Abstract
The goal of individualized drug therapy requires physicians to be able to accurately predict an individual’s response to a drug. Both genetic and environmental factors are known to influence drug response. ‘Pharmacogenetics’ is the study of the role of inheritance in variation in drug response phenotypes. Pharmacogenetics is now moving genome-wide to become ‘pharmacogenomics’, resulting in the recognition of novel biomarkers for individual variation in drug response. This article reviews the development, promise and challenges facing pharmacogenomics, using examples of drugs used to treat or prevent cardiovascular disease.
Introduction
Advances in DNA sequencing and genotyping have made it possible to rapidly and accurately identify variation in DNA sequence and structure. As a result, we can now correlate genomic variation with drug response, which helps us to predict individual variation in responses to specific drugs, to optimize drug selection and dose, and to avoid adverse effects associated with drug therapy.
In 1931, Sir Archibald Garrod outlined the basic concepts underlying genetically programmed individual variation to drugs in his book ‘Inborn Factors of Disease’.1 However, it was not until the 1950s and 1960s that there were data in support of an effect of inheritance on drug response.2,3 These data arose from observations indicating that individual variation in drug and drug metabolite concentrations in blood or urine could be related to variation in response to standard drug doses.2,3 For example, some patients treated with the muscle relaxant succinylcholine experienced prolonged paralysis and apnea as a result of genetic variation in the activity of butyrylcholinesterase, an enzyme that metabolizes this drug.4,5 Subsequently, these adverse reactions were shown to result from an atypical form of the enzyme encoded by a variant allele for the butyrylcholinesterase gene. This allele contained a 209G>A single nucleotide polymorphism (SNP) that altered the encoded amino acid (that is, a nonsynonymous SNP).6
Researchers now use both genotype-to-phenotype and phenotype-to-genotype strategies to correlate genetic variation with drug response. The genotype-to-phenotype approach consists of first determining genetic variation and then evaluating its phenotypic effects, whereas the phenotype-to-genotype approach involves beginning with a drug-dependent phenotype and then trying to link the phenotype to variation in DNA sequence or structure. The genotype-to-phenotype approach is now more commonly used because of the availability of DNA sequencing information made possible with the advent of the Human Genome Project.
Traditionally, these studies have been performed by analyzing candidate genes in candidate pathways, selected on the basis of current knowledge—that is, pharmacogenetics (Box 1). The effect of genetic variation that results in altered drug response can be classified as being either pharmacokinetic or pharmacodynamic. Pharmacokinetic effects influence the drug’s absorption, distribution, metabolism or excretion, thereby influencing the concentration of the drug that reaches target. Pharmacodynamic effects influence the drug’s target itself and/or signaling pathways downstream from the drug’s target.
Box 1 . Glossary of terms.
Types of analysis
Pharmacogenetics: analysis of candidate genes in candidate pathways (selected on the basis of current knowledge) to find genetic variation that correlates with drug response
Pharmacogenomics: analysis of DNA sequence variation across the entire genome to find genetic variation that correlates with drug response
Types of effect
Pharmacodynamic effect: influence on the drug target itself and signaling pathways downstream of the drug target
Pharmacokinetic effect: influence on the concentration of a drug reaching its target via effects on drug absorption, distribution, metabolism and excretion
The advent of genome-wide association studies has made it possible to move beyond just performing pharmacogenetic studies and has allowed us to query the effect of DNA sequence variation across the entire genome on drug response—that is, pharmacogenomics. The traditional classification of the effects of polymorphisms identified by pharmacogenetics can also be applied to genetic effects on drug response that have been identified via pharmacogenomics. Although genome-wide association studies can generate many false positive results, usually require large numbers of samples, and are currently extremely expensive,7 they promise to help dramatically increase our understanding of the contribution of inheritance to variation in drug response.
In this Review, we discuss the development of pharmacogenetics and its evolution into pharmacogenomics using examples of polymorphisms that affect the treatment and/or prevention of cardiovascular disease. The aim of this Review is not to describe all cardiovascular examples known, but to provide examples that will facilitate cardiologists’ understanding of this important field and the impact it will have on prescribing practices.
Cardiovascular pharmacogenetics
Inherited pharmacokinetic effects
First described in the early to mid 1970s, a classic example of a pharmacogenetically identified pharmacokinetic effect is that of inherited variation in the N-acetyltransferase-2-catalyzed N-acetylation of drugs, such as the vasodilator hydralazine and the antiarrhythmic agent procainamide.8 ‘Fast acetylators’ had lower plasma concentrations of these drugs and, as a result, were relatively resistant to therapy, whereas ‘slow acetylators’ had higher levels and were at risk for drug-related toxicity like lupus erythematosus.9,10
In the late 1970s, genetic variation was described for cytochrome P450, family 2, subfamily D, polypeptide 6 (CYP2D6), which is an important microsomal drug-metabolizing enzyme involved in the biotransformation of many cardiovascular drugs, including the anti-arrhythmic agents flecainide and propafenone, and the β-blocker metoprolol.11 Patients who were ‘poor metabolizers’ of CYP2D6, 5–10% of the white population, had exaggerated responses to standard doses of CYP2D6 substrate drugs, such as metoprolol.12 The CYP2D6 gene was cloned and sequenced in the late 1980s,13,14 resulting in the identification of gene deletion and nonsynonymous SNPs associated with being a poor metabolizer, and gene duplication associated with being an ‘ultrarapid metabolizer’.15 Notably, CYP2D6 gene duplication can vary from a frequency of 1% in Europe to 24% in Northern Africa.16 Indeed, striking ethnic variations in allele types and frequencies have been identified for many polymorphisms of pharmacogenetic importance.
A more-recent example of the discovery of genetic variation resulting in pharmacokinetic effects is a study of polymorphisms in cytochrome P450—in this case cytochrome P450, family 2, subfamily C, polypeptide 19 (CYP2C19)—that affect response to clopidrogel. When treated with clopidogrel, patients with reduced-function CYP2C19 alleles (30% of the study population) had lower plasma concentration of the active drug metabolite and higher risk for adverse events such as death, myocardial infarction, stroke (a 53% increase in this composite end point) and coronary stent thrombosis (hazard ratio of 3.09) compared with noncarriers,17 thus identifying a high-risk group that should be targeted with alternative antiplatelet agents.
Inherited pharmacodynamic effects
The identification of inherited variation in the concentration of drug reaching its target (that is, polymorphisms that had pharmacokinetic effects) laid the foundations for the application of pharmacogenetics in the identification of genetic effects on drug targets themselves (that is, of polymorphisms with pharmacodynamic rather than pharmacokinetic effects). Several examples of identified pharmacodynamic effects of polymorphisms involve drugs that are used to treat cardiovascular disease.
Heart failure results in over a million hospitalizations each year in the US.18 Despite advances in medical therapy, the mortality rate for patients with advanced heart failure approaches 50% at 5 years.19 The evidence is now clear that inheritance contributes to variation in response to drugs used to treat heart failure. For example, β-blockers have been a mainstay in the therapy of systolic heart failure; however, despite many clinical trials showing a distinct reduction in mortality with β-blocker therapy, the Beta-Blocker Evaluation of Survival Trial (BEST) failed to show a significant survival benefit for bucindolol treatment in patients with class 3–4 heart failure.20 Response to β-blockers is primarily mediated by an effect on the β1-adrenergic receptor encoded by ADRB1. At least 13 nonsynonymous SNPs have been described in ADBR1, one of which results in an Arg389Gly mutation, shown to have functional implications that affect outcome after β-blocker therapy; this polymorphism alters the intracellular carboxy terminus of ADRB1, which couples with the stimulatory G protein, Gs.21 The allele encoding Gly389 has a frequency of 30% in white and Chinese populations and 40% in African-Americans.22 Individuals homozygous or heterozygous for this minor allele exhibit reduced myocardial contractility in response to isoproterenol compared with patients homozygous for Arg389.23 This observation suggests that ADRB1 with Arg389 might have better coupling efficacy with Gs than the Gly389 allozyme. Bucindolol caused a greater decrease in cAMP generation in Arg389 carriers, and patients homozygous for Arg389 displayed a 38% reduction in mortality (P = 0.03) compared with placebo, whereas Gly389 carriers (heterozygous or homozygous patients) had a 10% reduction that was not statistically significant (P = 0.57). The improved survival associated with the allele encoding Arg389 was independent of race.23
Although the Arg389 allele was not shown to be a risk factor for heart failure, black individuals homozygous for Arg389 and for an inherited deletion of four amino acids in the alpha-2-C adrenergic receptor (ADRA2C), ADRA2C del322–325—which occurs ten times more frequently in African-Americans than in the white population, and is associated with impaired agonist binding and coupling of the receptor to multiple effector molecules24—had a tenfold increase in risk for the development of heart failure, compared with a fivefold increase in risk in those only homozygous for the latter variant.25 This finding illustrates the potential importance of a polygenic approach, that is, taking multiple genes into account in a pharmacogenetic analysis.
This concept was further highlighted by the discovery of a nonsynonymous SNP that generated a Gln41Leu polymorphism in the G-protein-coupled receptor kinase GRK-5—an enzyme that phosphorylates ADRB1—to result in uncoupling of the receptor from G proteins, thus desensitizing it and simulating pharmacological β-blockade.26 In African-Americans, a population in which this polymorphism is found in approximately 40% of individuals (it is rarely found in white individuals), patients with the Leu41 GRK-5 allozyme who were not treated with β-blockers had a survival benefit similar to those with the Gln41 allozyme who were treated with β-blockers. Furthermore, no survival difference was noted between individuals with the Leu41 allozyme who were and were not treated with β-blockers, which emphasizes the intrinsic β-blockade-like effect of this genetic polymorphism and its protective effect against death or time to heart transplantation.
Influence of both types of inherited effect
Both pharmacokinetic and pharmacodynamic effects should be considered for any one drug that is associated with inherited variation in a patients’ response to its administration, as some drugs have been shown to be influenced by both types of inherited effects. Warfarin is a good example of a drug affected by both inherited pharmacokinetic and pharmacodynamic effects on a patient’s response to its administration.
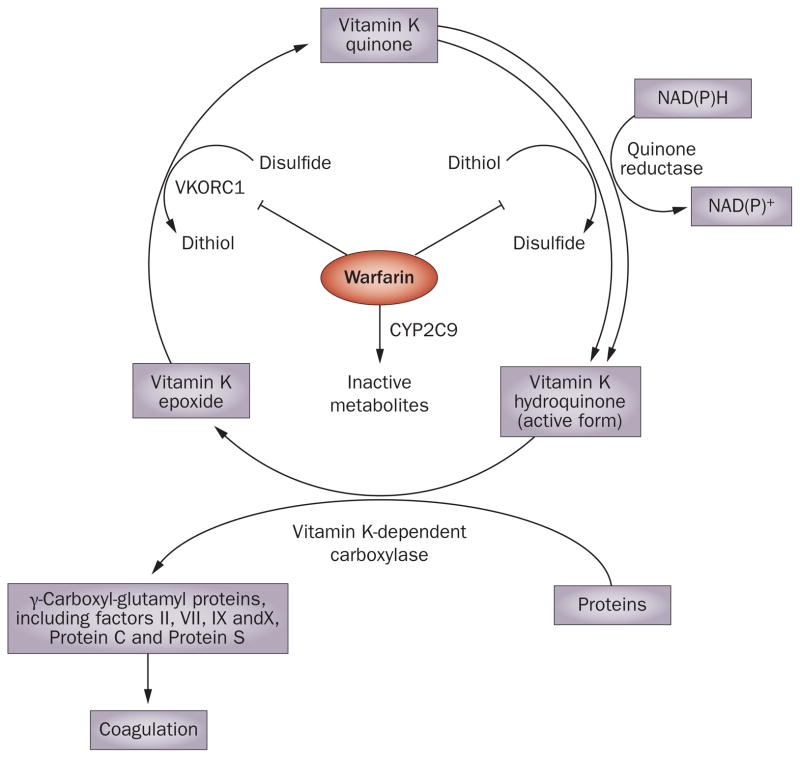
Warfarin is widely prescribed as an oral anticoagulant in patients with common cardiac disorders such as atrial fibrillation, and acts by inhibiting the vitamin K cycle required for the post-translational gamma-carboxylation of several blood coagulation factors (Figure 1). However, warfarin has a narrow therapeutic index and is one of the most common causes of emergency room visits for adverse drug reactions.27 The initial loading dose of warfarin is empiric, and patients can be either resistant or sensitive to its anticoagulation effect, as monitored by the international normalized ratio (INR).
Figure 1.
Warfarin, which is metabolized by CYP2C9, inhibits the vitamin K cycle via actions on thiol-dependent enzymes, such as vKORC1, that are required for regeneration of active vitamin K. The vitamin K cycle is essential for post-translational gamma-carboxylation of several blood coagulation factors and, therefore, coagulation. Abbreviations: vKORC1, vitamin K epoxide reductase complex 1; CYP2C9, cytochrome P450, family 2, subfamily C, polypeptide 9.
As early as the 1960s, this variability was suggested to be due, in part, to inherited differences in drug metabolism.28 Warfarin is a racemic mixture, with R and S stereoisomers. S-Warfarin is three to five times more potent than the R isomer, and is largely responsible for the therapeutic effects of the drug.29 S-Warfarin is metabolized by cytochrome P450, family 2, subfamily C, polypeptide 9 (CYP2C9), a genetically polymorphic, microsomal drug-metabolizing enzyme with common nonsynonymous SNPs that result in Arg144Cys (CYP2C9★2) and Ile358Leu (CYP2C9★3) alterations in amino acid sequence. Patients heterozygous for the CYP2C9★2 and CYP2C9★3 alleles are at increased risk for hemorrhagic complications during warfarin therapy.30 However, the SNPs in CYP2C9 account for only 6–18% of the variance in final warfarin dose. Subsequently, after the discovery of vitamin K epoxide reductase complex 1 (VKORC1), one of the thiol-dependent enzyme targets of warfarin,31 VKORC1 haplotypes were found to independently explain 15–30% of the variance in final warfarin dose required.32
The discovery of VKORC1 haplotypes demonstrates the importance of considering both inherited mutations resulting in pharmacodynamic effects for warfarin and those resulting in pharmacokinetic effects (that is, those that involve CYP2C9), with both having important roles in the identification of patients at risk for the development of INR values greater than four.33 VKORC1 was not cloned until 2004,31 which substantially limited our ability to apply pharmacogenetics to warfarin dosing. The combined use of CYP2D9 and VKORC1 genotyping—a ‘pathway approach’ that uses both pharmacokinetic and pharmacodynamic information—has clearly advanced the clinical utility of warfarin pharmacogenetics.
Genetic testing for CYP2C9★2, CYP2C9★3, and VKORC1 -1639G>A (the major SNP in the haplotype that indicates ‘sensitivity’) is now available for clinical use. However, it is important to note that clinical covariants such as age, sex, race, height, weight, tobacco use, liver disease and concomitant medication also contribute to the final warfarin dose, and these factors must be included in estimations of appropriate dose. These clinical factors account for 17–21% of the variability in warfarin dosing, while genetic variation alone accounts for 30–35% of the variability. A combination of these factors, as demonstrated in dosing algorithms, could predict up to 55% of warfarin dosing variability without measuring INR values.34 In 2009, the International Warfarin Pharmacogenetics Consortium published a report showing the utility and success of using a genotype-based approach for predicting warfarin dosing, demonstrating that a pharmacogenetic approach was better in predicting required warfarin dose than a clinical algorithm alone or a fixed dose approach.35 The greatest beneficial effect was seen among patients ultimately requiring at least 49 mg per week and those needing 21 mg or less per week, that is 46% of the population treated. As the clinical value of genotype-based dosing for warfarin remains controversial and has not been examined in a prospective manner on a large scale, the National Heart Lung and Blood Institute has initiated a large clinical trial designed to determine the potential value of the addition of genotype data to the determination of INR, particularly during the initiation of warfarin therapy.36
The example of warfarin pharmacogenetics illustrates the importance of looking for genetic variation that results in pharmacokinetic effects, and that which results in pharmacodynamic effects, when considering any one therapy.
Pharmacogenomics
As demonstrated by warfarin pharmacogenetics, a major limitation of the ‘candidate gene’ pharmacogenetic strategy is that it focuses on what we already know, but it is unable to query genes involved in pathways and mechanisms that remain to be discovered. Genome-wide association studies potentially provide an opportunity to go beyond our current knowledge by assaying hundreds of thousands of SNP markers across the entire genome. A good example of where this type of approach has been invaluable is in the treatment of patients using statins. Indeed, one of the first genome-wide association studies used to identify genetic variation that might contribute to risk for an adverse drug reaction involved statin-induced skeletal myopathy.37
Statins are routinely prescribed to millions of patients for prevention of myocardial infarction and stroke. There is large individual variation in the ability of statins to reduce LDL cholesterol, an important therapeutic end point. Individual variation in genes encoding apolipoprotein E, cholesterol ester transfer protein and lipoprotein lipase do not accurately predict response to statin therapy.38 Furthermore, known genetic variants of CYP3A4, an enzyme that catalyzes the metabolism of many statins, do not predict risk for the occurrence of adverse response such as myopathy.39
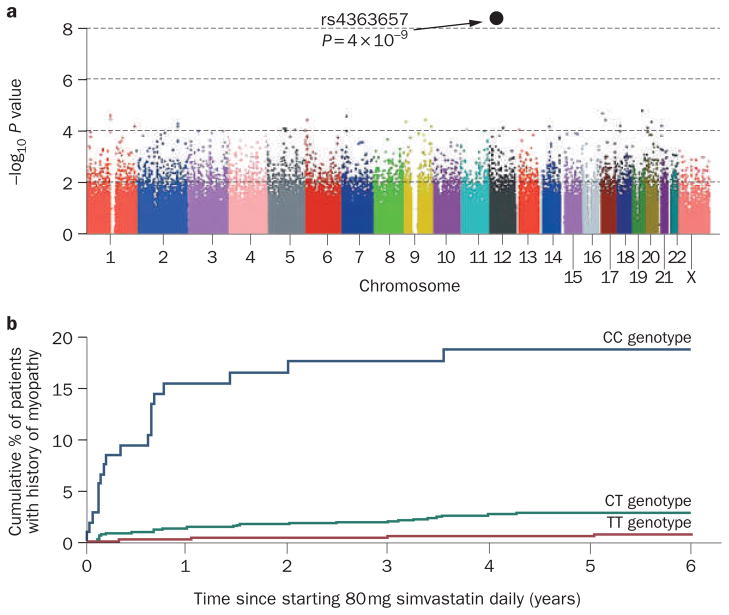
The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) was a randomized trial of high dose simvastatin.37 Patients in the SEARCH trial who developed skeletal myopathy while being treated with 80 mg simvastatin daily, and matched controls, were genotyped using a genome-wide association platform that contained approximately 300,000 SNPs scattered across the genome. A strong association with severe myopathy was seen for non-coding SNP rs4363657, a polymorphism in intron 11 of the SLCO1B1 gene on chromosome 12 (Figure 2a). The odds ratio for the development of myopathy was 16.9 among CC homozygotes and 4.5 for CT heterozygotes as compared with TT homozygotes (Figure 2b). SLCO1B1 encodes OATP1B1, a solute carrier organic transporter responsible for the active transport of statins into hepatocytes and, thus, plasma clearance and subsequent metabolism of the drug. Individuals with the C allele had elevated statin blood concentrations, probably resulting in a greater risk for myopathy. About 30% of the population in SEARCH was either heterozygous or homozygous for this SLCO1B1 allele, and this SNP could account for approximately 60% of statin-induced myopathy cases in this population. Conceivably, identification of this SNP before starting statin therapy could lead to the recommendation that these patients be prescribed water-soluble and hence more ‘muscle-safe’ statins like pravastatin and rosuvastatin. This example is almost certainly only the beginning of a series of genome-wide association studies involving drugs that are used to treat cardiovascular disease.
Figure 2.
Genome-wide association study data for statin-induced myopathy.
a | Association between myopathy and each SNP assayed during the genome-wide association study. The x axis shows the location of each SNP in the gene and the y axis is −log10 of this P value (for example, 6 is P = 10−6). b | Estimated cumulative risk of myopathy associated with 80 mg of simvastatin daily. Abbreviation: SNP, single nucleotide polymorphism. Copyright © 2008 Massachusetts Medical Society. All rights reserved. The SEARCH Collaborative Group et al. N. Engl. J. Med. 359, 789–799 (2008).37
Future directions
Pharmacogenetics and pharmacogenomics represent an important facet of attempts to individualize drug therapy in the present ‘genomic’ era. The application of genetic information during the clinical therapeutic encounter could potentially decrease costs by directing the most appropriate care to each individual patient rather than a ‘blanket approach’ based on the average response observed during clinical trials. Even though many examples of the successful application of pharmacogenetics now exist, the traditional candidate gene approach has limited us to studies of candidates that we already know. Genome-wide association studies may help to address the limitations of that approach. Genomic screens also allow for identification of multiple variants that might incrementally affect response to a particular drug at the same time. What is the foreseeable future of this exciting field?
Potential application problems
Even after the identification of a functionally important genetic variant, major regulatory, economic and ethical issues will influence the eventual clinical acceptance and application of pharmacogenomic data. There are concerns that coverage and payment decisions by insurance companies might be affected by such information. The additional value of these tests in clinical care over routine clinical practice will have to be evaluated. By not taking into account individual differences in patients and by promoting effectiveness of therapeutic strategies in groups of patients, comparative effectiveness research supported by the American Reinvestment and Recovery Act might also impede progress in the field.
Although genome-wide association studies promise to help make it possible to identify gene variations that predict risk for adverse drug reactions or lack of drug efficacy, acquiring appropriate clinical samples in large numbers, the difficulty of obtaining clearly defined phenotypes and the expense of genotyping a large number of samples, currently limits the application of this approach. Furthermore, the low frequency of certain serious adverse drug reactions may also limit its applicability.
Because of the large number of false positives that are generated when hundreds of thousands or millions of SNPs are tested, a standard quality control measure for genome-wide association studies is the requirement to replicate findings in an independent population,7 often a practically difficult and expensive process. In addition, although genome-wide association studies can point to unanticipated biological pathways or genes, it is often difficult to determine functional mechanisms responsible for the clinical effects of these pathways and/or genes. For that reason, pharmacogenomic model systems that complement clinical genome-wide association studies have been developed.
Pharmacogenomic model systems
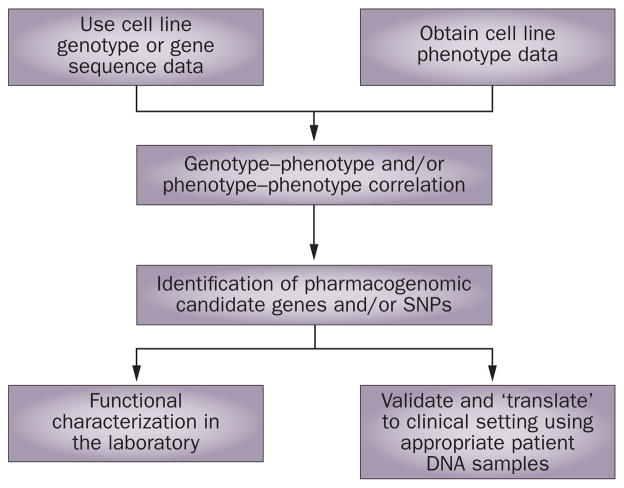
Clinical genome-wide association studies can be complemented by the use of model systems such as cell-line-based in vitro systems. One example involves Epstein-Barr-virus-transformed lymphoblastoid cell lines from hundreds of unrelated individuals of different ethnic groups, which are available through the NIH-supported Coriell Institute. Panels of these cell lines have been used as one pharmacogenomic model system. Genome-wide SNP and expression array data are available for these cell lines, so they can be used to determine drug-related phenotypes (for example, cytotoxicity) and can then be used to study the role of inheritance in drug response.40 Specifically, phenotype–genotype analysis can be performed by correlating drug response with genome-wide SNPs so novel genes can be identified. Functional studies of SNPs or genes in which the SNPs are found can then be performed in the laboratory and, subsequently, they can be validated by clinical translational studies. A schematic outline of this type of approach is shown diagrammatically in Figure 3. An example of the successful use of this approach has been demonstrated with two cancer drugs, gemcitabine and cytosine arabinoside. In that case, a variation in the gene for FK506-binding protein 5 (FKBP5), a protein not involved in known pharmacokinetic and pharmacodynamic pathways for these two antineoplastic cytidine analogs, was found to alter tumor sensitivity to both drugs, in addition to a variation in the gene that encodes cytosolic 5′-nucleotidase 3 (NT5C3), a protein that is involved in a known cytidine analog metabolism pathway.41 Subsequent studies demonstrated that FKBP5 has an important role in response to a variety of antineoplastic agents.42
Figure 3.
Diagrammatic representation of the use of a cell-line-based model system to identify and validate, both functionally and clinically, pharmacogenomic candidate genes. Permission obtained from Oxford University Press © Wang, L. & Weinshilboum, R. M. Hum. Mol. Genet. 17, R174–R179 (2008).40
Another example of the use of a cell-line-based model system for drug discovery and pharmacogenomics research is the NCI-60 cell line collection. This set of cancer cell lines has been extensively profiled at the molecular level more than any other set of cells in existence,43 complementing the HapMap sample set and the ‘Human Variation Panel’ cell line system.
Clinical translation
A series of important questions must be addressed before the clinical ‘translation’ of pharmacogenomics. Obviously, pharmacogenomic tests must demonstrate clinical utility and their use must help to predict outcomes and provide tangible benefit to the patient. This type of diagnostic test must be shown either to be better than existing clinical strategies or to supplement them (for example, genotyping added to the determination of INR for warfarin). The test must also be cost-effective and have an appropriate turnaround time in the clinical laboratory. In a value-based, cost-conscious environment, cost-effectiveness will be a major determinant of whether payers will be willing to bear the cost of genetic testing to guide drug therapy. However, by one analysis, the use of CYP2C9 and VKORC1 genotyping to help determine initial warfarin dose could potentially result in US$1.1 billion in savings by reducing hemorrhagic complications and thromboembolic strokes.44 Obviously, the validity of this type of estimate will have to be determined in real-life clinical practice settings. One analysis of cost-effectiveness used a Markov state transition decision model based on three randomized trials and concluded that genotype-guided warfarin therapy for patients with nonvalvular atrial fibrillation might only be cost effective in a high-risk subgroup during initiation of therapy.45
What is clear, however, is that the rapid development of genomic technology will accelerate the expansion of our understanding of the role of inheritance in variation in drug response. The large volume of data that will be made available will challenge our ability to integrate this knowledge into clinical practice.
Conclusions
The rapid progress made in genomic medicine, especially after completion of the Human Genome Project, has helped us move closer to the goal of individualized drug therapy. Pharmacogenetics has evolved into pharmacogenomics as we have developed the ability to apply genome-wide techniques to study the role of inheritance in drug response phenotypes. These developments will eventually make it possible for physicians to more-accurately predict individual variation in drug response. Implementation of the Genetic Information Nondiscrimination Act of 200846 in the US will help to address patients’ concerns with regard to the use of this type of clinical genetic information.
Clinical genotyping of individual genes will almost certainly be a transitory phase in the clinical application of genomics. We have already entered an era when direct-to-consumer companies, such as 23andMe and Navigenics, offer screening of patients’ DNA for over 1 million SNPs at a cost as low as $399, and the day when complete gene sequencing (3 billion nucleotides for under $1,000) will be available for individual patients is possibly as close as 2015.47 The challenge to physicians will involve integrating this tidal wave of genetic information into the clinical decision-making process to better individualize drug therapy.
Acknowledgments
Supported in part by HL 84904 (Heart Failure Clinical Research Network), a Marie Ingalls Cardiovascular Career Development Award (N. L. Pereira), a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award (R. M. Weinshilboum), and NIH grants UL1RR24150 (N. L. Pereira), R01 GM28157 (R. M. Weinshilboum), R01 CA132780 (R. M. Weinshilboum) and U01 GM61388 (The Pharmacogenetics Research Network). We thank L. Wussow for her assistance with the preparation of this manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Scriver CR, Childs B, editors. Garrod’s Inborn Factors in Disease. Oxford University Press; New York: 1989. [Oxford Monographs on Medical Genetics vol. 16] [Google Scholar]
- 2.Motulsky AG. Drug reactions enzymes, and biochemical genetics. JAMA. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 3.Vesell ES, Page JG. Genetic control of drug levels in man: antipyrine. Science. 1968;161:72–73. doi: 10.1126/science.161.3836.72. [DOI] [PubMed] [Google Scholar]
- 4.Kalow W, Gunn DR. The relationship between dose of succinylcholine and duration of apnea in man. J Pharmacol Exp Ther. 1957;120:203–214. [PubMed] [Google Scholar]
- 5.Kalow W, Gunn DR. Some statistical data on atypical cholinesterase of human serum. Ann Hum Genet. 1959;23:239–250. doi: 10.1111/j.1469-1809.1959.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 6.Lockridge O. In: Pharmacogenetics of Drug Metabolism: International Encyclopedia of Pharmacology and Therapeutics. Kalow W, editor. Pergamon Press; New York: 1992. pp. 15–50. [Google Scholar]
- 7.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 8.Reidenberg MM, Drayer DE, Levy M, Warner H. Polymorphic acetylation procainamide in man. Clin Pharmacol Ther. 1975;17:722–730. doi: 10.1002/cpt1975176722. [DOI] [PubMed] [Google Scholar]
- 9.Perry HM, Jr, Tan EM, Carmody S, Sakamoto A. Relationship of acetyl transferase activity to antinuclear antibodies and toxic symptoms in hypertensive patients treated with hydralazine. J Lab Clin Med. 1970;76:114–125. [PubMed] [Google Scholar]
- 10.Price Evans DA. In: Pharmacogenetics of Drug Metabolism: International Encyclopedia of Pharmacology and Therapeutics. Kalow W, editor. Pergamon Press; New York: 1992. pp. 95–178. [Google Scholar]
- 11.Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: a new pharmacogenetic defect. Eur J Clin Pharmacol. 1979;16:183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- 12.Lennard MS, et al. Oxidation phenotype—a major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–1560. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez FJ, et al. Human debrisoquine 4-hydroxylase (P450IID1): cDNA and deduced amino acid sequence and assignment of the CYP2D locus of chromosome 22. Genomics. 1988;2:174–179. doi: 10.1016/0888-7543(88)90100-0. [DOI] [PubMed] [Google Scholar]
- 14.Kimura S, Umeno M, Skoda RC, Meyer UA, Gonzalez FJ. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989;45:889–904. [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson I, et al. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci USA. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akulli E, et al. Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 17.Mega JL, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2008;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 18.Roger VL, et al. Trends in heart failure incidence and survival in a community-based population. JAMA. 2004;292:344–350. doi: 10.1001/jama.292.3.344. [DOI] [PubMed] [Google Scholar]
- 19.Bardy GH, et al. Amiodarone or an implantable cardioverter-defibrillator for congestive heart failure. N Engl J Med. 2005;352:225–237. doi: 10.1056/NEJMoa043399. [DOI] [PubMed] [Google Scholar]
- 20.The Beta-Blocker Evaluation of Survival Trial Investigators. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. N Engl J Med. 2001;344:1659–1667. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- 21.Taylor MR. Pharmacogenetics of the human beta-adrenergic receptors. Pharmacogenomics J. 2007;7:29–37. doi: 10.1038/sj.tpj.6500393. [DOI] [PubMed] [Google Scholar]
- 22.Moore JD, Mason DA, Green SA, Hsu J, Liggett SB. Racial differences in the frequencies of cardiac beta(1)-adrenergic receptor polymorphisms: analysis of c145A>G and c1165G>C. Hum Mutat. 1999;14:271. doi: 10.1002/(SICI)1098-1004(1999)14:3<271::AID-HUMU14>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Liggett SB, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small KM, Forbes SL, Rahman FF, Bridges KM, Liggett SB. A four amino acid deletion polymorphism in the third intracellular loop of the human alpha 2C-adrenergic receptor confers impaired coupling to multiple effectors. J Biol Chem. 2000;275:23059–23064. doi: 10.1074/jbc.M000796200. [DOI] [PubMed] [Google Scholar]
- 25.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 26.Liggett SB, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med. 2007;147:755–765. doi: 10.7326/0003-4819-147-11-200712040-00006. [DOI] [PubMed] [Google Scholar]
- 28.O’Reilly RA, Aggeler PM, Hoag MS, Leong LS, Kropatkin ML. Hereditary transmission of exceptional resistance to coumarin anticoagulant drugs. The first reported kindred. N Engl J Med. 1964;271:809–815. doi: 10.1056/NEJM196410152711602. [DOI] [PubMed] [Google Scholar]
- 29.Daly AK, King BP. Pharmacogenetics of oral anticoagulants. Pharmacogenetics. 2003;13:247–252. doi: 10.1097/00008571-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 31.Rost S, et al. Mutations in vKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 32.Rieder MJ, et al. Effect of vKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz UI, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesko LJ. The critical path of warfarin dosing: finding an optimal dosing strategy using pharmacogenetics. Clin Pharmacol Ther. 2008;84:301–303. doi: 10.1038/clpt.2008.133. [DOI] [PubMed] [Google Scholar]
- 35.International Warfarin Pharmacogenetics Consortium et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.http://clinicaltrials.gov/ct2/show/NCT00839657
- 37.The SEARCH Collaborative Group et al. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 38.Mangravite LM, Krauss RM. Pharmacogenomics of statin response. Curr Opin Lipidol. 2007;18:409–414. doi: 10.1097/MOL.0b013e328235a5a2. [DOI] [PubMed] [Google Scholar]
- 39.Zuccaro P, et al. Tolerability of statins is not linked to CYP450 polymorphisms, but reduced CYP2D6 metabolism improves cholesteraemic response to simvastatin and fluvastatin. Pharmacol Res. 2007;55:310–317. doi: 10.1016/j.phrs.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Weinshilboum RM. Pharmacogenomics: candidate gene identification, functional validation and mechanisms. Hum Mol Genet. 2008;17:R174–R179. doi: 10.1093/hmg/ddn270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pei H, et al. FKBP51 acts as a scaffolding protein to regulate Akt phosphorylation. Cancer Cell. in press. [Google Scholar]
- 43.Weinstein JN. Spotlight on molecular profiling: ‘Integromic’ analysis of the NCI-60 cancer cell lines. Mol Cancer Ther. 2006;5:2601–2605. doi: 10.1158/1535-7163.MCT-06-0640. [DOI] [PubMed] [Google Scholar]
- 44.Hughes DA, Pirmohamed M. Warfarin pharmacogenetics: economic considerations. Pharmacoeconomics. 2007;25:899–902. doi: 10.2165/00019053-200725110-00001. [DOI] [PubMed] [Google Scholar]
- 45.Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 46.Hudson KL, Holohan MK, Collins FS. Keeping pace with the times—the Genetic Information Nondiscrimination Act of 2008. N Engl J Med. 2008;358:2661–2663. doi: 10.1056/NEJMp0803964. [DOI] [PubMed] [Google Scholar]
- 47.Jones M. Francis Collins Addresses State of Personalized Medicine. GenomeWeb. 2009 [online], http://www.genomeweb.com/dxpgx/francis-collins-addresses-state-personalized-medicine.