression on tumor cells (via ATM; refs. 8, 9). In these studies, inhibition of the ATM/ATR pathways using caffeine abrogated NKG2DL upregulation on fibroblasts in response to aphidicolin treatment (9). Therefore, we investigated the involvement of this pathway in stress-mediated Clr-b downregulation. Interestingly, caffeine blocked both Clr-b downregulation and NKG2DL upregulation on NIH3T3 fibroblasts in response to aphidicolin treatment (Fig. 4A and B). However, caffeine did not block aphidicolin-mediated loss of Clr-b on C1498 leukemia cells; rather, caffeine treatment alone promoted Clr-b down-regulation, and the combination of caffeine and aphidicolin promoted a more striking loss of Clr-b. As a control, constitutive NKG2DL expression and stress-mediated NKG2DL upregulation were abrogated on both C1498 leukemia cells and NIH3T3 fibroblasts by caffeine (Fig. 4A and B). These findings are consistent with differential effects observed previously: The ATR pathway seems to regulate aphidicolin-induced NKG2DL on fibroblasts, whereas the ATM pathway might regulate constitutive NKG2DL expression on some tumor cells (8, 9).
Figure 4.
Differential effects of caffeine versus shRNA-mediated ATR/ATM silencing on aphidicolin-mediated Clr-b downregulation. A and B, flow cytometric analysis of Clr-b or NKG2DL expression on C1498 leukemia cells and NIH3T3 fibroblasts treated overnight with aphidicolin (16 µmol/L) in the absence or presence of 1 h caffeine pretreatment (2 mmol/L, 10 mmol/L). Histograms show Clr-b or NKG2DL expression (shaded area) versus secondary reagent alone (dotted line). A vertical dashed line indicates the MFI on untreated cells; numbers indicate MFI values. Representative of three independent experiments. C and D, as in A and B, except cells were modified by retroviral shRNA-mediated knockdown of ATR, Chk1, ATM, or Chk2, or using a scrambled shRNA. Transduced cells were gated for GFP reporter expression. Histograms show gated GFP+ (transduced) cells versus GFP− (untransduced) cells analyzed for Clr-b or NKG2DL expression; untreated cells (solid line), treated cells (shaded area), or secondary reagent alone (dotted line). Plain numbers indicate MFI of untreated cells, boldfaced numbers indicate MFI of aphidicolin-treated cells. Representative of three independent experiments.
Because caffeine exhibits pleiotropic effects independent of ATR/ATM inhibition (30–33), previously characterized shRNA vectors (9) were used to specifically silence the ATM, ATR, Chk1, and Chk2 gene products, and then Clr-b modulation on C1498 and NIH3T3 cells was reassessed. Extending previous findings (9), aphidicolin-induced NKG2DL upregulation on NIH3T3 fibroblasts could be blocked by shRNA-mediated knockdown of Chk1 or ATR, but not ATM or Chk2 (Fig.4C and D; see also Supplementary Fig. S2). In contrast, none of the shRNA constructs prevented aphidicolin-mediated Clr-b downregulation on either NIH3T3 fibroblasts or C1498 cells. In fact, ATR/Chk1 silencing (but not ATM/Chk2 silencing) modestly enhanced aphidicolin-mediated downregulation of Clr-b, and somewhat lowered constitutive Clr-b expression, on NIH3T3 fibroblasts (Fig. 4C and D).
Collectively, although redundancy in the DDR pathway cannot be excluded, the failure of shRNA-mediated silencing of these kinases to inhibit aphidicolin-mediated Clr-b down-regulation (on both NIH3T3 and C1498 cells), suggests that loss of Clr-b in response to genotoxic stress might occur via a novel mechanism, independent of the ATR/ATM pathways (10). The lack of ATM involvement is further supported by the failure of γ-irradiation and the radiomimetic drugs, bleomycin and phleomycin, to promote Clr-b downregulation. In addition, studies using p53−/− MEF and inhibitors of p53 function (such as pifithrin-α; ref. 34) failed to reveal any involvement of p53 (a downstream effector of the ATM/ATR-mediated DDR signaling pathways; ref. 35) on stress-mediated Clr-b downregulation (data not shown). In turn, the partial ability of caffeine to abrogate aphidicolin-mediated Clr-b downregulation (on NIH3T3 but not C1498 cells) might be due to effects unrelated to the DDR pathway (36), such as cell cycle blockade (31), or inhibition of PI3K, DNA-PK, PAK1, or other caffeine-sensitive kinases (30, 32, 33, 37). To gain further insight, we examined Clr-b expression at the transcript level.
Genotoxic stress-mediated loss of Clr-b occurs at the transcript level
It was previously shown that Clr-b is rapidly lost at both the transcript and surface protein levels in response to CMV infection (21). However, it is not known whether Clr-b down-regulation following genotoxic stress occurs at the transcript level or whether the surface protein is being actively internalized. To address this question, Clr-b transcripts were analyzed by semiquantitative RT-PCR using primers spanning the entire coding sequence. As shown in Fig. 5A, Clr-b transcripts were rapidly lost in C1498 cells undergoing genotoxic stress. Moreover, the loss of Clr-b transcripts following stress induction correlates temporally with the loss of Clr-b protein at the cell surface (Fig. 1A, Fig. 5A), similar to previous observations following CMV infection (21). Thus, like viral infection, genotoxic stress directly regulates steady-state levels of endogenous Clr-b transcripts.
Figure 5.
Genotoxic stress promotes a loss of endogenous Clr-b transcripts, whereas ectopic expression of Clr-b transcripts abrogates Clr-b downregulation. A, semiquantitative RT-PCR analysis of Clr-b transcript expression (full-length coding sequence) in C1498 cells either untreated or treated with cisplatin (5 µmol/L) as indicated (using 3-fold serial cDNA dilutions). Inverse ethidium bromide gel images and quantitative signal intensities (using Quantity One software) are shown. B, flow cytometric analysis of endogenous or ectopically overexpressed Clr-b protein on C1498 cells following treatment as indicated. Retroviral C1498 transductants overexpressing a CMV promoter–driven full-length Clr-b coding sequence were gated according to IRES-GFP reporter gene expression. Histograms show Clr-b expression (shaded area) versus secondary reagent alone (dotted line); numbers indicate MFI values. A vertical dashed line indicates the median Clr-b expression on untreated cells. Representative of three independent experiments.
To investigate this further, the full-length Clr-b coding sequence was ectopically overexpressed in C1498 cells under the control of a heterologous CMV promoter (pMCIG vector; ref. 28), and then Clr-b surface levels were reassessed. As shown in Fig. 5B, stable overexpression of Clr-b transcripts prevents downregulation of Clr-b surface protein mediated by diverse agents. These data show that loss of Clr-b in response to genotoxic stress, like that promoted by transformation (16) or viral infection (21), is controlled at the transcript level.
Notably, transcript level regulation has been previously reported for at least some NKG2DLs that are upregulated upon transformation or genotoxic stress (10, 11). However, other NKG2DLs (e.g., Mult1) were recently shown to be posttranslationally regulated in response to cell stress (10). Therefore, we examined protein level regulation of Clr-b on cells exposed to genotoxic stress.
Regulation of cell surface Clr-b protein following genotoxic stress
Although Clr-b expression is regulated at the transcript level, it is still possible that Clr-b surface protein might also be posttranslationally internalized, either constitutively or following genotoxic stress (10). Consistent with this possibility, the Clr-b cytoplasmic tail contains a number of motifs that could affect its cell surface expression, including several lysine residues that may serve as substrates for ubiquitination (Supplementary Fig. S3). Indeed, several viruses have evolved E3 ubiquitin ligases that posttranslationally regulate cell surface expression of MHC-I, B7 family members, ICAM-1, or NKG2DL (38–41). A potential role for the ubiquitin-proteasome degradation pathway in Clr-b downregulation following genotoxic stress was investigated using MG132, a pharmacologic inhibitor of the 26S proteasome.
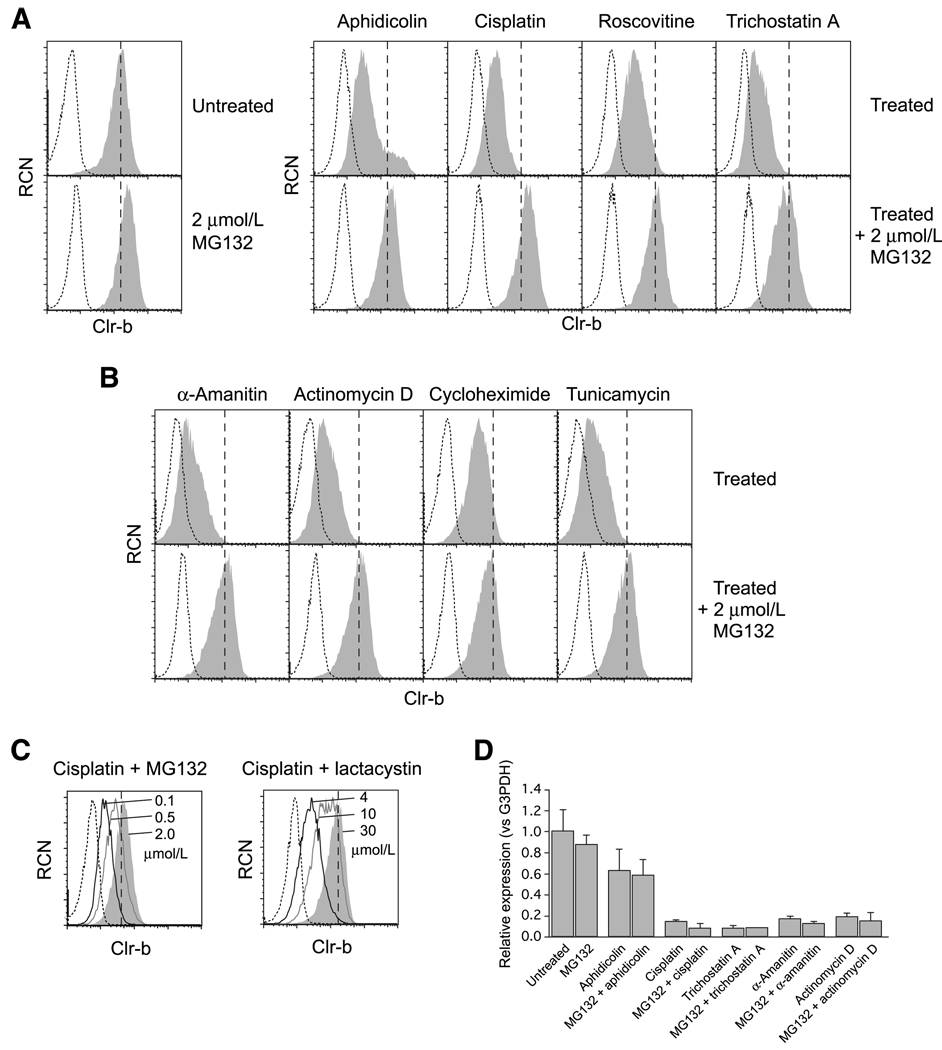
Interestingly, MG132 pretreatment abrogated Clr-b downregulation following treatment of C1498 cells with diverse agents, including aphidicolin, cisplatin, roscovitine, trichostatin-A, and UV-C irradiation, whereas MG132 treatment alone slightly (but consistently) increased steady-state Clr-b surface expression (Fig. 6A; data not shown). Furthermore, MG132 administration also blocked downregulation of surface Clr-b in response to generalized inhibitors of transcription, translation, and posttranslational processing, such as α-amanitin, actinomycin-D, cycloheximide, and tunicamycin (Fig. 6B), the latter of which also promotes endoplasmic reticulum-associated stress.
Figure 6.
Inhibition of the ubiquitin-proteasome pathway uncouples stress-mediated Clr-b surface protein downregulation from loss of endogenous transcripts. A, flow cytometric analysis of Clr-b expression on C1498 cells either untreated or treated overnight in the presence or absence of 1 h pretreatment with MG132 (2 µmol/L). Histograms show Clr-b expression (shaded area) versus secondary reagent alone (dotted line) on cells treated with aphidicolin (16 µmol/L), cisplatin (15 µmol/L), roscovitine (5 µmol/L), or trichostatin A (100 nmol/L). A vertical dashed line indicates median Clr-b expression on untreated cells. B, as in A, except cells were treated with α-amanitin (30 µmol/L), actinomycin D (5 nmol/L), cycloheximide (40 µg/mL), or tunicamycin (0.5 µg/mL). C, as in A, except cells were treated with cisplatin (15 µmol/L) with increasing concentrations of MG132 or lactacystin. Histograms show Clr-b expression at maximum inhibitor concentrations (shaded area), or decreasing concentrations (solid lines, as indicated), relative to median Clr-b expression on untreated cells (vertical dashed line), or secondary reagent alone (dotted line). D, real-time quantitative RT-PCR analysis of Clr-b expression in C1498 cells treated as in A with aphidicolin (8 µmol/L), cisplatin (15 µmol/L), trichostatin A (100 nmol/L), α-amanitin (30 µmol/L), or actinomycin D (5 nmol/L). Relative ΔΔCt values were calculated using G3PDH as internal control, normalized to untreated samples. Representative of three independent experiments.
Because MG132 can exert pleiotropic effects independent of blocking ubiquitin-dependent proteasomal degradation, including an ability to inhibit calpains and cathepsins, another proteasomal inhibitor of greater specificity, lactacystin, was also used (42). As shown in Fig. 6C, both MG132 and lactacystin prevented stress-mediated Clr-b downregulation in a dose-dependent manner. Similar results were observed using NIH3T3 fibroblasts and MEF cells (data not shown). Importantly, the administration of proteasomal inhibitors had no influence on the downregulation of Clr-b transcripts following administration of genotoxic agents or transcriptional inhibitors, as revealed by quantitative real-time RT-PCR (Fig. 6D). This shows that downregulation of Clr-b transcripts can be uncoupled from a loss of Clr-b surface protein.
Nonetheless, the MG132 and lactacystin data do not necessarily implicate the proteasome itself in direct degradation of Clr-b. Pharmacologic inhibition of the proteasome leads to the accumulation of nondegraded polyubiquitinated aggregates and a rapid depletion of free ubiquitin levels, resulting in general impairment of ubiquitin-dependent processes (43). One such proteasome-independent process, the endolysosomal trafficking pathway, targets endocytosed monoubiquitinated proteins for recycling and/or lysosomal degradation (44). Interestingly, chloroquine, an inhibitor of endolysosomal acidification (45), also impaired aphidicolin- and cisplatinmediated Clr-b downregulation (data not shown). Collectively, these results suggest that ubiquitin-dependent processes play an important role in normal Clr-b cell surface turnover, whereas stress-mediated Clr-b downregulation is ultimately controlled at the transcript level. Therefore, any perturbation in Clr-b transcript levels might rapidly influence Clr-b surface expression.
Discussion
It has been previously shown that stimulatory ligands for the NKG2D and DNAM-1 immunoreceptors are upregulated by the DDR pathway following genotoxic stress, an effect that enhances the susceptibility of target cells to NK cell–mediated lysis (9, 15). Here, for the first time, we show that genotoxic stress promotes the functional downregulation of an inhibitory NK ligand, Clr-b. The ability of numerous distinct chemotherapeutic and genotoxic agents to promote Clr-b downregulation suggests that cell stress may initiate a conserved and programmed cellular response governing Clr-b expression. Thus, pathways governing the cellular response to transformation (16), genotoxic stress (an initiating event in transformation), viral infection (21), and other pathologic states might share common features (46, 47).
Similar to the findings reported here for Clr-b downregulation, cell surface expression of the NKG2DL, Mult1, was upregulated independently of the ATR/ATM pathway in response to cell stress induced by heat shock and UV irradiation (10). Surface expression of Mult1 was shown to be dependent on inhibition of the ubiquitin-proteasome and endolysosomal degradation pathways responsible for retention of the protein within normal, nonstressed cells. These findings highlight a potentially conserved ubiquitindependent mechanism for Clr-b downregulation under stressed versus nonstressed conditions.
How Clr-b transcript and surface protein levels might be regulated requires further investigation. One possible mechanism is via the direct control of Clrb (Clec2d) promoter activity and the production of nascent Clr-b transcripts. A second potential mechanism is through the posttranscriptional regulation of mRNA stability. This intriguing possibility was recently suggested by the finding that the rodent Clr-b mRNAs (i.e., the mouse Clec2d8 and rat Clec2d11 gene products; refs. 17, 21) possess an embedded autocatalytic discontinuous hammerhead ribozyme sequence (48). Thus, cleavage of steady-state Clr-b transcripts by the internal ribozyme upstream of the poly-adenylation site could promote the loss of mRNA stability or impair protein translation. Whether protein factors normally antagonize autocatalysis mediated by the ribozyme sequence, or whether cofactors are recruited to the cleavage site upon genotoxic stress, requires further testing. On the other hand, preexisting Clr-b transcripts may also be regulated at the level of mRNA stability and/or protein translation by microRNA-mediated silencing mechanisms, such as that observed for NKG2DL expression (49, 50). Thus, Clec2d promoter activity, elements within the Clr-b untranslated regions, and endogenous or pathogen-encoded Clr-b–specific microRNAs are all relevant targets to warrant future studies on the missing-self control of Clr-b expression in response to pathologic processes (16, 17, 21).
In conclusion, how NK cells recognize abnormal targets remains incompletely understood. Understanding how a programmed cellular response mediates Clr-b downregulation in real time during genotoxic stress will enhance our knowledge of the events that lead to innate recognition of transformed and infected cells, how malignant NK-resistant tumors evolve, and how chemotherapeutic agents might influence NK recognition of normal and tumor cells. Future insight into this MHC-independent mode of missing-self ligand modulation will affect our views of NK cell–mediated recognition.
Supplementary Material
Acknowledgments
The authors thank Drs. A. Martin, C.J. Guidos, T.H. Watts, J.C. Zúñiga-Pflücker, and L. Horan for helpful comments and suggestions, and C. Kirkham for assistance with p53 genotyping and tissue culture treatments.
Grant Support
HFSP Career Development Award CDA-0037/2005, MRI Early Researcher Award ERA-07-04-071, CIHR Operating Grant FRN-74754, CIHR New Investigator Award, and BWF Investigator in the Pathogenesis of Infectious Disease Award (J.R. Carlyle); NSERC PGS-D Award (J.H. Fine); OGS Award (P. Chen); and CIHR Vanier Award (A. Mesci).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Beutler B. Innate immunity: an overview. Mol Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Mesci A, Ljutic B, Makrigiannis AP, Carlyle JR. NKR-P1 biology: from prototype to missing self. Immunol Res. 2006;35:13–26. doi: 10.1385/IR:35:1:13. [DOI] [PubMed] [Google Scholar]
- 4.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 5.MacFarlane AW, Campbell KS. Signal transduction in natural killer cells. Curr Top Microbiol Immunol. 2006;298:23–57. doi: 10.1007/3-540-27743-9_2. [DOI] [PubMed] [Google Scholar]
- 6.Raulet DH. Missing self recognition and self tolerance of natural killer (NK) cells. Semin Immunol. 2006;18:145–150. doi: 10.1016/j.smim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Gasser S, Raulet DH. The DNA damage response arouses the immune system. Cancer Res. 2006;66:3959–3962. doi: 10.1158/0008-5472.CAN-05-4603. [DOI] [PubMed] [Google Scholar]
- 8.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–142. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 9.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nice TJ, Coscoy L, Raulet DH. Posttranslational regulation of the NKG2D ligand Mult1 in response to cell stress. J Exp Med. 2009;206:287–298. doi: 10.1084/jem.20081335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK- cell lysis. Blood. 2007;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Rabinovich BA, Hurren R, Cosman D, Miller RG. Survival versus neglect: redefining thymocyte subsets based on expression of NKG2D ligand(s) and MHC class I. Eur J Immunol. 2005;35:439–448. doi: 10.1002/eji.200425621. [DOI] [PubMed] [Google Scholar]
- 13.Rabinovich BA, Shannon J, Su RC, Miller RG. Stress renders T cell blasts sensitive to killing by activated syngeneic NK cells. J Immunol. 2000;165:2390–2397. doi: 10.4049/jimmunol.165.5.2390. [DOI] [PubMed] [Google Scholar]
- 14.Tang KF, Ren H, Cao J, et al. Decreased Dicer expression elicits DNA damage and up-regulation of MICA and MICB. J Cell Biol. 2008;182:233–239. doi: 10.1083/jcb.200801169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soriani A, Zingoni A, Cerboni C, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 16.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlyle JR, Mesci A, Fine JH, et al. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 19.Plougastel B, Dubbelde C, Yokoyama WM. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics. 2001;53:209–214. doi: 10.1007/s002510100319. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H, Kartsogiannis V, Hu YS, et al. A novel osteoblast-derived C-type lectin that inhibits osteoclast formation. J Biol Chem. 2001;276:14916–14923. doi: 10.1074/jbc.M011554200. [DOI] [PubMed] [Google Scholar]
- 21.Voigt S, Mesci A, Ettinger J, et al. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, Zuniga-Pflucker JC. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol. 1999;162:5917–5923. [PubMed] [Google Scholar]
- 23.Cerwenka A, Bakker AB, McClanahan T, et al. Retinoic acid early in-ducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 25.LaBelle JL, Truitt RL. Characterization of a murine NKT cell tumor previously described as an acute myelogenous leukemia. Leuk Lymphoma. 2002;43:1637–1644. doi: 10.1080/1042819021000002974. [DOI] [PubMed] [Google Scholar]
- 26.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 27.Ryan JC, Niemi EC, Nakamura MC, Seaman WE. NKR-P1A is a target-specific receptor that activates natural killer cell cytotoxicity. J Exp Med. 1995;181:1911–1915. doi: 10.1084/jem.181.5.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlyle JR, Mesci A, Ljutic B, et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol. 2006;176:7511–7524. doi: 10.4049/jimmunol.176.12.7511. [DOI] [PubMed] [Google Scholar]
- 29.Aust JG, Gays F, Mickiewicz KM, Buchanan E, Brooks CG. The expression and function of the NKRP1 receptor family in C57BL/6 mice. J Immunol. 2009;183:106–116. doi: 10.4049/jimmunol.0804281. [DOI] [PubMed] [Google Scholar]
- 30.Block WD, Merkle D, Meek K, Lees-Miller SP. Selective inhibition of the DNA-dependent protein kinase (DNA-PK) by the radiosensitizing agent caffeine. Nucleic Acids Res. 2004;32:1967–1972. doi: 10.1093/nar/gkh508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez D. Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J Biol Chem. 2003;278:37139–37145. doi: 10.1074/jbc.M307088200. [DOI] [PubMed] [Google Scholar]
- 32.He Z, Ma WY, Hashimoto T, Bode AM, Yang CS, Dong Z. Induction of apoptosis by caffeine is mediated by the p53, Bax, and caspase 3 pathways. Cancer Res. 2003;63:4396–4401. [PubMed] [Google Scholar]
- 33.Gabrielli B, Chau YQ, Giles N, Harding A, Stevens F, Beamish H. Caffeine promotes apoptosis in mitotic spindle checkpoint-arrested cells. J Biol Chem. 2007;282:6954–6964. doi: 10.1074/jbc.M610104200. [DOI] [PubMed] [Google Scholar]
- 34.Komarov PG, Komarova EA, Kondratov RV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 35.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 36.Sarkaria JN, Busby EC, Tibbetts RS, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 37.Foukas LC, Daniele N, Ktori C, Anderson KE, Jensen J, Shepherd PR. Direct effects of caffeine and theophylline on p110 δ and other phosphoinositide 3-kinases. Differential effects on lipid kinase and protein kinase activities. J Biol Chem. 2002;277:37124–37130. doi: 10.1074/jbc.M202101200. [DOI] [PubMed] [Google Scholar]
- 38.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 39.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155:1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coscoy L, Ganem D. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J Clin Invest. 2001;107:1599–1606. doi: 10.1172/JCI12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas M, Boname JM, Field S, et al. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 43.Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. Rapid deu-biquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- 44.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 45.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo MH, Rosenke K, Czornak K, Fortunato EA. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J Virol. 2007;81:1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiebusch L, Neuwirth A, Grabenhenrich L, Voigt S, Hagemeier C. Cell cycle-independent expression of immediate-early gene 3 results in G1 and G2 arrest in murine cytomegalovirus-infected cells. J Virol. 2008;82:10188–10198. doi: 10.1128/JVI.01212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stern-Ginossar N, Gur C, Biton M, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.