Abstract
Background. Histologic evaluation of baseline kidney biopsies is an inconsistent tool to predict graft outcomes, which might be assisted by gene expression analysis.
Methods. We evaluated 49 consecutive kidney graft biopsies obtained post-reperfusion in 18 deceased donors (DD) and 31 living donors (LD) at our center. Biopsies were evaluated and scored using Banff criteria. Low-density real-time polymerase chain reaction arrays were used to measure intragraft expression of 95 genes associated with programmed cell death, fibrosis, innate and adaptive immunity and oxidative stress signaling. A pool of 25 normal kidney biopsies was used as control. We applied a stepwise forward selection procedure to build a multiple regression model predicting estimated glomerular filtration rate (eGFR) at 1 year after transplant using baseline clinical characteristics and gene expression levels.
Results. DD grafts displayed a pattern of gene expression remarkably different from LD, including an increased expression of complement protein C3, and chemokines, CXCL1 and CXCL2, consistent with the proinflammatory setting of ischaemia–reperfusion injury. There was no association between any of the reperfusion biopsy histological features and either renal function at 1 year post-transplant or risk of acute rejection. Conversely, older donor age (R2 = 0.17, P < 0.001) and higher integrin β2 gene expression levels (incremental R2 versus Donor Age-only model = 0.23, P < 0.001) jointly predicted lower eGFR at 1 year after transplant (multiple regression R2 = 0.40). Patients with higher ITGβ2 expression levels in baseline biopsies showed lower eGFR, higher levels of proteinuria and more transplant glomerulopathy on the 1-year per-protocol biopsies.
Conclusion. ITGβ2 gene expression in reperfusion biopsies may represent a prognostic marker for kidney transplant recipients, potentially helpful in shaping patients’ treatment. Further studies are needed to confirm our findings.
Keywords: gene expression, integrin β2, kidney transplant, predictor, reperfusion biopsy
Introduction
At the time of organ donation, histological evaluation of the kidney has been used to assess the quality of a donor graft and often used as a predictor of graft outcome. However, correlation between morphologic parameters in reperfusion biopsies and graft outcomes has not been a uniform finding [1]. While graft survival has been associated with the degree of glomerulosclerosis [2,3], stronger correlations between interstitial scarring or vascular changes and outcome have been demonstrated [4,5]. Thus, there is growing interest in the identification of potential predictors of allograft function, not only to allow for personalization of clinical management but also to provide an opportunity to identify new targets for more specific immunosuppressive therapies [6].
Gene expression analysis of reperfusion biopsies has been proposed as an instrument to better predict graft function than histological evaluation [7]. Indeed, altered expression of genes encoding for mediators of inflammatory and immune responses have been implicated in allograft injury and may precede histological abnormalities [7,8]. Moreover, analysing gene expression is objective and quantitative and may provide further insights on the pathogenesis of graft injury [7]. Gene microarrays have been increasingly utilized in transplantation, allowing for the evaluation of thousands of genes simultaneously [9]. Through complex statistical analyses, this technique has defined patterns of gene expression in different transplant conditions, such as acute rejection [10] or delayed graft function [8]. However, it has yet to identify single genes that predict graft outcome. An alternative approach is to identify smaller groups of genes potentially implicated in graft outcome on the basis of available evidence and to evaluate the predictive value of the expression levels of each single gene by real-time polymerase chain reaction (RT-PCR) [9]. When comparing microarray versus RT-PCR assessment of gene expression in renal allograft biopsies, both techniques gave similar results in abnormal kidneys, but RT-PCR was more sensitive for small changes of gene expression when no clear pathological changes were seen [9]. Of note, RT-PCR is also a less expensive technique than microarray; thus, it could be more easily implemented on a wider, clinical scale.
To begin to identify potential gene signatures associated with allograft preservation and their potential impact on allograft outcomes, we used a RT-PCR approach with a customized low-density array (LDA) of 95 genes. These gene primer/probe sets were chosen to evaluate injury and repair signals associated with organ procurement and reperfusion including genes associated with programmed cell death, fibrosis, innate and adaptive immunity and oxidative stress signaling. Not unexpectedly, there was substantial difference in transcript signatures between deceased and living kidney donors, highlighting the significant cellular activation and stress during procurement. Interestingly, these biopsies showed relatively little ischaemic damage or other histological features of injury. To further test the role of RT-PCR using LDA, we also analysed the relationship of transcripts and allograft outcomes and identified associations with changes in allograft function. Thus, while histological features of acute and chronic injury may be absent, RT-PCR analysis of reperfusion biopsies demonstrates significant cellular activity at a molecular level.
Materials and methods
Patients and biopsy acquisition
All recipients of a first kidney graft referred to the renal transplant center of the National Institutes of Health (NIH) from December 2002 to April 2007 and providing written informed consent both to the reperfusion biopsy procedure and to the genetic and histological analyses performed on the kidney samples were included in the study. Reperfusion biopsies (16-gauge) were obtained 30–60 min after renal transplantation and revascularization of living (n = 34) or deceased (n = 20) allografts. All deceased donors met standard criteria and were not considered extended. Biopsies from 25 control kidneys were obtained from living renal donors during open nephrectomy (‘normal kidney pool’). Normal renal cortical tissue was obtained before renal extirpation or vascular cross-clamp.
Immunosuppressive therapy consisted of induction with lymphocyte-depleting antibodies (alemtuzumab or rabbit antithymocyte globulin) or non-depleting CD25-specific antibodies (basiliximab or daclizumab) followed by maintenance treatment with a calcineurin inhibitor (cyclosporine or tacrolimus), mycophenolate mofetil and/or sirolimus and/or prednisone. Target cyclosporine and tacrolimus levels were 150–200 and 5–12 ng/mL during the first year after transplantation, respectively. Target sirolimus levels were 8–15 ng/mL during the same period after transplantation.
After discharge, patients were regularly followed up at the NIH renal transplant outpatient clinic. Individuals with acutely increased serum creatinine levels of 15% or more above baseline without other causes of graft dysfunction (e.g. urinary tract infection, obstructive uropathy) or individuals with proteinuria >1 g per day) underwent percutaneous graft biopsy. If rejection was present on the biopsy, it was treated with bolus corticosteroids (500 mg methylprednisolone) for 3 days then a corticosteroid taper. Glomerular filtration rate (eGFR) at 1 year after transplant was estimated according with the simplified Modification of Diet in Renal Disease (MDRD) formula [11]. In the majority of recipients (85%), surveillance biopsies were performed in the context of protocol requirements.
Reperfusion biopsy preparation
Biopsy cores were frozen in liquid nitrogen within 1 min from procurement. Biopsy samples for RNA extraction were kept at −80°C or on dry ice until processing [12]. For total cellular RNA extraction, samples were homogenized (Power Gen 35 homogenizer) in 780 µl Trizol reagent (Life Technologies, Grand Island, NY) and 20 µl Glycogen (Roche, Mannheimer, Germany) and incubated for 5 min on ice. Samples were extracted with chloroform and spun at 14,000 g in a microfuge for 5 min. Samples were then precipitated with isopropanol and washed with 70% ethanol. Total cellular total RNA (2.5 µg) was resuspended in 30 µl diethylpyrocarbonate (DEPC)-treated dH2O and converted to cDNA with random hexamers for 60 min at 37°C according to manufacturer’s instruction (Roche AMV kit, random primers).
All biopsies for histology were fixed in buffered formalin, and 5 µm paraffin sections were stained with haematoxylin–eosin and periodic acid–Schiff (PAS) and scored according to Banff criteria [13]. Biopsy findings were classified as glomerulitis, interstitial inflammation, tubulitis, intimal arteritis, glomerulopathy, interstitial fibrosis, tubular atrophy, fibrous intimal thickening, arteriolar hyaline thickening and mesangial matrix increase. Changes in each of these parameters were scored from 0 to 3. Additional sections were pretreated for 10 min with phosphate-buffered saline (PBS) containing 0.03% H2O2 and stained with monoclonal antibodies (mAb) for C4d, leukocyte common antigen (LCA), total (CD3), CD4 and CD8 T cells as we have previously described [14]. A transplant pathologist evaluated all patient biopsies in a masked fashion. Infiltrating cell phenotype was semiquantitatively scored from 0 to 6 based on immunohistochemical staining in the renal cortex, as previously described [14,15]. In this schema, scoring is based on evaluation of the largest collection and most severe involvement, in cortical areas, where 0 is no cells seen, and 6 represents confluence of positive cells involving the full width of the biopsy or >1 mm of biopsy length.
Quantification of transcripts by real-time polymerase chain reaction
cDNA samples (200 ng) from each biopsy were run on low-density arrays cards containing primers-and-probe sets for 95 targets chosen based on potential relevance to the study of allograft biology (Applied Biosystems, Foster City, CA) (Table 1). 18S was used as the house-keeping gene, as its expression does not change in the clinical conditions under evaluation [15]. Samples were run in duplicates and compared with a pooled sample of cDNA in the normal kidney pool as described above. In 37 patients with available cDNA, single PCRs for CD11a (ITGAL), CD11b (ITGAM) and CD11c (ITGAX) genes were performed (all from Applied Biosystems). cDNA (10 ng) was diluted in a total volume of 25 µl of RT-PCR reaction buffer with 2.5 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphate, 0.5 µM of each target or external control (18s) 3′ and 5′ primer pair and 3 U of Taq DNA polymerase (Gibco-BRL, Carlsbad, CA).
Table 1.
Gene targets in low-density array cards
| Gene name | Gene symbol | Gene name | Gene symbol |
|---|---|---|---|
| Actinin, alpha 4 | ACTN4 | Hypoxia-inducible factor 1, alpha subunit | HIF1A |
| Angiogenin | ANG | Hypoxia-inducible protein 2 | HIG2 |
| Annexin A5 | ANXA5 | Heme oxygenase (decycling) 1 | HMOX1 |
| Beta-2-microglobulin | B2M | Heat shock transcription factor 1 | HSF1 |
| BCL2-associated X protein | BAX | Heat shock 27 kDa protein 2 | HSPB2 |
| B-cell CLL/lymphoma 2 | BCL2 | Intercellular adhesion molecule 1 (CD54) | ICAM1 |
| Baculoviral IAP repeat-containing 5 | BIRC5 | Interleukin 13 | IL13 |
| Complement component 3 | C3 | Interleukin 6 | IL6 |
| Complement component 4A, 4B | C4A,C4B | Interleukin 8 | IL8 |
| Calpain 1 | CAPN1 | Integrin, alpha V (vitronectin receptor, Alpha polypeptide, antigen CD51) | ITGAV |
| Caspase 3 | CASP3 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | ITGB2 |
| Catalase | CAT | Integrin, beta 6 | ITGB6 |
| Chemokine (C–C motif) ligand 2 | CCL2 | Lectin | LGALS9 |
| Chemokine (C–C motif) receptor 2 | CCR2 | Lymphotoxin alpha (TNF superfamily, member 1) | LTA |
| CD2-associated protein | CD2AP | Lymphotoxin beta (TNF superfamily, member 3) | LTB |
| CD3e molecule, epsilon (CD3-TCR complex) | CD3E | Membrane metallo-endopeptidase | MME |
| CD4 molecule | CD4 | Myeloperoxidase | MPO |
| CD40 molecule, TNF receptor superfamily member 5 | CD40 | Myocilin, trabecular meshwork inducible glucocorticoid response | MYOC |
| CD40 ligand (TNF superfamily, member 5, hyper-IgM syndrome) | CD40LG | Neutrophil cytosolic factor 1 | NCF1,NCF1B,NCF1C |
| CD68 molecule | CD68 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 1 (p105) | NFKB1 |
| CD80 molecule | CD80 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 2 (p49/p100) | NFKB2 |
| CD86 molecule | CD86 | Nitric oxide synthase 2A (inducible, hepatocytes) | NOS2A |
| CD8a molecule | CD8A | Notch homolog 1, translocation-associated | NOTCH1 |
| Cell death-inducing DFFA-like effector b | CIDEB | Neuronal pentraxin II | NPTX2 |
| Clusterin | CLU | Paired box 2 | PAX2 |
| Collagen, type IV, alpha 5 | COL4A5 | Platelet-derived growth factor beta Polypeptide | PDGFB |
| Colony stimulating factor 3 (granulocyte) | CSF3 | Podoplanin | PDPN |
| Cytotoxic T-lymphocyte-associated protein 4 | CTLA4 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and Cyclooxygenase) | PTGS2 |
| Cathepsin G | CTSG | Protein tyrosine phosphatase, receptor type, O | PTPRO |
| Cortactin | CTTN | S100 calcium binding protein B | S100B |
| Chemokine (C-X3-C motif) receptor 1 | CX3CR1 | Selectin P | SELP |
| Chemokine (C-X-C motif) ligand 1 | CXCL1 | Superoxide dismutase 2, mitochondrial | SOD2 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | Superoxide dismutase 3, extracellular | SOD3 |
| Chemokine (C-X-C motif) ligand 11 | CXCL11 | Synaptopodin | SYNPO |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | Transcription factor 21 | TCF21 |
| Chemokine (C-X-C motif) receptor 3 | CXCR3 | Toll-like receptor 2 | TLR2 |
| Desmin | DES | Toll-like receptor 4 | TLR4 |
| DNA fragmentation factor, 45 kDa, alpha polypeptide | DFFA | Tumour necrosis factor (TNF superfamily, member 2) | TNF |
| Endothelin converting enzyme 1 | ECE1 | Tumour necrosis factor receptor superfamily, member 1A | TNFRSF1A |
| Endothelin 1 | EDN1 | Tumour necrosis factor (ligand) superfamily, member 10 | TNFSF10 |
| Erythropoietin | EPO | Tumour necrosis factor (ligand) superfamily, member 12–13 | TNFSF12–TNFSF13 |
| Fas | FAS | Tumour protein p53 | TP53 |
| Fas ligand (TNF Superfamily, member 6) | FASLG | Translocator protein (18 kDa) | TSPO |
| Fibronectin 1 | FN1 | Vascular endothelial growth factor A | VEGFA |
| GATA binding protein 3 | GATA3 | Villin 2 | VIL2 |
| Growth hormone receptor | GHR | Vimentin | VIM |
| Glutathione peroxidase 1 | GPX1 | Wilms tumour 1 | WT1 |
| Hepatocyte growth factor | HGF |
Normal kidney samples were obtained from another group of 25 living renal donors during open nephrectomy before renal extirpation or vascular cross-clamp. The pool was prepared by combining equal amounts of cDNA from each individual within the group to establish a homogenous reference for normal kidney. RT-PCR data were analysed using Sequence Detection System version 2.2.1 software included with the ABI PRISM 7900 Sequence Detector (Applied Biosystems, Foster City, CA). Final quantification was derived using the comparative threshold cycle (Ct) method and was reported as the n-fold difference of either post-reperfusion or stable allograft biopsies relative to the pool of normal biopsies. Five samples had 18s cDNA Ct values greater than 15 and were excluded. Thus, presented data refer to the 31 living donor and the 18 deceased donor recipients with cDNA levels lower than 15. All data calculations were performed with Microsoft Excel software (Microsoft, Redmond, WA).
Sample size estimation
We estimated that 50 patients would achieve about 85% power to find as statistically significant a correlation coefficient of 0.40 (i.e. the R2 being 0.16) between the baseline predictor and eGFR at 1-year post-transplant, using a two-sided alpha level of 0.05. The correlation coefficient is identical to the regression coefficient whenever both the dependent and independent variables are expressed as standard deviation units. We also estimated that 50 patients (25 per group) achieve about 80% power to detect an effect size of 0.8 for the comparison of gene expression between deceased and living donor grafts using a two-sided (uncorrected) alpha level of 0.05. An effect size of 0.8 means that the two populations being compared have almost half (exactly 47.4%) of their combined areas which is not overlapped and that the R2 (i.e. the proportion of variance accounted for by group membership) is about 0.14 [16]. For the purpose of achieving a sample size of 50 patients, we included all patients with reperfusion biopsies that had not been previously studied [12]. However, five patients could not be considered for the analyses, as they had 18s cDNA Ct values greater than 15, indicating unreliable quality of the RNA. This reduced the number of patients from 54 to 49. Moreover, due to the higher rate of transplants from living donors performed at the NIH, the final numbers of patients in two cohorts were unbalanced, which reduced the power of the study.
Statistical analyses
Expression levels of the 95 genes present in the microarray cards, along with ITGAL, ITGAM and ITGAX genes, were transformed using logarithm to base 10 before data analysis. Two-tailed P-values for the difference in gene expression level between living and cadaveric kidneys were adjusted with the Bonferroni–Holm method to allow for multiple comparisons.
Two-sample differences in continuous variables were examined by t-test or Mann–Whitney test whenever appropriate; differences in categorical variables were examined by Fisher’s exact test. A two-tailed P-value of less than 0.05 was regarded as statistically significant.
We aimed at generating a regression model which could predict eGFR at 1 year post-transplant using baseline patients’ characteristics and gene expression levels. All statistically significant variables from the univariate analysis were selected in a stepwise forward selection procedure to build a multiple regression model, which we carefully tested for goodness of fit and model assumptions [17]. The following demographic, clinical and laboratory variables were considered for univariate analysis in the 47 patients with complete data: donors’ and recipients’ ages and gender, presence of diabetes, immunosuppressive protocol, occurrence of acute rejection, presence of acute tubular necrosis (ATN) and Banff score of reperfusion biopsies and expression levels of the 95 genes present in the microarray cards, along with ITGAL, ITGAM and ITGAX genes.
Lacking new patient data to perform ‘external validation’ of the model, we used internal validation to obtain an honest estimate of performance for patients that are similar to those in the development sample. We used the bootstrap method for internal validation of the multiple regression model [18]. With the bootstrap method model, validation is evaluated by examining the difference between the bootstrap samples and original sample concerning which variables are retained by stepwise selection, through goodness-of-fit measures such as the R2 and the calibration curve (i.e. the regression line of observed versus predicted eGFR) and by the area under the receiving operating characteristics curve (AUC, also known as the c statistic or c index) which is a measure of model discrimination [18]. The AUC is computed after having separated patients into two classes according to whether their observed eGFR 1 year post-transplant is below or above a pre-specified value (e.g. 50 mL/min/1.73 m2) and is equivalent to the probability that the predicted eGFR is higher in patients with higher observed eGFR (e.g. ≥50 mL/min/1.73 m2) compared with patients with lower observed eGFR (e.g. below 50 mL/min/1.73 m2). To obtain stable results, the bootstrap procedure was repeated 1000 times, and the differences between each of the 1000 bootstrap samples and original sample were averaged [18]. The Bootstrap internal validation procedure was performed with validate.ols program (Validation of an Ordinary Linear Model) from the Design library using the R package 2.9.2 (2009, R Development Core Team, URL http://www.R-project.org). We used Stata Release 11 software (2009; StataCorp, College Station, TX, USA) for all the remaining analyses.
Results
Baseline patient characteristics
Table 2 shows baseline recipients’ and donors’ characteristics, both in the whole study group and in the two cohorts of recipients of deceased and living donor kidneys. Recipients’ age and gender distribution were similar between the two groups, as well as the causes of end-stage renal disease (ESRD). Panel reactivity antibody (PRA) was 0 for all patients. As expected, cold ischaemia time for grafts from deceased donors was significantly longer than in living donor grafts (P = 0.001; Table 2). No recipient developed delayed graft function, defined as dialysis requirement within the first week after transplant, which prevented any analysis on the impact of histology lesions or gene expression levels on the risk of developing this complication.
Table 2.
Baseline patient characteristics
| Overall (n = 49) | Deceased donor | Living donor | |
|---|---|---|---|
| Recipients | Recipients | ||
| (n = 18) | (n = 31) | ||
| Age (years) | 44.8 ± 15.1 | 48.6 ± 16.9 | 42.6 ± 13.7 |
| Male gender (n) | 34 | 12 | 22 |
| Presence of diabetes (n) | 6 | 4 | 2 |
| Cause of end-stage renal disease (n) | |||
| Glomerulonephritis | 18 | 8 | 10 |
| Diabetes mellitus | 6 | 3 | 3 |
| Pyelonephritis or interstitial nephritis | 6 | 3 | 3 |
| Polycystic kidney disease | 10 | 2 | 8 |
| Other | 6 | 1 | 5 |
| Unknown | 3 | 1 | 2 |
| Donor age (years) | 39.4 ± 11.6 | 35.2 ± 12.9 | 41.8 ± 10.3 |
| Donor male gender (n) | 24 | 11 | 13 |
| Cold ischaemia time (h) | 8.0 ± 9.6 | 16.6 ± 8.4* | 1.2 ± 0.6 |
| HLA Mismatches | 3.8 ± 1.4 | 4.1 ± 1.1 | 3.4 ± 1.5 |
Continuous variables are presented as mean ± standard deviation.
*P < 0.05, **P < 0.001 for the comparison between deceased and living donor recipients.
Histological findings
All kidney biopsies appeared substantially normal (Table 3). Glomerular sclerosis was negligible in all the samples (1.6 ± 5.2% versus 3.4 ± 7.2% in deceased versus living donor graft, respectively). Leukocyte infiltrates were rare and not different between deceased and living donor kidneys. Vascular changes were not present, nor were there significant interstitial abnormalities. Acute tubular injury resulting after ischaemic injury was noted in biopsies from eight (44%) deceased and two (6%) living donors (P = 0.003), though severity of these changes was mild. Thus, in our center, donor biopsies did not reveal significant pathological injury or abnormality.
Table 3.
Banff and immunohistochemical scoring of reperfusion biopsies and acute rejection episodes
| Overall (N = 49) | Deceased donor recipients (n = 18) | Living donor recipients (n = 31) | |
|---|---|---|---|
| Reperfusion biopsies | |||
| Banff scoring | |||
| Glomerulitis (g) | 0.16 ± 0.37 | 0.11 ± 0.33 | 0.19 ± 0.43 |
| Interstitial inflammation (i) | 0.32 ± 0.56 | 0.06 ± 0.25 | 0.47 ± 0.60 |
| Tubulitis (t) | 0.11 ± 0.31 | 0.0 ± 0.0 | 0.20 ± 0.39 |
| Intimal arteritis (v) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Glomerulopathy (cg) | 0.20 ± 0.40 | 0.25 ± 0.46 | 0.17 ± 0.39 |
| Interstitial fibrosis (ci) | 0.15 ± 0.36 | 0.06 ± 0.25 | 0.20 ± 0.26 |
| Tubular atrophy (ct) | 0.19 ± 0.40 | 0.06 ± 0.25 | 0.20 ± 0.14 |
| Fibro-intimal thickening (cv) | 0.09 ± 0.28 | 0.12 ± 0.33 | 0.07 ± 0.26 |
| Arteriolar hyalinosis (ah) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Mesangial matrix increase (mm) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Immunoperioxidase staininga | |||
| LCA | 1.71 ± 0.76 | 1.94 ± 0.70 | 1.59 ± 0.74 |
| CD3 | 1.73 ± 0.75 | 1.81 ± 0.67 | 1.69 ± 0.75 |
| CD4 | 2.02 ± 0.66 | 2.06 ± 0.70 | 2.00 ± 0.43 |
| CD8 | 1.73 ± 0.62 | 1.69 ± 0.70 | 1.76 ± 0.51 |
| Acute rejection episodes | |||
| Banff scoring | |||
| Borderline | 2 | 0 | 2 |
| 1A | 9 | 5 | 4 |
| 2A | 1 | 1 | 0 |
| 1B | 3 | 2 | 1 |
Continuous variables are reported as mean ± standard deviation.
Immunoperoxidase staining maximum score is 6; for details, see text.
*P < 0.05, **P < 0.001 for the comparison between deceased and living donor recipients (there were no statistically significant differences).
Transcriptional profiles in deceased and living donor grafts
Compared with normal kidney tissue, 21 of 95 genes were differentially expressed in living donor kidney allografts by at least 2-fold compared with 30 genes in deceased donor biopsies, despite the absence of significant cold ischaemia. Not surprisingly, when comparing living versus deceased donor kidneys, 13 of the 95 genes initially assayed were differentially expressed. The details of these results are shown in Figure 1. In living donors, apoptotic gene transcripts CAPN1 and CIDEB were both significantly suppressed compared with normal kidney and differed significantly from deceased donor grafts (Figure 1A). These results indicate a substantial regulatory response to structural injury due to hypoxia that is better preserved in living kidneys than deceased donor allograft.
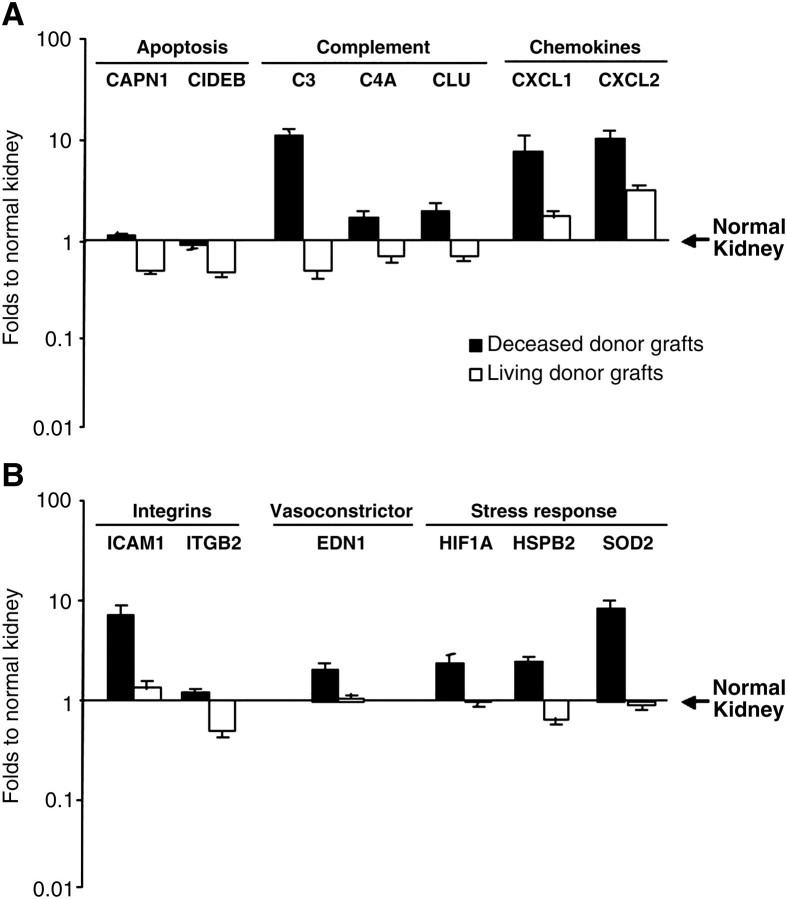
Fig. 1.
Expression data of the 13 genes whose levels resulted significantly different (P < 0.05 after Bonferroni–Holm adjustment for multiple tests) between deceased and living donor grafts within the pool of 95 genes initially tested. Results are expressed relative to normal kidneys. Genes involved in apoptosis and those encoding for complement proteins and chemokines are showed in panel A. Integrin, vasoconstrictor, and stress response genes are showed in panel B.
In spite of the absence of significant histological evidence of inflammation, deceased donor kidneys had significantly higher transcript levels for chemokines CXCL1 (7.6 ± 3.0 versus 1.7 ± 0.2-fold; P < 0.001) produced by macrophages and neutrophils and the neutrophil chemoattractant CXCL2 (10.2 ± 2.1 versus 3.1 ± 0.3; P < 0.001) compared with living donor grafts (Figure 1A). This expression was paralleled by increased expression of integrin beta 2 (ITGβ2), a leukocyte associated integrin (1.2 ± 0.1 versus 0.5 ± 0.1, respectively; P < 0.001), and ICAM-1 typically expressed on endothelial surfaces (6.6 ± 1.5 versus 1.3 ± 0.2-fold; P < 0.001) (Figure 1B). Finally, innate immune activation was also up-regulated in deceased grafts (Figure 1A) with significantly higher expression of genes encoding for complement proteins of the alternative (C3; 10.8 ± 1.9-fold versus 0.5 ± 0.1-fold; P < 0.001) and classical (C4a; 1.7 ± 0.2-fold versus 0.7 ± 0.1-fold; P < 0.001) activation pathways as well as clusterin (CLU; 1.9 ± 0.4 versus 0.7 ± 0.1-fold; P < 0.001), a lipoprotein with complement inhibitory activity. Thus, deceased donor grafts, even in the absence of histological features of tubular necrosis and death and inflammation, have marked transcriptional profiles of acute injury.
The up-regulation of genes involved in the inflammatory response in deceased donor grafts was further accompanied by significant induction of transcripts for stress response compared with living kidney donor grafts (Figure 1B). These genes included hypoxia-inducible factor 1 (2.3 ± 0.4 versus 0.9 ± 0.1-fold; P < 0.001), heat shock protein beta 2 (2.4 ± 0.3 versus 0.7 ± 0.1-fold; P < 0.001) and superoxide dismutase (6.0 ± 1.0 versus 0.9 ± 0.1-fold; P < 0.001). Not unexpectedly, these grafts also showed significantly higher expression levels of endothelin-1, a vasoconstrictive protein (P < 0.001) (Figure 1B). These profiles again underlie the severity of molecular and biochemical events that occur in deceased donor allografts, even in the absence of histological features.
Correlations of reperfusion biopsy transcriptional profiles with subsequent outcomes
With such substantial expression in the absence of histological features of injury, we hypothesized that these early molecular events might predict subsequent clinical outcomes and, in particular, affect the development of the alloimmune response and allograft function. Of 49 recipients, 15 developed an acute rejection episode during the first year of follow-up on protocol immunosuppression (Table 3). Incidence and severity of acute rejection episodes were similar between recipients of deceased and living donor grafts. All patients responded to steroid therapy and did not require any polyclonal or monoclonal antibody therapy. We analysed histological features and the gene transcript profile in the reperfusion biopsies, comparing those recipients with rejection and those without. We did not find associations between any of the reperfusion biopsy histological variables and the risk of acute rejection. Also, the majority of transcripts were not different in reperfusion biopsy transcript profile between rejectors and non-rejectors. However, in patients who subsequently experienced acute rejection episodes, reperfusion biopsy expression of CD152 (0.43 ± 0.16-fold), a negative regulator of T-cell function, was remarkably although not significantly depressed compared with non-rejectors (3.78 ± 1.68-fold), as well as NOS2A (0.31 ± 0.1-fold versus 0.87 ± 0.22-fold, respectively), the inducible form of nitric oxide synthase. These results suggest that higher expression of negative regulatory molecules may exert a protective effect toward rejection.
eGFR at 1 year after transplant in the whole cohort of patients was 52.1 ± 13.0 ml/min/1.73 m2. Not unexpectedly, deceased donor graft tended to have lower eGFR levels than living donor graft (46.7 ± 14.5 versus 55.2 ± 11.2 ml/min/1.73 m2, P = 0.029).
Table 4 shows the various genes and the clinical characteristics which were significantly correlated with graft function at 1 year after transplant in univariate analysis. However, on multivariate analysis, only donor age (−5.43 mL/min decrease in eGFR per 10-year increase in donor age; R2 = 0.17) and level of ITGβ2 gene (−21.60 mL/min decrease in eGFR per 10-fold increase in gene expression; incremental R2 versus Donor Age-only model=0.23) were independent predictors of eGFR at 1 year (multiple regression R2 = 0.40). The final multiple regression equation was the following:
Table 4.
Variables that resulted as significantly associated to eGFR at univariate and multivariate analysis
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| β | 95% CI | P-value | β | 95% CI | P-value | |
| Living donor | 8.56 | 0.92 to 16.21 | 0.029 | – | – | – |
| Donor age | −4.66 | −7.73 to −1.59 | 0.004 | −5.43 | −8.10 to −2.77 | <0.001 |
| CAPN1 | −14.49 | −28.11 to −0.87 | 0.038 | – | – | – |
| CXCL1 | −11.56 | −20.25 to −2.86 | 0.010 | – | – | – |
| ICAM1 | −8.52 | −15.71 to −1.32 | 0.021 | – | – | – |
| ITGB2 | −18.55 | −30.67 to −6.42 | 0.004 | −21.60 | −32.14 to −11.07 | <0.001 |
| MME | 12.33 | 3.04 to 21.63 | 0.010 | – | – | – |
β, regression coefficient; CI, confidence interval. The regression coefficient for Donor Age is the difference in eGFR per 10-year increase in donor age. As to gene expression level variables (which were transformed using logarithm to base 10 before analyses), the regression coefficient is the difference in eGFR per 10-fold increase in gene expression. None of the continuous variables showed a significant non-linear trend. The final multiple regression equation was the following:
47.77 − 5.43 * [(Donor Age − 39.4)/10] − 21.60 * ITGβ2.
eGFR 1 year post-transplant = 47.77 − 5.43 * [(Donor Age − 39.4)/10] − 21.60 * ITGβ2.
After having artificially dichotomized the observed eGFR into values below and greater than or equal to 50 mL/min/1.73 m2 and into values below and greater than or equal to 30 mL/min/1.73 m2, the AUC for the prediction of eGFR ≥ 50 mL/min/1.72 m2 was 0.85 (i.e. good discrimination), the AUC for the prediction of eGFR ≥ 30 mL/min/1.73 m2 was 0.92, demonstrating excellent discrimination for this range of eGFR values.
The above multiple regression equation showed evidence of good internal validity. In fact, fitting the multiple regression model to each bootstrap sample, donor age and ITGβ2 was statistically significant (P < 0.05) in 984 of the 1000 samples (98.4%). The multiple regression model which included the variables donor age and ITGβ2 gene demonstrated low bias for the calibration curves (i.e. the regression line in the plot of observed versus predicted eGFR; data not shown) and for the R2 (the average difference in R2 between the original samples and bootstrap sample being +0.06) and for the AUC (the average difference in the AUC between the original samples and bootstrap sample being +0.01).
To get a deeper insight into the role of ITGβ2 in reperfusion biopsies, we stratified patients according to expression levels of this gene above (n = 12) or below (n = 37) the normal kidneys. According to this stratification, patients with higher ITGβ2 levels had lower eGFR (45.8 ± 13.2 versus 55.1 ± 12.6 ml/min; P = 0.016) and higher urinary protein/creatinine ratio (0.41 ± 0.36 versus 0.17 ± 0.13; P = 0.037) than those with lower levels at 1 year after transplant. Among 42 recipients who underwent surveillance biopsy at 1 year post-transplantation, glomerular changes were more severe in those with higher ITGβ2 levels at reperfusion (Table 5).
Table 5.
Banff and immunohistochemical scoring in per-protocol biopsies at 1 year after transplant.
| Overall (N = 42) | ITGβ2 gene expression in reperfusion biopsies |
||
|---|---|---|---|
| Above normal kidneys (n = 9) | Below normal kidneys (n = 33) | ||
| Banff scoring | |||
| Glomerulitis (g) | 0.15 ± 0.57 | 0.11 ± 0.33 | 0.16 ± 0.63 |
| Interstitial inflammation (i) | 0.39 ± 0.70 | 0.56 ± 0.88 | 0.34 ± 0.65 |
| Tubulitis (t) | 0.37 ± 0.89 | 0.67 ± 1.32 | 0.28 ± 0.73 |
| Intimal arteritis (v) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Glomerulopathy (cg) | 0.17 ± 0.58 | 0.56 ± 1.01* | 0.06 ± 0.35 |
| Interstitial fibrosis (ci) | 0.37 ± 0.58 | 0.44 ± 0.53 | 0.34 ± 0.60 |
| Tubular atrophy (ct) | 0.78 ± 0.57 | 0.89 ± 0.60 | 0.75 ± 0.57 |
| Fibrous intimal thickening (cv) | 0.23 ± 0.42 | 0.22 ± 0.44 | 0.23 ± 0.43 |
| Arteriolar hyalinosis (ah) | 0.02 ± 0.15 | 0.11 ± 0.33 | 0.0 ± 0.0 |
| Mesangialmatrix increase (mm) | 0.07 ± 0.26 | 0.11 ± 0.33 | 0.06 ± 0.25 |
| Immunoperioxidase staininga | |||
| LCA | 3.54 ± 1.10 | 3.67 ± 1.12 | 3.50 ± 1.11 |
| CD3 | 3.38 ± 0.75 | 3.50 ± 0.93 | 3.35 ± 0.71 |
| CD4 | 3.50 ± 0.93 | 3.75 ± 0.89 | 3.44 ± 0.95 |
| CD8 | 2.75 ± 0.67 | 2.75 ± 0.46 | 2.75 ± 0.72 |
Continuous variables are reported as mean ± standard deviation.
aImmunoperoxidase Staining maximum score is 6; for details, see text.
*P < 0.05, **P < 0.001 for the comparison between patients with ITGβ2 levels above or below level of normal kidneys.
Expression levels of single ITGβ2 subchains
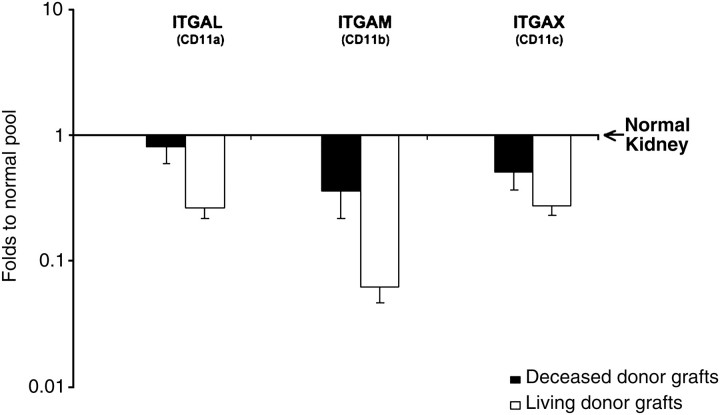
The relationship between intensity of ITGβ2 and eGFR at 1 year after transplant led to additional evaluation of integrin expression within reperfusion biopsies. Consequently, we evaluated expression of all three subchains that belong to ITGβ2 to form the family of β2 integrins. In line with ITGβ2 results, genes encoding for all three subchains composing CD11a (ITGAL), CD11b (ITGAM) and CD11c (ITGAX) genes were down-regulated in the whole cohort of grafts as compared with normal kidneys (0.19 ± 0.06, 0.37 ± 0.06, 0.50 ± 0.1, respectively). Moreover, there was numerically lower expression of all β2 integrins in living as compared with deceased donor grafts (Figure 2). When added to the multiple linear regression model, no individual β2 integrin supplanted the predictive value of donor age and overall ITGβ2 expression (data not shown).
Fig. 2.
Integrin β2 and IGTβ2 subchains (ITGAL, ITGAM, ITGAX) gene expression in deceased and living donor grafts. Results are expressed relative to normal kidneys. *P < 0.05 after Bonferroni–Holm adjustment for multiple tests (there were no statistically significant differences).
Discussion
Like previous reports, our study shows that the histological features of kidney allografts may appear virtually normal at baseline; thus, they are often of little avail in predicting graft outcome. Gene expression analysis of reperfusion biopsies may provide, instead, a useful tool for predicting graft function in renal transplant patients. In particular, the expression levels of ITGβ2, which were inversely correlated with eGFR at 1 year after transplant, emerged, along with donor age, as the best independent predictor of short-term graft function.
Not unexpectedly, reperfusion biopsies showed differential gene expression patterns according to the type of donor (deceased versus living donors). Notably, deceased donor grafts had increased apoptotic signaling and higher expression levels of genes for chemokines and integrins. This result may be a response of renal injury induced by brain death and prolonged cold ischaemia time and is in line with previous reports [19]. Intriguingly, deceased donor grafts showed also a marked increase in C3 gene expression, according with a major role of complement alternate pathway activation in the promotion of ischaemia/reperfusion (I/R) injury [20]. Indeed, C3-deficient mice are protected from damage induced by this noxious stimulus [21]. In addition, according with more recent evidence [22], we also found an up-regulation of C4a in deceased donor grafts, suggesting that also classical complement pathway may be implicated in I/R injury. This highlights the importance of complement cascade as a potential therapeutic target for kidney transplant patients, especially for recipients of organs more susceptible to I/R injury, such as those from older donor or with prolonged ischaemia times.
A major finding of the present study was that the expression level of ITGβ2 gene in baseline kidney biopsies is an independent predictor of graft function. The β2 integrins are heterodimers formed by the non-covalent binding of ITGβ2 chain with other unique subunits, such as ITGAL, ITGAM and ITGAX, which are restricted to different leukocyte subsets and act in cell–cell and cell–extracellular matrix adhesion [23]. They are restricted to leukocytes and function in both tethering and rolling of these cells on endothelium and in their firm arrest and transmigration through the vascular wall. In addition to the role in leukocyte migration, β2 integrins function in T-cell activation by stabilizing their interaction with antigen-presenting cells [23]. Importantly, the absence of β2 integrin on alloreactive T cells resulted in significantly less graft-versus-host-disease morbidity and mortality in murine allogeneic bone marrow transplantation models [24].
In line with the above evidence, we found that higher levels of ITGβ2 expression in reperfusion biopsies predicted worse eGFR at 1 year after transplant. This might be possibly related with increased leukocyte extravasation into grafts with up-regulated ITGβ2 expression levels that might therefore be more susceptible to immune and non-immune injury. Our relatively limited sample size, however, did not allow finding a significant correlation between acute rejection risk and ITGβ2 expression in reperfusion biopsies; thus, larger studies are needed to confirm this hypothesis. Similarly, the low incidence of delayed graft function in our cohort of patients made it difficult to assess whether higher ITGβ2 might result in an impaired graft function recovery. However, regardless of the mechanism involved, our present findings suggest that ITGβ2 may represent a prognostic marker potentially helpful in patients’ management. Consistently, patients with higher levels of ITGβ2 expression in reperfusion biopsies had higher proteinuria and more severe glomerular injury at 1 year per-protocol biopsies.
Conversely, gene expression levels of the other β2 integrin chains, such as ITGAL, ITGAM and ITGAX, were not significantly correlated with eGFR at 1 year. Though the small number of patients with this evaluation might account for this finding, it might also be hypothesized that expression levels of these chains has no predictive value because they are expressed by specific lymphocyte populations [23]. Alternatively, ITGβ2 is present on the membrane of all leukocytes; thus, it may provide a better picture of graft inflammatory state and its outcome.
Interestingly, our results, along with previous experimental evidence, would suggest that ITGβ2 integrin blockade might represent a valuable therapy to improve kidney graft outcomes. Induction with a mouse monoclonal antibody-targeting ITGAL (odulimomab) efficiently prevented acute rejection and remarkably (although not significantly) reduced incidence of delayed graft function over RATG in 101 renal transplant patients [25]. Subsequent to this study, a humanized form of this antibody has been successfully used to treat moderate-to-severe psoriasis [26]. Notably, a recent phase I/II randomized, multicenter trial showed that efalizumab was safe in renal transplant recipients on maintenance immunosuppression with cyclosporine, steroids and an antiproliferative agent [27]. Our findings support further studies testing this molecule as an alternative anti-rejection agent.
Donor age was the other major predictor of graft outcome. This is in line with the evidence that, even with similar histology patterns, grafts from older donors have a lower survival expectancy than younger ones [28]. Notably, despite the fact that donor age might affect gene expression pattern [29], in our study both variables predicted graft function in an independent manner, suggesting that chronological age of the kidney per se only partially accounts for transplant outcome. However, as older grafts will become increasingly more frequent in the future [>30] and are more susceptible to I/R injury than younger organs, it will be of utmost importance finding therapeutic strategies to limit the initial graft injury and, in this perspective, integrin blockade might represent a useful tool.
The study has some limitations. First, the relatively limited sample size reduces the impact of the present results. On the other hand, this is a relatively large collection of reperfusion biopsies that also includes follow-up biopsies at 1 year. Therefore, though further studies are needed to confirm these findings, our well-characterized cohort already provides important clinical information. Different immunosuppressive therapies in patients enrolled in the study might have biased the results, but lack of significant correlation between anti-rejection therapy and eGFR at 1 year suggests that the impact (if any) of this variable on graft outcome was marginal. Finally, the molecular data make biological sense as indicated by recent clinical trials blocking the integrin beta pathway.
In conclusion, ITGβ2 gene expression in reperfusion biopsy might represent a prognostic marker for kidney transplant patients potentially helpful in shaping patients’ treatment. Our data suggest that leukocyte recruitment through ITGβ2 blockade may mediate graft injury after ischaemia–reperfusion. Further studies into the blockade of this pathway may provide a novel strategy to ameliorate graft injury and improve long-term graft outcomes.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive, and Kidney Diseases of the National Institutes of Health (1Z01DK062008-05 DIR). Dr. R.B.M. is further supported by U01AI058013 and U01AI084150. The authors wish to thank Drs. Allan Kirk and David Kleiner for their clinical participation and thoughtful discussions.
Conflict of interest statement. R.B.M. is a PI for an Amgen Inc. transplant study, is a consultant for Amgen Inc. and has served on an advisory board for Genzyme Corporation. The results presented in this paper have not been published previously in whole or part, except in abstract format at the American Transplant Congress, 2008, and at the American Society of Nephrology, 2008.
References
- 1.El-Husseini A, Sabry A, Zahran A, et al. Can donor implantation renal biopsy predict long-term renal allograft outcome? Am J Nephrol. 2007;27:144–151. doi: 10.1159/000099944. [DOI] [PubMed] [Google Scholar]
- 2.Escofet X, Osman H, Griffiths DF, et al. The presence of glomerular sclerosis at time zero has a significant impact on function after cadaveric renal transplantation. Transplantation. 2003;75:344–346. doi: 10.1097/01.TP.0000044361.74625.E7. [DOI] [PubMed] [Google Scholar]
- 3.Randhawa PS, Minervini MI, Lombardero M, et al. Biopsy of marginal donor kidneys: correlation of histologic findings with graft dysfunction. Transplantation. 2000;69:1352–1357. doi: 10.1097/00007890-200004150-00024. [DOI] [PubMed] [Google Scholar]
- 4.Leunissen KM, Bosman FT, Nieman FH, et al. Amplification of the nephrotoxic effect of cyclosporine by preexistent chronic histological lesions in the kidney. Transplantation. 1989;48:590–593. [PubMed] [Google Scholar]
- 5.Seron D, Carrera M, Grino JM, et al. Relationship between donor renal interstitial surface and post-transplant function. Nephrol Dial Transplant. 1993;8:539–543. doi: 10.1093/ndt/8.6.539. [DOI] [PubMed] [Google Scholar]
- 6.Lachenbruch PA, Rosenberg AS, Bonvini E, et al. Biomarkers and surrogate endpoints in renal transplantation: present status and considerations for clinical trial design. Am J Transplant. 2004;4:451–457. doi: 10.1111/j.1600-6143.2004.00386.x. [DOI] [PubMed] [Google Scholar]
- 7.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85:626–635. doi: 10.1097/TP.0b013e318165491f. [DOI] [PubMed] [Google Scholar]
- 8.Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8:78–85. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 9.Allanach K, Mengel M, Einecke G, et al. Comparing microarray versus RT-PCR assessment of renal allograft biopsies: similar performance despite different dynamic ranges. Am J Transplant. 2008;8:1006–1015. doi: 10.1111/j.1600-6143.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 10.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 11.Levey AS GT, Kusek JW, Beck GJ. A simplified equation to predict glomerular filtration rate from serum creatinine. J Am Soc Nephrol. 2000;11 A0828. [Google Scholar]
- 12.Hoffmann SC, Kampen RL, Amur S, et al. Molecular and immunohistochemical characterization of the onset and resolution of human renal allograft ischemia-reperfusion injury. Transplantation. 2002;74:916–923. doi: 10.1097/00007890-200210150-00003. [DOI] [PubMed] [Google Scholar]
- 13.Racusen LC, Halloran PF, Solez K. Banff 2003 meeting report: new diagnostic insights and standards. Am J Transplant. 2004;4:1562–1566. doi: 10.1111/j.1600-6143.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- 14.Mannon RB, Hoffmann SC, Kampen RL, et al. Molecular evaluation of BK polyomavirus nephropathy. Am J Transplant. 2005;5:2883–2893. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann SC, Hale DA, Kleiner DE, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. Am J Transplant. 2005;5:573–581. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 16.Cohen J. Statistical power analysis in the behavioral sciences. Hillsdale, New Jersey: Laurence Erlbaum Associates; 1988. [Google Scholar]
- 17.Stata Base Reference Manual Release 11. College Station, TX, USA: Stata Press; 2009. Stata Corp. [Google Scholar]
- 18.Harrell F. Regression modeling strategies with application to linear models, logistic regression, and survival analysis. New York: Springer-Verlag; 2001. [Google Scholar]
- 19.Kainz A, Mitterbauer C, Hauser P, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4:1595–1604. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 20.Damman J, Schuurs TA, Ploeg RJ, et al. Complement and renal transplantation: from donor to recipient. Transplantation. 2008;85:923–927. doi: 10.1097/TP.0b013e3181683cf5. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Farrar CA, Abe K, et al. Predominant role for C5b-9 in renal ischemia/reperfusion injury. J Clin Invest. 2000;105:1363–1371. doi: 10.1172/JCI8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Csencsits K, Burrell BE, Lu G, et al. The classical complement pathway in transplantation: unanticipated protective effects of C1q and role in inductive antibody therapy. Am J Transplant. 2008;8:1622–1630. doi: 10.1111/j.1600-6143.2008.02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pribila JT, Quale AC, Mueller KL, et al. Integrins and T cell-mediated immunity. Annu Rev Immunol. 2004;22:157–180. doi: 10.1146/annurev.immunol.22.012703.104649. [DOI] [PubMed] [Google Scholar]
- 24.Liang Y, Liu C, Djeu JY, et al. Beta2 integrins separate graft-versus-host disease and graft-versus-leukemia effects. Blood. 2008;111:954–962. doi: 10.1182/blood-2007-05-089573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hourmant M, Bedrossian J, Durand D, et al. A randomized multicenter trial comparing leukocyte function-associated antigen-1 monoclonal antibody with rabbit antithymocyte globulin as induction treatment in first kidney transplantations. Transplantation. 1996;62:1565–1570. doi: 10.1097/00007890-199612150-00006. [DOI] [PubMed] [Google Scholar]
- 26.Hodulik S, Hadi S. Efalizumab: a biological agent for the treatment of psoriasis. Rev Recent Clin Trials. 2006;1:165–168. doi: 10.2174/157488706776876436. [DOI] [PubMed] [Google Scholar]
- 27.Vincenti F, Mendez R, Pescovitz M, et al. A phase I/II randomized open-label multicenter trial of efalizumab, a humanized anti-CD11a, anti-LFA-1 in renal transplantation. Am J Transplant. 2007;7:1770–1777. doi: 10.1111/j.1600-6143.2007.01845.x. [DOI] [PubMed] [Google Scholar]
- 28.Remuzzi G, Cravedi P, Perna A, et al. Long-term outcome of renal transplantation from older donors. N Engl J Med. 2006;354:343–352. doi: 10.1056/NEJMoa052891. [DOI] [PubMed] [Google Scholar]
- 29.Kainz A, Perco P, Mayer B, et al. Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation. 2007;83:1048–1054. doi: 10.1097/01.tp.0000259960.56786.ec. [DOI] [PubMed] [Google Scholar]
- 30.Chavalitdhamrong D, Gill J, Takemoto S, et al. Patient and graft outcomes from deceased kidney donors age 70 years and older: an analysis of the Organ Procurement Transplant Network/United Network of Organ Sharing database. Transplantation. 2008;85:1573–1579. doi: 10.1097/TP.0b013e31817059a1. [DOI] [PubMed] [Google Scholar]