Abstract
Kinetochores are macromolecular machines that couple chromosomes to dynamic microtubule tips during cell division, thereby generating force to segregate the chromosomes1,2. Accurate segregation depends on selective stabilization of correct ‘bi-oriented’ kinetochore-microtubule attachments, which come under tension due to opposing forces exerted by microtubules3. Tension is thought to stabilize these bi-oriented attachments indirectly, by suppressing the destabilizing activity of a kinase, Aurora B4,5. However, a complete mechanistic understanding of the role of tension requires reconstitution of kinetochore-microtubule attachments for biochemical and biophysical analyses in vitro. Here we show that native kinetochore particles retaining the majority of kinetochore proteins can be purified from budding yeast and used to reconstitute dynamic microtubule attachments. Individual kinetochore particles maintain load-bearing associations with assembling and disassembling ends of single microtubules for >30 min, providing a close match to the persistent coupling seen in vivo between budding yeast kinetochores and single microtubules6. Moreover, tension increases the lifetimes of the reconstituted attachments directly, via a catch bond-like mechanism that does not require Aurora B7-10. Based on these findings, we propose that tension selectively stabilizes proper kinetochore-microtubule attachments in vivo through a combination of direct mechanical stabilization and tension-dependent phosphoregulation.
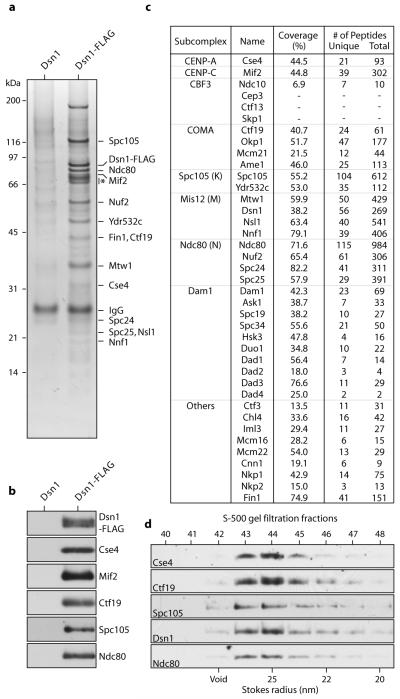
To isolate native yeast kinetochores, we modified a method that we previously developed to purify minichromosomes containing centromere-bound kinetochores11. We affinity purified the Dsn1-FLAG epitope-tagged kinetochore protein under physiological concentrations of salt to maintain kinetochore structure11. Although Dsn1 is a component of the four-member Mis12 kinetochore subcomplex (also called Mtw1/MIND complex12), silver-staining (Fig. 1a) and immunoblotting (Fig. 1b) of the detergent-eluate after SDS-PAGE revealed co-purification of components from nearly all core subcomplexes. In contrast, purifications via tags on subcomplexes other than Mis12 did not isolate the majority of kinetochore subcomplexes (Fig. S1, and 12,13).
Figure 1. Kinetochore particles can be isolated by affinity purification of Dsn1-FLAG.
a, Core kinetochore proteins co-purify with Dsn1-FLAG as visualized by silver-stained SDS-PAGE. Asterisk indicates non-specific co-purifying proteins. b, Immunoblot confirms co-association of DNA- and microtubule-binding components with Dsn1-FLAG. c, Mass spectrometry identifies all components of kinetochore subcomplexes except CBF3. Identities, percent sequence coverage, and number of identified peptides of core kinetochore proteins are shown. See Table S1 for all proteins identified by mass spectrometry. d, Eluted kinetochore particles were subjected to S-500 size exclusion chromatography and analyzed by immunoblots. The kinetochore proteins analyzed co-migrate as a complex (Stokes radius ~25 nm). Fig. S3 shows additional fractions.
Kinetochore components were the most abundant proteins in the Dsn1-FLAG-purified sample. Most bands on the silver-stained gels could be unambiguously assigned to core kinetochore proteins by gel shifts after epitope tagging (Fig. S2), and their relative abundance was consistent between preparations (Figs. 1a, 2a, S2, S4, and S6). Similarly, mass spectrometry indicated that core kinetochore proteins were the most abundant (Fig. 1c, Table S1 and Supplementary Note S1). Spindle checkpoint and other kinetochore regulatory proteins also co-purified, although motor proteins and the Aurora B protein kinase were not detected (Table S1 and Supplementary Note S2). To test whether the kinetochore proteins stably associate with Dsn1-FLAG, we performed gel filtration after FLAG peptide elution. A peak fraction (~25 nm Stokes radius) containing DNA- and microtubule-binding kinetochore components was detected (Figs. 1d, S3 and Supplementary Note S3). Taken together, these data show that stable, large assemblies spanning the entire kinetochore can be isolated from budding yeast.
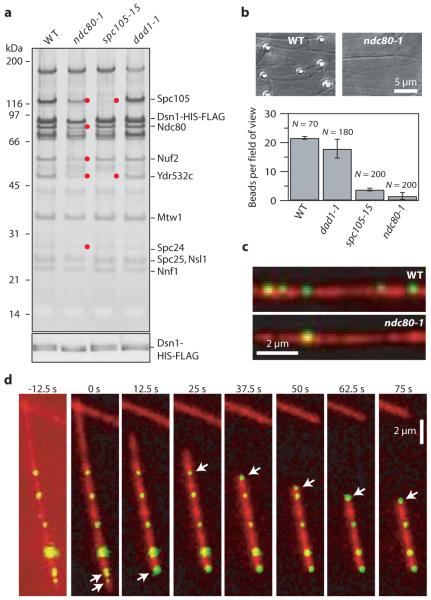
Figure 2. Purified kinetochore particles bind microtubules in vitro.
a, Silver-stained SDS-PAGE of Dsn1-HIS-FLAG kinetochore material from wild-type (WT), ndc80-1, spc105-15 and dad1-1 mutants. Red dots indicate reduced proteins in the mutant preparations (see Fig. S4). Bottom: Anti-FLAG immunoblot against Dsn1-HIS-FLAG. b, Binding of beads prepared with material from indicated strains to taxol-stabilized microtubules (mean ± s.d., from N fields, as indicated). c, Fluorescence images of Cse4-GFP kinetochore particles (green) from wild-type and ndc80-1 strains bound to taxol-stabilized microtubules (red). d, Selected frames from Movie S1 showing movement of Cse4-GFP particles (arrows) driven by the disassembling ends of a microtubule (red; see also, Fig. S7 and Movie S2).
To ask whether the purified kinetochore particles are functional, we developed several bead- and fluorescence-based assays. First, we double-tagged Dsn1 so the particles could be coupled via anti-penta-HIS antibodies to polystyrene microbeads. Beads prepared with kinetochore particles from wild-type cells bound densely along taxol-stabilized microtubules (22 ± 1 beads per field; Fig. 2b). If instead the beads were prepared using particles from ndc80-1 or spc105-15 mutant strains (Figs. 2a and S4), binding was severely reduced (2 ± 1 or 4 ± 1 beads per field, respectively; Fig. 2b). Fluorescent kinetochore particles from strains containing the centromeric histone variant Cse4 fused to green fluorescent protein (Cse4-GFP) behaved similarly (Figs. 2c, S5 and S6). Previous work using recombinant Ndc80 and Spc105 has suggested that both subcomplexes contribute synergistically to microtubule binding14. The dramatic loss of binding in our assays with either ndc80-1 or spc105-15 is consistent with this hypothesis. Notably, kinetochore particles from dad1-1 mutants (Dam1 complex) supported binding at near wild-type levels (18 ± 3 beads per field; Fig. 2b). This observation is consistent with previous analyses suggesting that the initial attachment of kinetochores to the sides of microtubules does not require the Dam1 complex15.
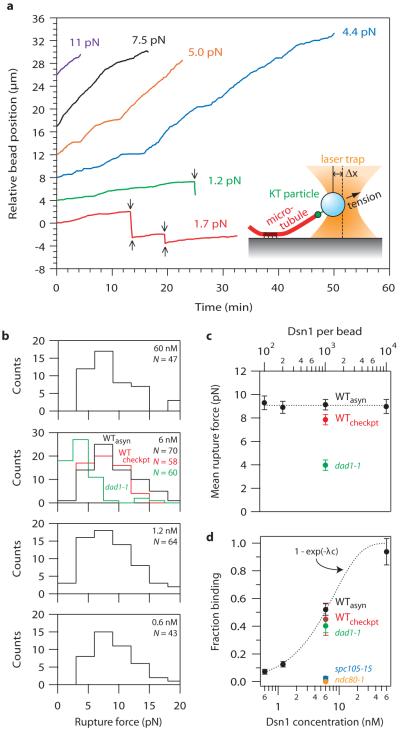
During mitosis in vivo, kinetochores persistently attach to the assembling and disassembling ends of microtubules, and they withstand tensile forces ranging from about 0.4 to 8.0 pN3,16. Time-lapse fluorescence imaging confirmed that the kinetochore particles track with disassembling ends in vitro (Fig. 2d, S7, Movies S1 and S2, and Supplementary Notes S4 and S5). Their disassembly-driven movement was highly processive, usually continuing until the filament depolymerized completely back to the seed. To test whether they also withstand physiological forces, we adapted a recently-developed bead motility assay16-19. Beads prepared with kinetochore particles from wild-type and various mutant strains were attached near the growing ends of individual microtubules and constant tension was applied using a servo-controlled laser trap. Bead-bound wild-type particles remained associated with the microtubule end, supporting continuous loads up to 11 pN (Figs. 3a, S8, and Supplementary Notes S4 and S5). These wild-type attachments were long-lived, with a mean duration comparable to that of mitosis in yeast (Fig. S9)6. They often persisted through multiple ‘catastrophe’ and ‘rescue’ events, where the filament switched from assembly to disassembly and vice versa (Figs. 3a, red trace, S8 and Movie S3), a behavior exhibited by kinetochores in vivo. During disassembly, the attachments also moved in a direction opposite the trapping force, demonstrating that they can harness energy stored in the microtubule lattice to produce mechanical work. Notably, their coupling behavior was more robust than simpler couplers based on recombinant Ndc80 and Dam1 complexes16-18 (Fig. S9), and they formed strong attachments even when the Dsn1:bead ratio was reduced below 200:1 (Figs. 3b, 3c, and S10). Particles from the dad1-1 mutant strain formed weaker attachments (Figs. 3b and 3c), while those from ndc80-1 or spc105-15 strains usually failed to interact detectably with the filaments (similar to the results with taxol-stabilized microtubules) (Fig. 3d).
Figure 3. Single kinetochore particles suffice for robust coupling.
a, Records of position versus time for native kinetochore-based attachments at indicated tensile loads. Arrows mark transitions (catastrophes and rescues). b, Rupture force distributions for beads prepared with particles at indicated concentrations from wild-type (WTasyn), dad1-1, and checkpoint-activated wild-type cells (WTcheckpt). c, Mean rupture force (± s.d., from N ruptures, indicated in b) versus labeling density, expressed as the Dsn1-HIS-FLAG concentration (bottom scale) and the corresponding Dsn1:bead ratio (top scale). d, Fraction of beads that bound a growing microtubule end (mean ± s.d., N = 11-396). Dotted curve shows Poisson fit (see text and Supplementary Information for details).
The robust behavior of the wild-type kinetochore particles at low Dsn1:bead ratios indicates that few particles, perhaps just one, may be required to form a load-bearing coupler (Supplementary Note S6). If single particles suffice, then under conditions of limiting particle density the strength of the interaction should remain invariant as the density of particles on the beads is reduced. Consistent with this prediction, the force required to rupture attachments associated with growing microtubule ends was statistically indistinguishable, averaging 9.1 ± 0.2 pN, across a 100-fold range of Dsn1:bead ratios (Figs. 3b and 3c). A second prediction is that the fraction of active beads, A(c), should vary according to the Poisson probability that a bead carries one or more active particles, A(c) = 1 - exp(−λc), where c is the relative Dsn1 concentration and λ is a fitting parameter. Indeed, this form of A(c) matches closely the fraction of beads that formed attachments to growing ends (Fig. 3d). The observation that a measurable event becomes rarer upon dilution while its properties remain unchanged is the hallmark of a ‘single molecule’ experiment. Analogous observations first demonstrated that single motor enzymes such as kinesin and myosin V are processive, for example20-22. Here it indicates that robust coupling is an intrinsic and stable property of the purified kinetochore particles – artificial oligomerization on the bead surface is not required. It also demonstrates a close match to the physiological situation in budding yeast, where each individual kinetochore is coupled to the tip of a single microtubule23.
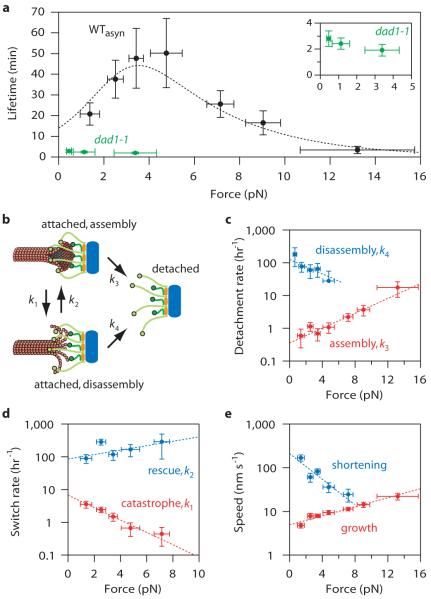
The basis for accurate chromosome segregation is thought to be tension-dependent stabilization of bi-oriented kinetochore-microtubule attachments24. This selective stabilization has been attributed to an indirect mechanism where tension inhibits phosphorylation of kinetochores by the Aurora B kinase4,5. As a first step toward reconstituting this mechanism, we measured the effect of tension on attachment lifetimes for individual end-associated kinetochore particles. Considering that the particles lacked detectable Aurora B (Table S1), that there was no ATP present to allow phosphorylation, and that protein-protein interactions are typically destabilized by force25,26, we expected a monotonic decrease in lifetime with increasing tension. Surprisingly, increasing tension between 1 and 5 pN enhanced the stability of the attachments (Fig. 4a), raising their mean lifetime from 21 ± 5 to 50 ± 17 min (p = 0.0012; based on N = 15 and 9 events observed during 5.2 and 7.5 hrs, respectively). This result shows that physiological levels of tension can stabilize kinetochore-microtubule attachments directly, by a mechanism that does not require Aurora B. The result is also reminiscent of ‘catch bonds’ between receptor-ligand pairs, which enhance cell adhesion in the presence of mechanical force7-10.
Figure 4. Tension stabilizes attachments between kinetochore particles and dynamic microtubules.
a, Measured attachment lifetimes for wild-type (WTasyn) and dad1-1 particles. Tension initially prolongs and then shortens lifetimes for wild-type attachments. Dotted curve shows prediction of the two-state model (see text). b, Schematic of two-state model with detachment during assembly and disassembly (rates k3 and k4, respectively), and interconversion between states (k1 and k2). c - e, Measured rates and exponential fits for detachment during assembly (c, red), detachment during disassembly (c, blue), catastrophe (d, red), rescue (d, blue), growth (e, red) and shortening (e, blue). Errors represent (a-d) counting uncertainty (N = 24-65) and (e) s.e.m. (N = 78-360).
Catch bonds are often explained using two-state kinetic models in which the receptor-ligand pair can switch between a strongly- and a weakly-bound state, and tension promotes adoption of the strong state7,9. Considering that microtubule tips also switch between two states, assembly and disassembly, we thought a similar model might apply to our reconstituted attachments (Fig. 4b). To test this, we measured independently how the rates of the following four events varied with tension: detachment during assembly, detachment during disassembly, catastrophe, and rescue. The detachment rate during assembly was low (~1 hr−1) and it increased gradually with tension (Fig. 4c, red). By comparison, detachment during disassembly was much faster (10- to 100-fold) but less sensitive to tension, decreasing with force (Fig. 4c, blue). We speculate that this suppression of detachment may arise from the force-dependent slowing of disassembly (Fig. 4e, blue). Together, the two detachment rates are consistent with a two-state catch bond-like model in which the strongly- and weakly-bound states correspond to tip assembly and disassembly, respectively. Tension also inhibited catastrophes (Fig. 4d, red) and promoted rescues (Fig. 4d, blue), similar to our previous findings with Dam1-based attachments17. The lower catastrophe and higher rescue rates imply that filaments under tension spend less time in the disassembling state, when the kinetochore particles are most vulnerable to detachment. We fit the force-dependence of all four rates with exponential curves (Figs. 4c and 4d, Table S2) and, without further fitting, used these to predict the lifetime versus force relationship for the two-state catch bond-like model (Supplementary Note S7). The excellent quantitative agreement with measured lifetimes (see Fig. 4a) confirms that this model can explain the tension-dependent stabilization effect. If an analogous effect occurs at kinetochore-microtubule junctions in vivo, it could make an important contribution to the selective stabilization of bi-oriented attachments (see Fig. S11).
In summary, our purification of active kinetochore particles has enabled the first direct measurements of the coupling strength between individual native kinetochore particles and dynamic microtubules. Robust coupling at the single particle level depends on the outer microtubule-binding subcomplexes from the budding yeast core kinetochore. Strikingly, tension enhances the stability of these attachments in a manner independent of Aurora B. On this basis we propose that selective stabilization of correct kinetochore-microtubule attachments occurs in vivo through a combination of at least two mechanisms, the canonical tension-dependent phosphoregulation, plus a more primitive mechanism based on tension-dependent modulation of tip dynamics.
Methods Summary
All yeast strains and plasmids used in this study are listed in Supplemental Tables 3 and 4. Media and genetic and microbial techniques were as described27. Immunoblotting and SDS-PAGE were as described28. Kinetochore particles were isolated by affinity-purifying Dsn1-FLAG or Dsn1-HIS-FLAG protein using a modified minichromosome purification protocol11 (see full methods). A typical concentration of Dsn1-FLAG or Dsn1-HIS-FLAG was ~4 μg/ml (60 nM). Size-exclusion chromatography was carried out on a Sephacryl S-500 HR column (Amersham). Estimation of Stokes radii was obtained using a high-molecular weight calibration kit (BioRad) and the void volume of the column was determined using 500 nm polystyrene beads (Polysciences Inc.). Mass spectrometry was performed as described11. TIRF microscopy and flow cell preparation were performed as previously described16,18. Purified Dsn1-HIS-FLAG kinetochore particles were linked to polystyrene beads via biotinylated anti-penta-HIS antibody, essentially as described16. The laser trap has also been described previously16-19.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank A. Desai for antibodies, and J. Kilmartin, G. Barnes, D. Pellman, R. Tsien and the Yeast Resource Center for strains and plasmids. We also thank M. Press for constructing the Cse4-GFP strain and the Seattle Mitosis Club for comments. We are grateful to T. Davis, W. Thomas, B. Zagotta, T. Tsukiyama, J. Stumpff, F. Rieke, S. Gordon, and the Biggins lab for comments on the manuscript. This work was supported by an NSF IGERT fellowship (DGE-0504573) and NIH traineeship (T32GM07270) to AFP, an NIH Cardiovascular Pathology traineeship (T32HL007312) to KKS, a Beckman Young Investigator grant to SB, NIH grants (GM078069 and GM064386) to SB, an NCI Cancer Center Support grant (CA015704) and an NIGMS grant (PM50 GM076547/Center for Systems Biology) to JAR, a Searle Scholar Award (06-L-111) to CLA, a Packard Fellowship for Science and Engineering (2006-30521) to CLA and an NIGMS grant (R01GM79373) to CLA. SB is a Scholar of the Leukemia and Lymphoma Society and TG is a Howard Hughes Medical Institute Early Career Scientist. The authors declare that none have a financial interest related to this work.
Appendix
Methods
Yeast strains, plasmids and microbial techniques
Media and genetic and microbial techniques were essentially as described27. Mitotic cultures were prepared with benomyl as described11. For temperature sensitive mutants, cells were shifted to 37 °C for 3 hours. Yeast strains and plasmids used in this study are listed in Supplemental Tables S3 and S4. The ndc80-129, spc105-1513, SPC105-GFP13, YDR532c-GFP13, dad1-130 alleles were crossed to make strains for this study. Strains containing NUF2-3GFP:HIS3 and CSE4-GFP:URA3 were made by integrating plasmid pSB897 digested with XcmI at the NUF2 locus and pSB1617 digested with StuI at the URA3 locus, respectively. Deletions, as well as 3FLAG, 13Myc and mCherry epitope tags were made using a PCR-based integration system and confirmed by PCR31-33. The 6HIS-3FLAG epitope tagging of the endogenous DSN1 gene was performed using a PCR-based integration system using primers SB2434-SB2435 and plasmid pSB1590 as a template. All tagged strains we constructed are functional in vivo and do not cause any detectable growth defects or temperature sensitivity. Specific primer sequences are available upon request.
Plasmid Construction
pSB1590 (DSN1-6HIS-3FLAG, URA3, integrating vector) was constructed in multiple steps as follows. First, pSB1110 (DSN1-12Myc, URA3, integrating vector) was made by amplification of DSN1 from pSB624 (DSN1, CEN, URA3)34 using primers SB1675 and SB1676. The PCR product was digested with XhoI and EcoRI and ligated into the same sites in pSB167 (12Myc, URA3, integrating vector)28. Second, a DNA fragment encoding 3FLAG with 5′ EcoRI and 3′ XmaI sites engineered was made by annealing SB1698 and SB1699. The fragment was then integrated just prior to the 12myc ORF of pSB1110 (DSN1-12Myc, URA3, integrating vector) that was digested with EcoRI and XmaI to make pSB1113 (DSN1-3FLAG, URA3, integrating vector). Finally, to make pSB1590 (DSN1-6HIS-3FLAG, URA3, integrating), a DNA fragment encoding 6HIS tag with EcoRI sites at both ends was made by annealing SB2432 and SB2433, which was then integrated just prior to 3FLAG of pSB1113 using EcoRI. The Nuf2-3GFP (pSB897) was made by PCR amplification of the carboxy-terminal 853 bp of NUF2 using primers SB1124 and SB1125 that have EcoRI and BamHI restriction sites engineered, respectively. The resulting PCR product was digested with EcoRI-BamHI and ligated into the same sites of pSB623 (kind gift of D. Pellman). To make Cse4-GFP (pSB1617), GFP was amplified by PCR from pSB623 using primers SB2443 and SB2444, digested with XbaI, and integrated into the XbaI site of pSB241 (CSE4, URA3, integrating vector)35. pSB1265 (3FLAG, TRP1) was made by replacing KAN marker gene of pSB812 (3FLAG, KAN) with TRP1 marker gene from pSB450 (TRP1) using SpeI and SalI. pSB1643 (GST-N-Spc105 expression vector) was made by PCR amplification of the amino-terminal 798 bp of SPC105 using primers SB2590 and SB2591 that have BamHI and EcoRI sites engineered, respectively. The resulting PCR product was digested with BamHI-EcoRI and ligated into the same sites of pGEX-2T (Amersham).
Isolation of kinetochore particles
Kinetochore particles were isolated by affinity-purifying Dsn1-FLAG or Dsn1-HIS-FLAG protein using a minichromosome purification protocol11 with the following modifications. Briefly, cells were grown in complete YPD media and extract was prepared by breaking cells in a blender with dry ice, followed by ultracentrifugation. Beads conjugated with anti-FLAG antibodies were incubated with extract for three hours with constant rotation, followed by four washes with BH/0.15 (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.1 % NP-40, 150 mM KCl, 15 % glycerol) containing protease inhibitors, phosphatase inhibitors and 2 mM DTT. Beads were further washed twice with BH/0.15 with protease inhibitors. Associated proteins were eluted from the beads by gentle agitation of beads in elution buffer (0.5 mg/ml 3FLAG peptide in BH/0.15 with protease inhibitors) for 25 min at room temperature. A typical concentration of Dsn1-FLAG or Dsn1-HIS-FLAG was ~4 μg/ml (60 nM) as determined by comparing the purified material with BSA standards on silver-stained SDS-PAGE gels. Similar results were obtained using SYPRO Ruby dye. Aliquots were made and stored at −80 °C. Typically, 2 L of asynchronously growing culture were used for microtubule-binding experiments, 24 L of mitotic culture for S-500 gel filtration experiments. Based on silver-stained SDS-PAGE, the composition of the kinetochore particles purified from mitotically arrested cells does not differ detectably from particles purified from asynchronous cultures. This finding is consistent with the observation that budding yeast kinetochores bind microtubules throughout most of the cell cycle36. To identify co-purifying proteins, associated proteins were eluted with detergent and analyzed by mass spectrometry as described11.
Protein and immunological techniques
Immunoblotting was performed as described28. Anti-Spc105 (1-266 amino acid) polyclonal antibodies were raised and affinity purified using pSB1643 (GST-N-Spc105) as previously described34 and used at a 1:1,000 dilution. Anti-FLAG antibodies (Sigma-Aldrich) were used at 1:3,000 and anti-Cse4 antibodies at 1:50034. Anti-Ndc80 (OD4, 1:10,000), anti-Ndc10 (OD1, 1:5,000), anti-Mif2 (OD2, 1:6,000), and anti-Ctf19 (OD10, 1:15,000) antibodies were generous gifts from Arshad Desai11. To compare microtubule binding activity between different kinetochore mutants, the concentration of Dsn1-FLAG was normalized by quantifying its signal intensity using the Odyssey infrared imaging system (Li Cor Bioscience). Silver-staining was performed on 4-12 % NuPAGE Novex Bis-Tris gels (Invitrogen) using a SilverQuest silver-staining kit according to instructions (Invitrogen). Size-exclusion chromatography was carried out on a Sephacryl S-500 HR column (Amersham) equilibrated in BH/0.15 at 4 °C. Estimation of Stokes radii was obtained using a high-molecular weight calibration kit (BioRad) and the void volume of the column was determined using 500 nm polystyrene beads (Polysciences Inc.).
TIRF microscopy
We utilized a custom TIRF microscope and flow cell preparation that has been previously described16,18,37. After an initial rinse with 0.3 ml ddH2O, ‘rigor’ kinesin38 diluted in BRB80 (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA at pH 6.9) containing 1 mg ml−1 κ-casein and 10 μM taxol (BCT) was introduced and allowed to bind for 5 minutes. Unbound kinesin was removed with 100 μl BCT, and Alexa 647-labeled taxol microtubules diluted in BCT plus oxygen scavengers (BCTscavs; 200 μg ml−1 glucose oxidase, 25 mM glucose, 35 μg ml−1 catalase and 5 mM DTT) were introduced and allowed to bind to the desired density. Flow cells were then washed with 50 μl BCTscavs, and fluorescent kinetochore particles (diluted in BCTscavs) were introduced and given 5 minutes to bind.
For experiments using dynamic microtubules, Alexa 647-labeled GMPCPP microtubules were bound to coverslip-adsorbed rigor kinesin, washed with 50 μl of warm growth buffer (BRB80, 1 mg ml−1 κ-casein, 1 mM GTP, 200 μg ml−1 glucose oxidase, 25 mM glucose, 35 μg ml−1 catalase and 5 mM DTT) and then incubated with growth buffer supplemented with 2 mg ml−1 Alexa 647 tubulin (1% labeled) and fluorescent kinetochore particles. Dynamic extensions were grown for ~5 minutes at 30 °C, after which depolymerization was triggered by exchanging for tubulin-free buffer.
Assay for bead binding to taxol-stabilized microtubules
Purified Dsn1-HIS-FLAG kinetochore particles were diluted in BRB80 plus 1 mg ml−1 κ-casein and linked to polystyrene beads via biotinylated anti-penta-HIS antibody, essentially as described in Powers et al. (2009). Flow cells (described in19) were treated with taxol-stabilized microtubules, which nonspecifically adsorb to the coverslip surface, and then blocked with BCT for 10 minutes prior to the introduction of kinetochore particle-coated beads. After a 10 minute incubation to allow for binding, the flow cells were imaged in DIC and the number of microtubule-bound beads per field of view was counted. Binding was negligible (2 beads in 150 fields) in negative controls using beads lacking the anti-penta-HIS antibody, prepared with WT kinetochore material at equivalent Dsn1:bead ratio (100:1).
Constant-force laser trap assays
To determine if bead-bound kinetochore particles can couple physiologically relevant forces to dynamic microtubule tips, we utilized a laser trapping-based motility assay16-19,39. Dynamic microtubule extensions were grown from coverslip-anchored GMPCPP-stabilized microtubule seeds in a buffer consisting of BRB80, 1 mg ml−1 κ-casein, 1 mM GTP, 250 μg ml−1 glucose oxidase, 25 mM glucose, 30 μg ml−1 catalase, 1 mM DTT and 1.5 mg ml−1 purified bovine brain tubulin. Assays were performed at 23 °C.
The laser trap has been described previously19. Position sensor response was mapped using the piezo stage to raster-scan a stuck bead through the beam, and trap stiffness was calibrated along the two principle axes using the drag force, equipartition, and power spectrum methods. Force feedback was implemented with custom LabView software. During clamping of the force, bead-trap separation was sampled at 40 kHz while stage position was updated at 50 Hz to maintain the desired load. Bead and stage position data were decimated to 200 Hz before storing to disk.
All the wild-type kinetochore particle data shown in Figs. 3a, 4, and S9 were recorded using beads prepared at a Dsn1:bead ratio of 200:1 (1.2 nM Dsn1-HIS-FLAG, 5.6 pM beads), well below the single particle limit. The statistics for kinetochore particles presented in Figs. S9a and S9b were calculated from a set of 40 individual events, lasting a total of 11.5 hrs, during which the particles were subjected to a constant tensile force of 1.9 ± 0.4 pN (mean ± s.d.). Event duration and travel distance were computed from the instant an attachment was fully loaded until the event ended, often due to bead detachment but sometimes for other reasons (e.g. when another bead fell into the trap). All individual event durations and travel distances were averaged, irrespective of how the events ended.
The data for wild-type particles in Figs. 4a and 4c – 4e were calculated from a set of 170 individual events, lasting a total of 42.5 hrs, during which the kinetochore particles were subjected to constant tensile forces between 0.3 and 18 pN. This dataset included all events used for Figs. 3a and S9, plus an additional 130 events recorded to investigate microtubule dynamics and attachment lifetimes as functions of force. Lifetimes (Fig. 4a) were computed by summing the total time of all events in a given force range and dividing by the number of detachments in that range. Rates of detachment during assembly (Fig. 4c, red) and catastrophe (Fig. 4d, red) were computed by counting the numbers of these events in a given force range and dividing by the total assembly time in that range. Likewise, rates of detachment during disassembly (Fig. 4c, blue) and rescue (Fig. 4d, blue) were computed by counting events and dividing by the total disassembly time in each force range.
Rupture force measurements
Beads were prepared with kinetochore material at molar ratios ranging from 100 to 10,000 Dsn1 molecules per bead. Individual beads were attached to the ends of growing microtubules and preloaded with a constant tension of 1.1 ± 0.1 pN for the dad1-1 mutants, or 3.8 ± 0.2 pN for the wild-type kinetochore particles. The laser trap was programmed to subsequently ramp the force at a defined rate (0.25 pN s−1) until the linkage ruptured or the load limit of the trap was reached (20 pN) and the bead escaped from the trap. At all but the highest densities of kinetochore material (i.e., below 60 nM Dsn1-HIS-FLAG), a small fraction (~14%) of the beads escaped the trap. Escape was ~3-fold more likely at 60 nM Dsn1-HIS-FLAG, suggesting that load might be shared by multiple particles at high densities. Note that for the Poisson probability curve in Fig. 4c, the fitting parameter λ accounts for both the number of Dsn1 molecules per particle and the proportion of particles that are geometrically inaccessible to the microtubule, or otherwise inactive.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 3.Nicklas RB. The forces that move chromosomes in mitosis. Ann. Rev. Biophys. Biophys. Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 4.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 7.McEver RP, Zhu C. Rolling Cell Adhesion. Annu Rev Cell Dev Biol. 2010;26:34. doi: 10.1146/annurev.cellbio.042308.113238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarangapani KK, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 9.Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 10.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 11.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 16.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franck AD, Powers AF, Gestaut DR, Davis TN, Asbury CL. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51:242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 21.Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 22.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 23.Winey M, et al. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 26.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 27.Rose MD, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 28.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods Additional References
- 29.Wigge PA, et al. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enquist-Newman M, et al. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell. 2001;12:2601–2613. doi: 10.1091/mbc.12.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- 35.Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gestaut DR, Cooper J, Asbury CL, Davis TN, Wordeman L. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. 2010;95:641–656. doi: 10.1016/S0091-679X(10)95032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 39.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.