Abstract
All cells employ ATP-powered proteases for protein-quality control and regulation. In the ClpXP protease, ClpX is a AAA+ machine that recognizes specific protein substrates, unfolds these molecules, and then translocates the denatured polypeptide through a central pore and into ClpP for degradation. Here, we use optical-trapping nanometry to probe the mechanics of enzymatic unfolding and translocation of single molecules of a multidomain substrate. Our experiments demonstrate the capacity of ClpXP and ClpX to perform mechanical work under load, reveal very fast and highly cooperative unfolding of individual substrate domains, suggest a translocation step size of 5–8 amino acids, and support a power-stroke model of denaturation in which successful enzyme-mediated unfolding of stable domains requires coincidence between mechanical pulling by the enzyme and a transient stochastic reduction in protein stability. We anticipate that single-molecule studies of the mechanical properties of other AAA+ proteolytic machines will reveal many shared features with ClpXP.
In all organisms, AAA+ family molecular machines harness the energy of ATP binding and hydrolysis to power macromolecular movement, disassembly, remodeling, or degradation (Hanson and Whiteheart, 2005; White and Lauring, 2007). A prominent subfamily of these machines function as ATP-dependent proteases, which are essential for protein quality control, help sculpt and maintain the proteome, and play important regulatory roles (Baker and Sauer, 2006; Schrader et al., 2009). AAA+ proteases include ClpXP, ClpAP, ClpCP, HslUV, Lon, FtsH, PAN/20S, and the 26S proteasome. Each of these enzymes contains a hexameric ring of AAA+ ATPases and a barrel-like peptidase, in which the proteolytic active sites are concealed within an interior chamber. Degradation is initiated when loops within the axial pore of the AAA+ ring engage an unstructured region of a protein substrate. ATP-fueled conformational changes in the ring are then thought to produce pulses of pulling that unfold the protein substrate and translocate it through the pore and into the degradation chamber.
In the ClpXP protease, ClpX recognizes, unfolds, and translocates protein substrates into the chamber of ClpP for degradation (Fig. 1A; for review, see Sauer et al., 2004). Short peptide sequences, such as the ssrA tag, mediate the initial binding of substrates to loops within the axial pore of the ClpX ring (Gottesman et al., 1998; Siddiqui et al., 2004; Martin et al., 2007; 2008a; 2008b). Each subunit of ClpX consists of an N domain, which is dispensable for degradation of ssrA-tagged substrates, and large and small AAA+ domains, which bind ATP and form the hexameric ring (Singh et al., 2001; Wojtyra et al. 2003; Martin et al., 2005).
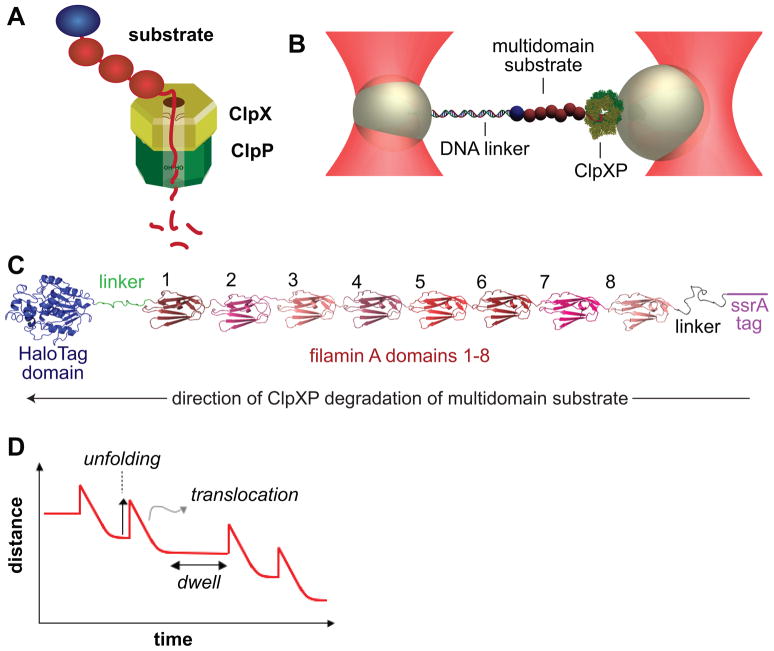
Figure 1. Experimental Design.
(A) Cartoon of the ClpXP protease midway through degradation of a multidomain substrate. The substrate is threaded through the pore of the ClpX hexamer (yellow-brown). Loops in the pore grip the substrate, and downward loop movements, powered by ATP binding and hydrolysis, unfold each domain and translocate the denatured polypeptide into the lumen of ClpP (green), where degradation to small fragments occurs (Ortega et al., 2000; Siddiqui et al., 2003; Martin et al., 2008a; 2008b; Glynn et al., 2009).
(B) Representation of an optical double-trap in a passive force-clamp geometry. ClpXP was attached to one polystyrene bead, a multidomain substrate was tethered to a second bead by double-stranded DNA, and connectivity between the beads was maintained by interactions between ClpXP and the substrate.
(C) Structural representation of the multidomain substrate.
(D) ClpXP unfolding of a domain of the substrate increases the bead-bead distance, whereas translocation of the unfolded polypeptide decreases this distance. The dwell is the time required to unfold the next domain.
Crystal structures of the ClpX hexamer suggest machine-like motions in which ATP binding and hydrolysis alter the orientation between the large and small AAA+ domains of a subunit, driving rigid-body motions that propagate around the ring and cause downward motions of axial-pore loops in contact with a substrate (Glynn et al., 2009). These movements are then thought to allow ClpX to pull on the degradation tag, resulting in translocation that wedges the native substrate against the narrow pore of the AAA+ ring, leading to eventual unfolding. Subsequent translocation of the denatured protein into ClpP results in degradation. Interestingly, translocation does not appear to depend on recognition of repeating features along the polypeptide “track”, as homopolymeric stretches of many chemically diverse amino acids in addition to unnatural sequences with extra methylene groups between successive peptide bonds are efficiently translocated by ClpXP (Barkow et al., 2009). Although the mechanism by which ClpX grips the translocating polypeptide remains incompletely understood, it seems likely that step size is determined by the molecular properties of the enzyme rather than the substrate.
ClpXP can hydrolyze as many as 500 ATPs during unfolding of a single very stable protein, although the average unfolding cost for less stable substrates can be as low as 10 ATPs (Kenniston et al., 2003; 2004). Each ATP-driven conformational cycle in ClpXP probably represents a new unfolding attempt, implying that most efforts to denature high-cost substrates are unsuccessful. Why are low-cost substrates denatured after a few attempts, whereas most attempts to denature high-cost substrates fail? Because the enzyme and substrate should have no “memory” of previous encounters, random factors, such as transient structural distortions caused by thermal fluctuations, are likely to change the probability of unfolding. For example, such fluctuations could weaken the substrate structure near the degradation tag, making unfolding easier when coincident with ClpXP pulling on the tag.
Single-molecule experiments can reveal aspects of molecular mechanism that are difficult to establish by experiments that measure the average properties of large populations of enzymes and substrates. Here, we use optical-trapping nanometry to assay the mechanics of unfolding and translocation during ClpXP degradation of single molecules of a model substrate containing multiple domains. We demonstrate that ClpXP sequentially unfolds and translocates each successive domain of this substrate in a highly localized and unidirectional fashion. Most unfolding events are very fast, with less than 1 ms between the start and finish of the reaction, and are highly cooperative, with intermediates detected in only ~15% of all cases. Subsequent ClpXP translocation of the unfolded substrate proceeds in a stepwise fashion, with the smallest resolved steps being 5–7 amino acids. In contrast to the very fast time of domain unfolding, ClpXP often spent much longer periods attempting to denature an engaged domain, supporting models in which successful enzyme-mediated denaturation requires mechanical pulling to coincide with transient reductions in substrate stability. Our results demonstrate that ClpXP can perform at least 5 kT of work per translocation step and support a power-stroke model of forcible denaturation of protein substrates.
Results
Substrate and experimental design
To assay single-molecule mechanical activity, we tethered a complex of ClpXP and a multidomain substrate between two polystyrene beads, held in a dual laser-trap configuration (Fig. 1B). As shown in Fig. 1C, the substrate contained an N-terminal HaloTag protein (Los and Wood, 2007), eight distinct immunoglobulin-like domains of the naturally occurring filamin-A protein (FLN; Fucini et al., 1997), and a C-terminal ssrA tag to target it to ClpXP (Gottesman et al., 1998). A 3500 base-pair DNA molecule, with a HaloTag ligand on one 5′ end and a biotin on the opposite 5′ end, was used to attach the substrate to a streptavidin-coated bead and to provide a spacer between the substrate and bead. Cecconi et al. (2005) first used DNA as a spacer and tether for force-induced studies of protein unfolding, linking these molecules by disulfide bonds. Because reducing agents are important for robust ClpXP activity but would cleave disulfides, we used the HaloTag domain to allow an alternative method of covalent linkage to the DNA. A biotinylated single-chain ClpXΔN hexamer complexed with ClpP was attached to a second streptavidin-coated bead (Martin et al., 2005; Shin et al., 2009).
To assemble this system, a glass-binding peptide aptamer (Aubin-Tam et al., 2010) was used to tether beads to a cover slip, substrate was attached to the beads, and a substrate-bound tethered bead was positioned close to an optically trapped ClpXP bead by moving the cover slip. Recognition of the ssrA tag and substrate engagement by ClpXP was verified by inter-bead tension. The substrate-bound bead was then captured by a second, stiffer trap, severing the aptamer connection to the glass surface. The stiffness and positions of the traps were adjusted to achieve a passive force-clamp geometry (Greenleaf et al., 2005), allowing movement of the ClpXP bead to maintain constant tension in response to the changes in substrate length that occur during degradation. The distance between the two beads should increase when ClpXP unfolds a domain, thereby increasing the observed substrate length, decrease when ClpXP moves along the substrate degrading the unfolded polypeptide, and remain constant while ClpXP works to unfold the next domain (Fig. 1D). Each experiment terminates when the connections between enzyme and substrate, enzyme and bead, or substrate and bead are lost.
Single-molecule unfolding and translocation
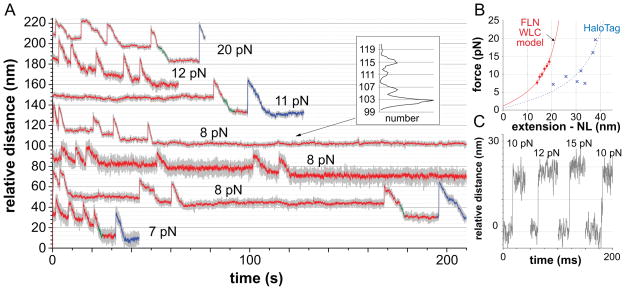
We used our assay to observe the events that occur during ClpXP degradation of single substrate molecules under different applied loads (Fig. 2A). Each trace consisted of abrupt upward steps, consistent with single-domain unfolding events, followed by slower downward movements, consistent with immediate translocation of the resulting unfolded polypeptide, followed by motionless periods or dwells, consistent with ClpXP working to unfold the next domain. At higher loads, a few unfolding events (less than 4%) were followed by a long dwell period rather than translocation, consistent with force-induced unfolding of a substrate domain that is not in contact with ClpXP (not shown). In Fig. 2A, events ascribed to ClpXP unfolding and translocation of FLN domains of the substrate are colored red, whereas those assigned to HaloTag domains are blue. We identified these individual events based on multiple observations. (i) Unfolding of the ~98-residue FLN domains resulted in 14–19 nm increases in bead separation, corresponding to the expected length difference between a denatured and a native FLN domain (Fig. 2B, red symbols), calculated using a worm-like chain (WLC) force-extension model to estimate the length of the unfolded domain (Bustamante et al., 1994; Schlierf et al., 2007; Ferrer et al., 2008). (ii) The substrate contains eight FLN domains (Fig. 1C), and multiple sequential FLN unfolding events were observed, with the intervening dwell positions spaced at ~4 nm intervals, the native domain length (Fig. 2A, inset). (iii) Unfolding of the larger HaloTag domain resulted in larger increases in bead separation (21–36 nm), again consistent with a denatured state described by a WLC model (Fig. 2B, blue symbols). (iv) When present, HaloTag-domain unfolding events were always terminal (Fig. 2A), as expected for ClpXP degradation progressing from the C-terminus to the N-terminus of the substrate. (v) A 47-residue linker separates FLN1 from the HaloTag domain (Fig. 1C, colored green). Consistently, the spacing between the dwell positions flanking FLN1-unfolding events was correspondingly larger than between other FLN-unfolding events (Fig. 2A). Taken together, the single-molecule traces support a processive degradation model in which ClpXP sequentially unfolds and translocates each successive domain of the substrate, with both reactions localized to the substrate domain in immediate contact with the enzyme (Lee et al., 2001; Kenniston et al., 2005; Martin et al., 2008c).
Figure 2. Single-Molecule Unfolding and Translocation by ClpXP.
(A) Representative extension-versus-time traces of unfolding and translocation events, accompanying ClpXP degradation of the multidomain substrate at different forces using 2 mM ATP. Traces are offset on the y-axis to avoid overlap. Gray lines represent raw data following decimation; colored lines represent averages over a 10-point sliding window. Red segments represent FLN domains; blue segments represent the HaloTag domain; green segments represent translocation of the 47-residue linker between the HaloTag domain and FLN1. Dwells between FLN subunits unfolding in the top 8 pN trace were spaced at ~4 nm (insert), which is the length of a folded FLN domain (Furuike et al., 2001). No traces contained eight FLN unfolding events, presumably because C-terminal FLN domains were unfolded and translocated before measurements began.
(B) Plots of force versus unfolding extension minus native length (NL) for FLN domains (red symbols; mean ± SEM) or the HaloTag domain (blue symbols). The red line is a WLC force-extension model (red line) with a contour length (37.3 nm) and a fitted persistence length (0.61 nm) consistent with reported values for FLN domains (Furuike et al., 2001). For the HaloTag domain, NL was 3.3 nm, the native distance between Asp106 and the HaloTag C-terminus; Asp106 forms a covalent bond with the HaloTag ligand (Los and Wood, 2007) on the DNA linker. The blue WLC curve was calculated with a persistence length of 0.61 nm and a fitted contour length of 62.1 nm, a value within 7% of previous measurements (Taniguchi and Kawakami, 2010).
(C) Raw data showing four independent FLN-unfolding transitions (offset on the x-axis for clarity). The three transitions on the left occurred without detectable intermediates and within 1 ms, as did ~85% of all unfolding transitions. The rightmost transition occurred in two stages, as expected for an unfolding intermediate, but was still complete within 5 ms; ~15% of FLN unfolding events behaved similarly.
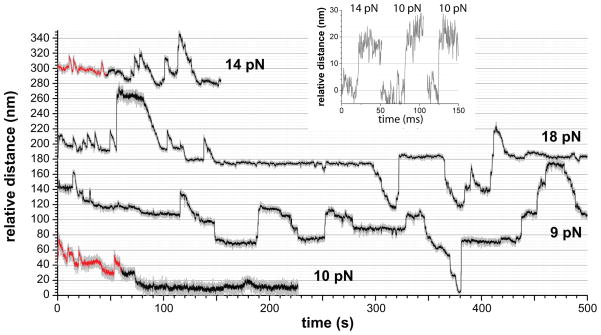
Biochemical experiments have shown that ClpX alone can unfold protein substrates, but at a rate 2–3 fold slower than ClpXP (Kim et al., 2000; Burton et al., 2001; Joshi et al., 2004; Martin et al., 2007). Indeed, in single-molecule experiments using single-chain ClpXΔN without ClpP, we also observed substrate unfolding and translocation (Fig. 3). Some unfolding and translocation events appeared similar to those observed for ClpXP, but many upward steps were far too large for single-domain unfolding and probably represent load-induced slipping of large segments of already unfolded and translocated substrate back through the ClpX pore. Engaged substrates are known to be released from ClpXP during attempts to denature very stable domains (Lee et al., 2001; Kenniston et al., 2005; Martin et al., 2008b; 2008c). We did not observe large-scale substrate slippage in the single-molecule ClpXP experiments, but such events are likely to sever the connection between ClpXP and the partially degraded substrate, ending the experiment. Because ClpXP unfolding and translocation events were better behaved and could be confidently assigned, the experiments and analyses below focus on these reactions.
Figure 3. ClpX Unfolding and Translocation.
Experiments with ClpXSC alone showed some upward steps consistent with single-domain FLN unfolding and subsequent translocation (red regions). Highly cooperative unfolding transitions were observed (insert) in these regions. In the black regions, many upward steps were too large to represent single-domain unfolding, were frequently not followed by translocation, and probably correspond to load-induced backward slipping of substrate segments that had already been unfolded/translocated by ClpX. Following translocation through the ClpX pore, portions of the substrate may interact with the bead, refold, or be engaged by another ClpXSC enzyme, further complicating analysis. The combined forward and backward motions result in a slow rate of net tracking along the substrate (e.g., in both middle traces there is no net forward movement between 150 and 500 s). Mean velocities from Gaussian fits (± 95% confidence interval) of the distribution of translocation velocities for ClpXSC were 21.1 ± 2.9 aa s−1 at 4–8 pN and 18.3 ± 2.2 aa s−1 at 8–12 pN.
Rapid and cooperative domain unfolding
The unfolding traces provide important information about the speed and cooperativity of domain denaturation by ClpXP. Notably, for ~85% of the FLN or HaloTag unfolding events, the increase in substrate length that accompanied denaturation occurred within the operational sampling time of less than 1 ms. The leftmost three traces in Fig. 2C show representative examples of fast, highly cooperative denaturation without detectable intermediates. For the remaining ~15% of unfolding events, denaturation occurred in two stages, but was still complete within 5 ms. The right trace in Fig. 2C shows an example of this class of event, which is expected for a reaction with a transiently populated unfolding intermediate. We also observed fast, highly cooperative unfolding of filamin domains in experiments using ClpX alone (Fig. 3, inset).
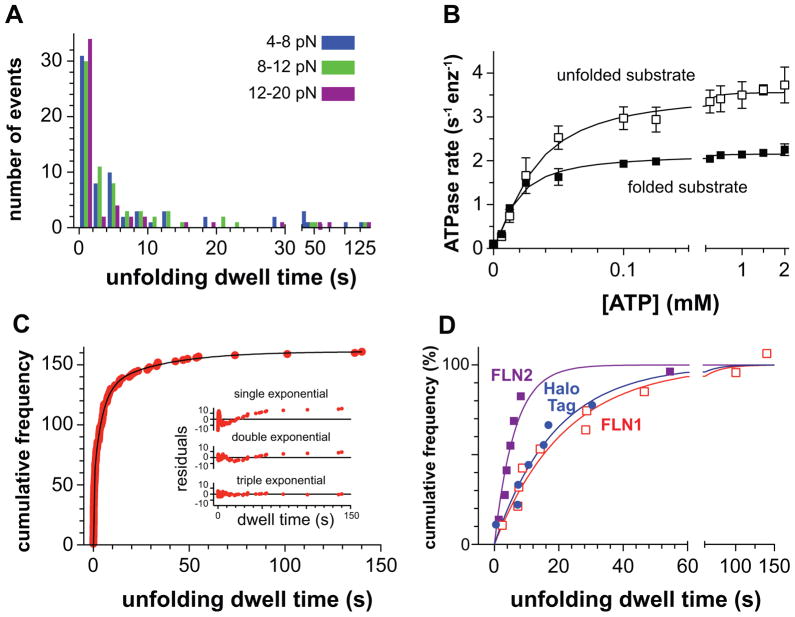
The dwell periods prior to abrupt unfolding represent the time required for successful ClpXP denaturation. Strikingly, some FLN-unfolding events closely followed completion of the prior translocation reaction, whereas others required more than 100 s (Figs. 2A & 4A). Importantly, large variations in pre-unfolding dwell times were observed between different experiments and within single experiments in which one enzyme tracks along the substrate (Fig. 2A). Many futile ATP-hydrolysis cycles are likely to occur during the long dwell periods. Indeed, the steady-state rate of ATP hydrolysis was ~2.2 s−1 enz−1 during ClpXP degradation of our multidomain substrate (a value that includes unfolding and translocation) and increased to 3.6 s−1 enz−1 during degradation of a model unfolded substrate (a value that should represent translocation alone; Fig. 4B). Depending on the force and specific experiment, analysis of the unfolding-translocation traces showed that ClpXP spent 66–75% of the time working to unfold individual domains of the substrate. Thus, the average ATPase rate during ClpXP unfolding should be 1.7–1.9 s−1 enz−1, indicating that an average of ~170–190 ATP molecules would be hydrolyzed during an unfolding dwell of ~100 s.
Figure 4. Distribution of Dwell Times Preceding FLN-Unfolding Events.
(A) Pre-unfolding dwell times (n=154) for ClpXP varied widely, irrespective of the force regime.
(B) Steady-state rates of ATP hydrolysis by ClpXSC/ClpP were determined at different ATP concentrations in the presence of the multidomain substrate (20 μM) or carboxymethylated titinI27-LAA (30 μM), a model unfolded substrate (Kenniston et al., 2003). Symbols are averages (± SD) based on independent replicates (n ≥ 3). The solid lines are non-linear least-squares fits to the Hill equation (rate = Vmax/(1+(KM/[ATP])h). Multidomain substrate; Vmax = 2.2 0.1 enz−1 sec−1; KM = 16.8 0.1 μM; h = 1.3 ± 0.1. Unfolded substrate; Vmax = 3.6 0.1 enz−1 sec−1; KM = 29.8 2.2 μM; h = 1.4 ± 0.1). No protein substrate; Vmax = 1.2 0.1 enz−1 sec−1; KM = 10.1 1.5 μM; h = 1.4 ± 0.3. Positive cooperativity (h > 1) in ATP hydrolysis by ClpX and ClpXP has been reported (Burton et al., 2003; Hersch et al., 2005).
(C) Dwell times were combined, plotted as cumulative frequencies, and fitted to a triple-exponential function, yielding rate constants of 3.4 ± 0.16 s−1 (amplitude 35 ± 1%), 0.23 ± 0.02 s−1 (amplitude 47 ± 2%) and 0.03 ± 0.01 s−1 (amplitude 18 ± 2%). The inset shows the residuals for single, double, and triple exponential fits.
(D) Single-exponential fits of cumulative frequencies versus pre-unfolding dwell times for FLN2 events (filled squares; k = 0.20 ± 0.04 s−1), FLN1 events (open squares; k = 0.04 ± 0.01 s−1), and HaloTag events (circles; k = 0.04 ± 0.02 s−1).
The very fast and highly cooperative nature of ClpXP-induced unfolding for the domains studied here rule out models in which unfolding requires a large number of ATP-hydrolysis events because the substrate is denatured a few amino acids at a time. However, our results support the idea that only some ClpXP pulling events result in successful denaturation because of stochastic fluctuations in the mechanical stability of the target domain or in the unfolding power of ClpXP, resulting in the hydrolysis of many ATPs and correspondingly long periods of time before domain unfolding occurs.
The distribution of dwell times preceding FLN unfolding events did not vary significantly with applied load (Fig. 4A). Dwell times were combined, arranged in ascending order, and assigned a number (cumulative frequency) representing the total number of events with that or a smaller value. A plot of cumulative frequency versus dwell time (Fig. 4C) was fit well by a triple-exponential function with rate constants of 3.4 ± 0.16 s−1 (amplitude 35 ± 1%), 0.23 ± 0.02 s−1 (amplitude 47 ± 2%) and 0.03 ± 0.01 s−1 (amplitude 18 ± 2%). Because the sequence of each FLN domain in the substrate is different (pairwise homology 20–40%), it seems likely that they also have different stabilities. Distinct stability classes could account for the different exponential classes. Indeed, for the small number of unfolding events that could be confidently ascribed to FLN1, FLN2, or the HaloTag-domain, the dwell distributions were fitted well (R2 0.93–0.97) by single-exponential functions with rate constants of 0.04 s−1 for FLN1 and the HaloTag domain and 0.2 s−1 for FLN2 (Fig. 4D). We cannot eliminate the possibility that more data would reveal some multi-exponential character for each domain, but the clear differences between the FLN1 and FLN2 distributions support a role for domain heterogeneity in the overall distribution. An exponential distribution of dwell times for a specific domain is consistent with a model in which stochastic fluctuations render only some engaged domains susceptible to ClpXP-induced unfolding at any given time. Notably, the fastest exponential phase for the pooled FLN domains had a rate constant (3.4 s−1) near the upper end of the range for ATP hydrolysis during unloaded ClpXP degradation (2.2–3.6 s−1 enz−1; Fig. 4B), suggesting that unfolding of some FLN domains requires hydrolysis of just one ATP molecule.
Translocation velocity and force dependence
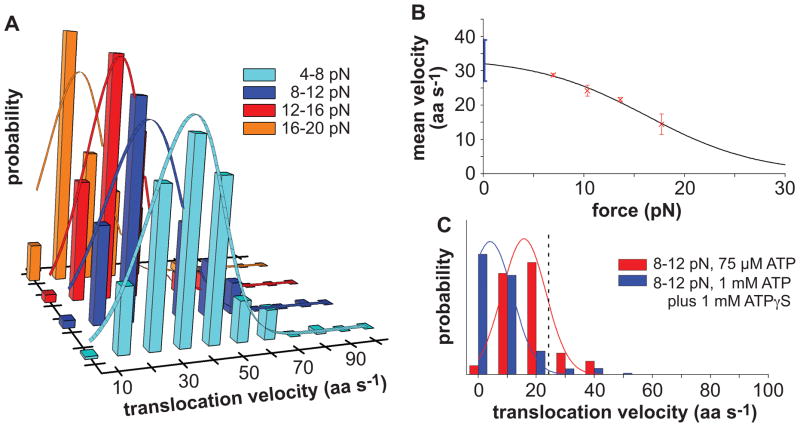
We calculated ClpXP translocation velocities from the slopes of traces following 232 individual unfolding events, resulting in the distributions shown in Fig. 5A. Slopes were initially calculated in units of nm s−1 and were then converted to amino acids (aa) s−1 using a WLC model. ClpXP translocation slowed as the resisting force increased, with mean rates of ~30 aa s−1 at loads of 4–8 pN and ~15 aa s−1 at loads of 16–20 pN (Fig. 5B).
Figure 5. Translocation Velocities at Different Forces and Nucleotide Concentrations.
(A) Histograms with distributions of ClpXP translocation velocities, weighted by the length translocated at that velocity, are shown for experiments using 2 mM ATP in four force regimes. The solid lines are Gaussian fits.
(B) Red symbols are mean velocities from Gaussian fits (± 95% confidence interval), calculated from the panel-A data. The solid line is a non-linear least-squares fit of the force dependence to a single-barrier Boltzmann model (see text). The blue bar represents a range of unloaded velocities from single-molecule and ensemble experiments using different substrates (Martin et al., 2008c; Shin et al., 2009).
(C) Translocation-velocity distributions at 8–12 pN for experiments performed using 75 μM ATP (frequent rupture of bead linkage occurred when less ATP was used) or 1 mM ATP plus 1 mM of ATP S. The dashed vertical line represents the mean velocity for 8–12 pN using 2 mM ATP. Solid lines are Gaussian fits.
In biochemical experiments, the rates of ClpXP translocation and ATP hydrolysis are linearly related (Martin et al., 2008c). Thus, slowing ATP turnover should slow single-molecule translocation. In fact, the average translocation rate decreased ~35% when the ATP concentration was reduced from 2 mM to 75 μM (Fig. 5C). This decrease is consistent with a KM of ~40 μM at a load of 8–12 pN; in unloaded experiments, KM for steady-state ATP hydrolysis was ~30 μM during degradation of an unfolded substrate (Fig. 4B). When ClpXP was fueled by a mixture of 1 mM ATP and 1 mM ATP S, which ClpXP hydrolyzes slowly (Burton et al., 2003; Martin et al., 2008c), the single-molecule translocation rate also dropped ~80% (Fig. 5C).
The force dependence of the average ClpXP translocation velocity fit well to a single-barrier Boltzmann equation (Fig. 5B; Wang et al., 1998)
where k is the Bolztmann constant, T is the absolute temperature,ν0 is the fitted unloaded velocity (32 ± 4 aa s−1), δ is the fitted distance between the enzyme ground state and transition state (0.7 ± 0.4 nm), and A is the fitted ratio of times associated with force-dependent versus force-independent reaction steps (0.05 ± 0.1). The small value of A suggests that a force-independent reaction is normally rate limiting for overall ClpXP translocation. Notably, ν0 was similar to unloaded ClpXP translocation rates (27–39 aa s−1) calculated from single-molecule and biochemical experiments using different substrates (Martin et al., 2008c; Shin et al., 2009). Extrapolation based on the single-barrier Boltzmann model predicts that translocation would reach 5% of ν0 at 33 pN, and ClpXP would stall at a slightly higher force.
Stepwise translocation
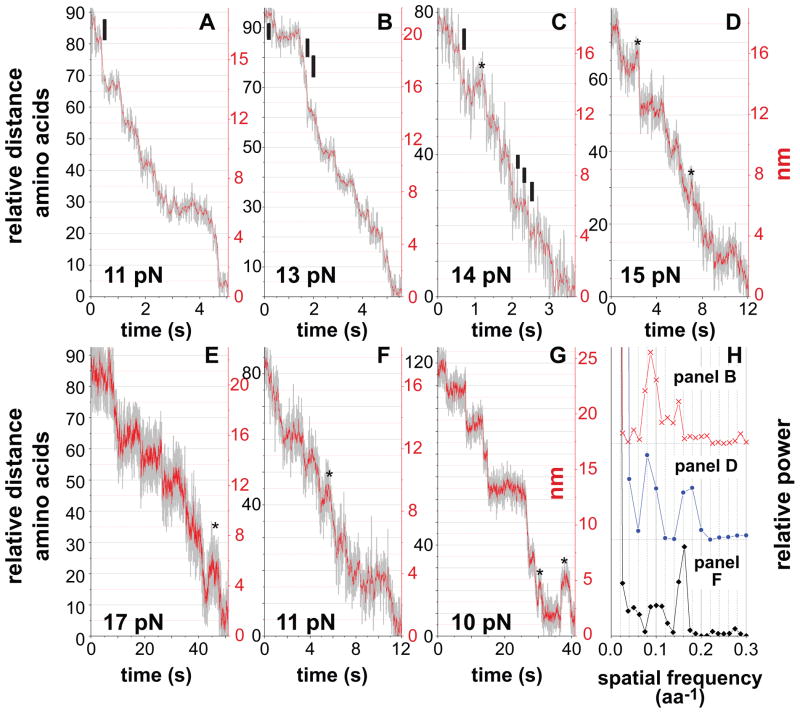
Prior studies indicate that the conformational change that allows ClpXP to translocate polypeptides is generated by hydrolysis of one ATP (Martin et al., 2005; 2008c) but also show that translocation does not require a fixed spacing of substrate side chains or peptide bonds (Barkow et al., 2009). The latter property, which is probably important in allowing translocation of polypeptide segments with highly diverse chemical and physical properties, suggests that the translocation step need not be an integral or even a constant value. In the clearest individual traces (Fig. 6A–6G), the smallest discernable ClpXP translocation steps were ~1 nm, corresponding to ~6 amino acids based on a WLC model. Analysis of pairwise distance distributions showed main peaks corresponding to steps of 5.7–7.1 and 10–13 residues (Figure 6H). Based on these results, each ClpXP translocation step would move 5–7 amino acids, with the longer 10–13 residue increments corresponding to sequential but unresolved steps. Dividing ν0 by the average ATP-hydrolysis rate during steady-state ClpXP degradation of an unfolded substrate yields 9 ± 1 residues per ATP hydrolyzed, a value intermediate between the shorter and longer steps. It is possible, therefore, that some hydrolysis events result directly in the longer 10–13 residue steps. For ClpX alone, the smallest physical steps ranged from 6.2–7.7 residues (Fig. 7).
Figure 6. Visualization of Individual ClpXP Translocation Steps.
(A)–(G) For the y-axis of each panel, the black numbers represent amino acids, assuming a WLC model with a persistence length of 0.61 nm (see Fig. 2B legend), and the red numbers represent changes in bead-bead distance. In panels A–C, the black vertical bars represent the smallest steps resolved. Experiments were performed using 2 mM ATP (A–E), 75 μM ATP (F), or 1 mM ATP plus 1 mM ATP S (G). Asterisks mark events that appear to be minor slipping of the substrate relative to ClpXP, which was more common during translocation under higher loads, at low ATP, or in ATP/ATP S mixtures.
(H) Power spectra (Svoboda et al., 1993) of pairwise-distance distribution functions of single translocation curves show main peaks near ~0.08 and ~0.16 aa−1, corresponding to step sizes of ~6.25 (range 5.7–7.1) and ~12.5 (range 10–13) amino acids.
Figure 7. Translocation Steps by ClpX alone.
(A)–(G) For the y-axis of each panel, the black numbers represent amino acids, assuming a WLC model with a persistence length of 0.61 nm (see Fig. 2B legend), and the red numbers represent changes in bead-bead distance.
(H) Power spectra of pairwise-distance distribution functions of single translocation curves show main peaks near ~0.095 and ~0.15 aa−1, corresponding to step sizes of ~6.8 (range 6.2–7.7) and ~10.5 (range 10–11) amino acids.
Discussion
The design of our substrate allowed identification of specific ClpXP unfolding and translocation events. For example, the spacing of the dwell periods flanking FLN1-domain unfolding is larger than for other FLN domains, because the FLN1 domain is separated from the HaloTag domain by a ~50-residue linker. Similarly, ClpXP unfolding of the HaloTag domain results in a larger increase in substrate length than unfolding of FLN domains. For future studies, additional natural or unnatural sequences or native protein domains could be placed between FLN1 and the HaloTag domain, allowing single-molecule studies of ClpXP unfolding and/or translocation of these elements.
We found that the velocity of ClpXP translocation decreases as the resisting force increased, indicating that at least one reaction in the overall translocation process is force dependent. However, the A value determined from fitting to the single-barrier Boltzmann model was far below 1, suggesting that a mechanical step is not rate limiting in the absence of an applied force. RNA polymerase also has a very low A value, and force-independent pyrophosphate release is known to be rate limiting for this enzyme (Wang et al., 1998). It remains to be determined what reaction or reactions limit the rate of translocation and ATP hydrolysis by ClpXP, but the force-dependent step probably occurs at a rate at least 20-fold faster based upon the A value of 0.05. Curiously, we did not observe a change in unfolding dwell times as the force was increased. Because the ATPase rate and translocation rate appear to be tightly coupled, as shown here and previously (Martin et al., 2008c), it seems very likely that the ATPase rate also slows at high forces, and one might therefore expect a corresponding increase in the average time required for unfolding. However, we suspect that force-induced destabilization of the substrate makes denaturation by ClpXP easier, offsetting any force-induced decrease in ATP-hydrolysis rates.
Motor proteins operate by power-stroke mechanisms, by Brownian-ratchet capture of random thermal motions, or by a combination of these mechanisms (Hwang and Lang, 2009). The smallest ClpXP and ClpX translocation steps are ~5–8 amino acids in length, a size small enough to be driven by plausible structural changes in the ClpX hexamer during a power stroke (Glynn et al., 2009) or by thermal motion. Although we cannot rule out some Brownian contributions, we favor a mechanism that relies predominantly on a power stroke for several reasons. First, the polypeptide substrate is threaded through the ClpX and ClpXP rings during translocation, an architecture that results in close enzyme-substrate contacts and favors a direct-drive mechanism. Second, translocation under load was highly unidirectional; ~150 consecutive forward steps were observed in some trajectories and reverse steps were extremely rare, except at the highest loads or at reduced rates of ATP hydrolysis. Thus, a Brownian mechanism would need to be exceptionally efficient at capturing thermal motion in the correct direction and preventing motion in the opposite direction to account for translocation. Notably, however, ClpXP can translocate substrates from the C-terminus to the N-terminus or in the opposite direction and does not require recognition of specific spacings of side chains or peptide bonds (Barkow et al., 2009). Thus, unidirectional capture of thermal motion by binding to repeating molecular features of the polypeptide track seems highly unlikely. Third, we observe pre-unfolding dwells that indicate that the folded domain is in intimate contact with the ClpX motor, without significant motions or slack that would allow a Brownian mechanism to operate. Finally, it is difficult to envision a strictly Brownian mechanism that could account for the ability of ClpXP to accelerate protein-unfolding reactions by factors as large as 106-fold (Kim et al., 2000), whereas a power-stroke mechanism has this potential. Consistently, ClpXP can perform significant mechanical work. Indeed, a ~1 nm step against 20 pN of applied load corresponds to ~5 kT of mechanical work per step, which would increase to ~7.5 kT at the extrapolated stall force.
How might the pulling activity of the ClpX motor lead to protein unfolding? Depending upon the stability and unfolding properties of a domain, ClpXP probably operates using three general denaturation mechanisms. (i) For meta-stable domains, one power stoke may result in cooperative denaturation. Indeed, the shortest unfolding dwells observed in our assays are consistent with events associated with ClpXP hydrolysis of a single ATP. (ii) For more stable domains, each independent power stroke appears to have some probability of success linked to transient destabilization of the domain resulting from fluctuations in thermal energy, although fluctuations in the enzyme could also contribute to the probabilistic nature of unfolding. Notably, the largest unfolding dwell times in our assays were longer than 100 s, a period in which ClpXP could hydrolyze ~200 ATP molecules. As expected for a model in which the stability of a domain followed a Boltzmann distribution, the unfolding dwell times for specific FLN domains or the HaloTag domain were roughly exponentially distributed. (iii) In principle, some ClpXP unfolding events could also occur in discrete steps. For example, if enzymatic pulling disrupted a small region of protein structure and translocation of that segment prevented refolding, then the partially unfolded protein might be sufficiently stable to persist. Under these circumstances, the energy barrier for unfolding of the remaining structure would be lowered, allowing subsequent power strokes to complete unfolding (Lee et al., 2001). Approximately 15% of the FLN-unfolding events did show an unfolding intermediate, but these events also occurred in less than 5 ms, which is far faster than the average time required for ATP hydrolysis (> 250 ms). We cannot eliminate models in which a few coordinated power strokes contribute to these non-cooperative unfolding events, but the most likely model is that they also result from hydrolysis of a single ATP with subsequent thermally driven denaturation of the remaining elements of native structure. Thus, the FLN and HaloTag unfolding events observed in our experiments appear to occur by one of the first two mechanisms discussed. Stepwise ClpXP unfolding, which requires multiple ATP-hydrolysis events, is more likely to be observed for proteins like GFP and ribonuclease H, in which unfolding intermediates have higher kinetic or thermodynamic stabilities than the domains studied here (Hollien and Marqusee, 1999; Dietz and Rief, 2004; Kenniston et al., 2004; Martin et al., 2008c).
Although protein unfolding and thus degradation can be very energetically costly (Kenniston et al., 2003), it is important to note that ClpXP and other AAA+ proteases must denature and degrade a highly diverse assortment of cellular proteins with radically different structures and stabilities. Thus, evolution is unlikely to have optimized ClpXP activity for any single substrate. Repeated pulling on a peptide tag is a simple mechanism that allows relatively low-cost degradation of metastable substrates but eventual high-cost degradation of hyperstable proteins as well. Some AAA+ family proteases appear have limited unfoldase activity and to specialize in degrading poorly structured proteins (Herman et al., 2003). Because the degradation of unfolded substrates requires energy consumption that is inversely proportional to translocation step size, it will be important to determine if these proteases employ larger translocation steps, which minimize the net cost of ATP-fueled degradation but also limit unfolding power.
ClpXP appears to use a relatively low gear (small step size), allowing it to work against substantial loads. By contrast, cytoplasmic dynein, a dimeric AAA+ machine, takes longer steps (typically 8 nm) as it moves along the microtubule cytoskeleton but has a lower stall force (~7 pN; Gennerich et al., 2007). Several molecular motors that track along nucleic acids also take relatively small steps and can work against substantial forces, including bacterial RNA polymerase (~25 pN; 1 base or ~0.4 nm steps; Wang et al., 1998; Abbondanzieri et al., 2005), the AAA+ 29 DNA-packaging motor (~50 pN; 2.5 base-pair or ~0.85 nm steps; Smith et al., 2001; Moffitt et al., 2009), and the ribosome (>20 pN; 3 base or ~1.3 nm steps; Wen et al., 2008). Moreover, each of these machines encircle the single or double-stranded nucleic acid. Likewise, the polypeptide substrates of ClpXP are threaded through the enzyme, allowing intimate contacts that maintain processivity even when working against a substantial resisting force. We anticipate that other AAA+ machines with relatively small step sizes will also be able to generate high forces, allowing them to carry out mechanical work suited to their specific biological tasks.
Experimental procedures
Biotinylated ClpXSC, a single-chain variant containing six repeats of E. coli ClpXΔN (residues 61–423), and E. coli ClpP-His6 were prepared as described (Kim et al., 2000; Shin et al., 2009). The multidomain substrate was constructed in pFN18A (Promega) and encoded an engineered Rhodococcus rhodochrous haloalkane dehalogenase (HaloTag) domain at the N-terminus (Los and Wood, 2007), a linker of 47 residues containing two TEV protease sites, immunoglobulin repeats 1–8 of human filamin A (accession CAI43198; residues 279–1066), residues 5303–5341 of human titin (accession CAA62188; corresponding to the C-terminal half of the I27 domain), a His6 tag, and a C-terminal AANDENYALAA ssrA tag. This substrate was expressed in E. coli strain ERL (Shin et al., 2009). Cells were initially grown to OD600 ~0.6 at 37 °C in 1 L of LB broth, chilled to 25 °C, and induced with 1 mM IPTG. Cells were harvested 3 h later, resuspended in 50 mM sodium phosphate buffer (pH 8), 500 mM NaCl, 10% glycerol, 20 mM imidazole, and 10 mM 2-mercaptoethanol, and frozen at −80 °C. For purification, cells were thawed and lysed using a French Press, before addition of 2 mM MgCl2 and 500 units of benzonase nuclease. After 30 min at 4 °C, lysates were centrifuged at 30,000 x g for 30 min, and the clarified lysate was passed over a Ni++-NTA affinity column (Qiagen). After washing, bound protein was eluted with buffer containing 250 mM imidazole and was further purified by anion-exchange chromatography (HiTrap Q XL, GE Healthcare) using a 0–0.5 M KCl gradient. Appropriate fractions were identified by SDS-PAGE, pooled, and concentrated by precipitation using 60% saturated ammonium sulfate. Concentrated protein (~1 mL) was loaded onto a S300 HR 16/60 size exclusion column (GE Healthcare) equilibrated in 20 mM HEPES (pH 7.6), 200 mM KCl, 10% glycerol, 0.1 mM EDTA, and 1 mM DTT. After chromatography, fractions were assayed by SDS-PAGE, pooled, concentrated (Amicon Ultra-30 kDa MWCO) after addition of 0.05% NP40 detergent, and were frozen at −80 °C. The concentration of the multidomain substrate was determined in 6 M guanidine HCl, 75 mM bicine (pH 8.3) using an extinction coefficient of 133620 M−1 cm−1 at 280 nm.
Steady-state rates of ATP hydrolysis by ClpXSC (110 nM) in the presence of ClpP-His6 (460 nM) were determined using an NADH enzyme-linked assay (Kim et al., 2000) in PD buffer (25 mM HEPES (pH 7.6), 10 mM MgCl2, 10% glycerol, 1 mM DTT, 100 mM KCl, and 0.1% tween-20) at room temperature (23 ± 1 °C). ATPase assays were performed without protein substrate or with near saturating concentrations (20–30 μM) of our multidomain HaloTag-FLN substrates or of a model unfolded substrate (carboxymethylated-titinI27-LAA).
A 3500 base-pair DNA linker was constructed by PCR using M13mp18 plasmid as template, one oligonucleotide primer with a 5′ biotin, and a second oligonucleotide primer containing a 5′ amino group. Following PCR, the 5′ amino group was crosslinked to HaloTag® thiol (O4) ligand (Promega P6761) using sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate, prior to conjugation to the HaloTag portion of the substrate.
A 10 μL flow cell was prepared with 1 μm streptavidin-coated beads (Polyscience Inc.), which were tethered to a glass cover slip via a glass-binding peptide aptamer as described (Aubin-Tam et al., 2010). The biotinylated DNA-HaloTag-filamin1–8-ssrA molecule was then introduced into the flow cell, incubated for 30 min, and washed with 5 mg/mL bovine serum albumin in PD buffer. Biotinylated ClpXSC was incubated with 1.26 μm streptavidin-coated polystyrene beads (Spherotech Inc.), and the mixture was centrifuged and resuspended once in PD buffer with 5 mg/mL BSA, 1 μM ClpP, 2 mM ATP, a creatine-phosphate based ATP-regeneration system, and an oxygen-scavenging system consisting of 5 mg/mL β-D-glucose, 0.25 mg/ml glucose oxidase, and 0.03 mg/mL catalase. After flowing 30 μL of the solution containing ClpXP-bound beads through the flow cell, it was sealed and loaded on the optical-trapping instrument. Experiments were performed at room temperature.
A dual optical trap with a dual detection system was implemented using a 1064 nm trapping laser (IPG Photonics) and a 975 nm detection laser (Avanex). Both beams were split using polarizing beam splitters before being focused at the sample plane with a 100 × 1.4 NA objective (Nikon). One trap branch was steered with acoustic optic deflectors (IntraAction). Nanometer scale positional resolution was achieved for each bead through isolation of orthogonal detection beams with a polarizer along the detection branch and steered towards two position-sensitive devices (PSDs; Pacific Silicon).
The ClpXP bead (1.26 μm) was held in a weak trap (0.01–0.08 pN/nm) and brought into the vicinity of a surface-bound, aptamer-tethered bead (1 μm) to allow ClpXP to initiate degradation. The 1 μm bead was then captured by a stiffer trap (~0.2 pN/nm), and the stage was moved to rupture aptamer linkages with the glass surface. Bead positions were sampled at 3 kHz, antialias-filtered at 1.5 kHz, and an acousto-optic deflector was used to position the 1.26 μm bead in the passive force-clamp region (Greenleaf et al., 2005), ~400 nm from the beam center. At the end of the motility record and after rupture of the bead-bead connection, each bead was raster scanned through an automated protocol to calibrate position and stiffness (Lang et al., 2002). Trap stiffness was obtained using equipartition methods. Bead separation was isolated from the lab frame by subtracting position on PSD1 from PSD2, removing common-mode noise. Applied load was determined from the position of the 1 μm bead in the stiffer trap.
The traces shown in Figures 2A, 3, 6, and 7 were obtained by decimating the differential position of beads by a factor of 25 using an 8th-order low pass Chebyshev Type I filter, and then averaged with an exponentially weighted 10-point moving average. When translocation of a domain was nearly constant, velocity was determined by a linear fit of displacement over time. When several apparent velocities could be distinguished, each region of relatively constant velocity was computed separately and weighted by the displacement that occurred at that velocity. Pairwise distance distribution functions (PDF) were calculated by binning distance differences in single translocation curves, with a bin width of 0.1 amino acids. Power spectra were calculated from the PDF by fast Fourier transformation (Svoboda et al., 1993).
Acknowledgments
We thank F. Nakamura for filamin-A plasmids and S. Glynn, J. Kardon, A. Nager, B. Stinson, Y. Shin, and other members of our labs for advice and discussions. This study was supported by National Science Foundation Career Award 0643745 (M.J.L.), the Howard Hughes Medical Institute, and National Institutes of Health grants AI-15706 and AI-82929 (A.O.O.). T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Tam M, Appleyard DC, Ferrari E, Garbin V, Fadiran OO, Kunkel J, Lang MJ. Adhesion through single peptide aptamers. J Phys Chem A. 2010 doi: 10.1021/jp1031493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BM, Williams TL, Baker TA. ClpX-mediated remodeling of mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol Cell. 2001;8:449–454. doi: 10.1016/s1097-2765(01)00307-0. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: Linkage between ClpXP-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Science. 2003;12:893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer JM, Lee H, Chen J, Pelz B, Nakamura F, Kamm RD, Lang MJ. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc Natl Acad Sci USA. 2008;105:9221–9226. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nature Struct Biol. 1997;4:223–230. doi: 10.1038/nsb0397-223. [DOI] [PubMed] [Google Scholar]
- Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP- 280) molecules detected by atomic force microscopy. FEBS Lett. 2001;498:72–75. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Crystal structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with C-terminal peptide tails added by the SsrA tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive All- Optical Force Clamp for High-Resolution Laser Trapping. Phys Rev Lett. 2005;95:208102–208106. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hollien J, Marqusee S. Structural distribution of stability in a thermophilic enzyme. Proc Natl Acad Sci USA. 1999;96:13674–13678. doi: 10.1073/pnas.96.24.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W, Lang MJ. Mechanical Design of Translocating Motor Proteins. Cell Biochem Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of Substrate Denaturation and Translocation by the ClpXP Degradation Machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two- dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008a;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008b;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease: critical dependence on ATP-hydrolysis rates and energy landscapes. Nat Struct Mol Biol. 2008c;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Sauer RT, et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlierf M, Berkemeier F, Rief M. Direct Observation of Active Protein Folding Using Lock-in Force Spectroscopy. Biophys J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nature Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Davis JH, Brau RR, Martin A, Kenniston JA, Baker TA, Sauer RT, Lang MJ. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc Natl Acad Sci USA. 2009;106:19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Kawakami M. Application of HaloTag protein to covalent immobilization of recombinant proteins for single molecule force spectroscopy. Langmuir. 2010;26:10433–10436. doi: 10.1021/la101658a. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Wojtyra UA, Thibault G, Tuite A, Houry WA. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem. 2003;278:48981–48990. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]