Abstract
Background
When the case-only study design is used to estimate statistical interaction between genetic (G) and environmental (E) exposures, G and E must be independent in the underlying population, or the case-only estimate of interaction (COR) will be biased. Few studies have examined the occurrence of G-E association in published control group data.
Methods
To examine the assumption of G-E independence in empirical data, we conducted a systematic review and meta-analysis of G-E associations in controls for frequently investigated DNA repair genes (XRCC1 Arg399Gln, Arg194Trp, or Arg280His, XPD Lys751Gln, and Asp312Asn, and XRCC3 Thr241Met) and smoking (ever/never smoking, current/not current smoker, smoking duration, smoking intensity and pack-years).
Results
Across the 55 included studies, SNP-smoking associations in controls (ORz) were not reliably at the null value of 1.0 for any SNP-smoking combinations. Two G-E combinations were too heterogeneous for summary estimates: XRCC1 399 and ever-never smoking (N=21), and XPD 751 and pack-years (N=12). ORz ranges for these combinations were: [ORz (95% confidence interval (CI)] 0.7 (0.4, 1.2) – 1.9 (1.2, 2.8) and 0.8 (0.5, 1.3) – 2.3 (0.8, 6.1), respectively). Estimates for studies considered homogeneous (Cochran’s Q p-value <0.10) varied 2- to 5-fold. No study characteristics were identified that could explain heterogeneity.
Conclusions
We recommend the independence assumption be evaluated in the population underlying any potential case-only study, rather than in a proxy control group(s) or pooled controls.
Impact
These results suggest that G-E association in controls may be population-specific. Increased access to control data would improve evaluation of the independence assumption.
Keywords: case-only, gene-environment interaction, DNA repair genes, smoking, controls
Introduction
The case-only study design as proposed by Prentice et. al (1) and promoted by Piegorsch et. al. (2) has been increasingly used to estimate the magnitude of statistical interaction between two measured exposures with respect to a given outcome, most commonly a genetic and an environmental exposure. This method requires only cases, no controls or defined cohort. Provided the relevant exposures are independent in the underlying source population, the case-only study can estimate a specific form of statistical interaction, but not main effects of the two exposures.
There are potential advantages to the case-only method in several settings. Estimation of the interaction parameter from case-only analyses is more efficient than for a traditional case-control study (i.e. fewer cases are required for similar precision of estimate) and with no need for controls, there are fewer participants overall (3). Not using controls may mitigate selection biases due to, for example, differential recruiting success between cases and controls, or differential recall of environmental exposures by case-control status. Invasive procedures that are part of cases’ diagnosis or treatment often cannot be done ethically in healthy volunteers, especially vulnerable groups such as pediatric populations (4). But these advantages come at a cost. A case-only study only estimates interaction on a multiplicative scale (deviation of the rate ratio for those having both the genetic and environmental exposures from the product of rate ratios for those with either the genetic or the environmental exposure, but not both). It cannot estimate the independent effect of either exposure, or interaction on the additive scale. This limits its use to situations in which the independent effects of the two exposures are not of interest, nor are synergism or antagonism of the exposures (5–6). Control-selection bias is the only validity threat the case-only design avoids, in comparison with the case-control design. Consequently, case-only studies have been proposed by several investigators as an initial screening method to identify candidate gene-environment or gene-gene interactions (7–9).
However, the increase in precision and avoidance of control-selection bias in the case-only method requires a major assumption: that the two exposures are independent in the source population (Z=1) (1–2). Although the constancy of rate ratios between different strata of exposure in the underlying source population is the true parameter of interest, control groups from case-control studies are frequently used to estimate Z using ORz. Data simulations have demonstrated that even when violations of the independence assumption are of small magnitude they can strongly bias the case-only interaction parameter (7). Chance can also play a role. Since the expectation that ORz=1 when Z=1 is a large sample asymptotic approximation, as sample size decreases, ORz will deviate from the null with increasing frequency through random error alone (7). Further, when control-group gene-environment (G-E) associations are of similar magnitude but opposite in direction to the interaction effect, a case-only study may not detect interaction effects, a Type II error (7, 10).
However, published control group data on the associations of interest for G×E interaction research are limited. Therefore, we undertook a systematic review and meta-analysis of selected DNA repair gene polymorphisms in XRCC1, XPD and XRCC3 and smoking behavior in control groups, using ORz to estimate Z. The purpose in estimating ORz was to determine the degree of bias in the COR, relative to the interaction estimate from a case-control analysis, assuming no control-selection bias. Heterogeneity was explored using stratified analysis and meta-regression of study characteristics. The primary aim of this project was to evaluate the independence assumption for selected SNPs and smoking behavior. This will enable investigators considering a stand-alone case-only study of gene-environment interaction with these exposures to evaluate the independence assumption more rigorously, potentially identifying situations in which case-only estimates may be more or less valid.
Methods
Data Abstraction
PubMed, ISI Web of Science and the CDC Genomics and Disease Prevention databases were searched up to March 6, 2007 for peer-reviewed literature likely to contain non-case data on the joint distribution of any of the polymorphisms of interest. Polymorphisms of interest were non-synonymous single nucleotide changes (SNPs) in XRCC1[Arg399Gln (rs25487), Arg194Trp (rs1799782), and Arg280His (rs25489)], XPD [Asp312Asn (rs1799793) and Lys751Gln (rs13181)], and XRCC3 [Thr241Met (rs861539)] (11–15).
Non-case groups were defined as any group not selected on disease status (e.g. cohorts, convenience samples and control groups from case-control studies). For simplification non-case groups will be referred to as controls throughout this article. There were no language restrictions on searches. A list of keywords for PubMed and the ISI Web of Science was developed in consultation with an information specialist from UNC Health Science Library to ensure that searches would be as inclusive as possible. Keywords for smoking were “smoking”, “tobacco”, “tobacco smoke”, “tobacco smoke pollution”, and “smoker”. The SNPs were searched by combining “polymorphism” and “polymorphism, genetic” with the SNP-specific keywords “XRCC1”, “XPD”, "xeroderma pigmentosum group d protein", “ERCC2” and “XRCC3”. ISI Web of Science keywords were “smok*” and “tobacco,” and “XRCC1”, “XPD”, “ERCC2” and “XRCC3”. GDPInfo was searched by limiting by factor menu terms to “smoking behavior”, “smoking (tobacco) passive”, “smoking (tobacco) bidi”, “smoking (tobacco)”, “smoking (tobacco) maternal”, “tobacco”, “indoor air pollution”, “nicotine (nasal spray)”, and “nicotine (transdermal)”, and gene menu terms to “XRCC1”, “XPD”, “ERCC2” and “XRCC3”.
Inclusion criteria were deliberately broad. To be included, an article had to contain original control group data on the joint distribution of any genotype of interest and any aspect of tobacco smoking behavior. Abstracts were excluded.
Abstracts were screened for controls with relevant genotype and smoking data. SNP designations considered equivalent are shown in Table 1. If an abstract passed the initial screening, the full paper was reviewed for data appropriate for construction of a 2×2 table for genotype-smoking association in controls (ORz). If a genotype-smoking ORz could be calculated, the following data were abstracted: SNP, genotype categories (3 level additive, dominant and/or recessive models), smoking status and dose categories [ever/never, current/not current, smoker/non-smoker, ever/former/current, pack-years (PY), duration and/or intensity], and cell counts for all genotype and smoking categories.
Table 1.
SNP designations for data abstraction
| XRCC1 | XRCC1 | XRCC1 | XPD (ERCC2) | XPD (ERCC2) | XRCC3 | |
|---|---|---|---|---|---|---|
| SNP designations | Arg399Gln | Arg194Trp | Arg280His | Lys751Gln | Asp312Asn | Thr241Met |
| G28152A | C26304T | G27466A | A35931C | G23591A | C18067T | |
| exon 10 | exon 6 | exon 9 | exon 23 | exon 10 | exon 7 | |
| rs25487 | rs1799782 | rs25489 | rs13181 | rs1799793 | rs861539 | |
| R399Q | R194W | R280H | K751Q | D312N | T241M | |
| ERCC2_18880_A>C | ERCC2_6540_G>A | |||||
| Minor allele | 399 Gln | 14 Trp | 280 His | 751 Gln | 312 Asn | 241 Met |
| MAF range | 0.13–0.39 | 0.05–0.37 | 0.03–0.09 | 0.07–0.45 | 0.06–0.44 | 0.02–0.43 |
MAF=minor allele frequency
The following study characteristics were also abstracted: year of publication, study design (case-control, cohort, cross-sectional, convenience, other), source of control group (for case-control: population, hospital, friends and non-blood related family, convenience, community, neighborhood, other; for cohorts: population, occupational, convenience, other), type of clinic that hospital- or clinic-based control groups were from (disease clinics, checkup clinics), study outcome (cancer [type], non-cancer disease, non-disease), full study/control group size (N), country, percent male participants, ethnicity, Hardy Weinberg equilibrium p-value, full study/control group size (N), and minor allele frequency (MAF). An estimate of central tendency for participants’ age in years (“average age”) was derived for each study using, in order of preference: median, mean, weighted average across study categories, midpoint of range. One non-English language article could not be evaluated.
Selection of Study Comparisons
No study population contributed to any analysis more than once, maintaining independence of observations. Analyses focused on associations with genotype categorized using a dominant model (i.e. homozygotes of the most common allele were the referent group, compared to heterozygotes plus homozygotes of the minor allele) due to the small number of studies that provided sufficient information to assess recessive or additive models. The minor allele did not vary across studies for any of the included SNPs. Smoking status was categorized as (1) ever/never (referent), and (2) current/not current (referent). Smoking amount was analyzed as (1) pack-years [PY, highest vs. lowest non-zero category (referent)], (2) duration [years, longest vs. shortest non-zero category (referent)] and (3) smoking intensity [cigarettes/day, heaviest vs. lightest non-zero category (referent)].
Studies that did not provide sufficient data to include ‘passive only’ smoking in the never smoking group were excluded. For analyses of current/not current smoking, never+former smokers and “non-smokers” (unless identified as never smokers) were considered not current smokers. Pack-years of smoking (PY = pack-years = number of packs smoked per day multiplied by years smoked; 20 cigarettes=1 pack) were collected as categorical variables with different cutpoints. We analyzed PY as relative categories [heaviest vs. lightest smokers regardless of PY cutpoints] and absolute categories [high PY (all categories with a cutpoint above a specified minimum) vs. low PY (all categories with a cutpoint below a specified maximum)]. The minimum and maximum cutpoints varied by SNP but were all chosen to maximize the number of included studies while keeping the range of cutpoints small enough that no study would have >1 cutpoint between the minimum and maximum cutpoints. Similar to PY, smoking intensity was categorized by relative (heaviest vs. lightest smokers regardless of cigarette/day cutpoints) and absolute (>= 20 vs. <20 cigarettes/day) measures. The smoking duration cutpoints were 20 and 40 years, inclusive, for all SNPs.
Statistical analyses
Crude ORs and 95% confidence limits were calculated from cell counts (Stata 9.2, using metan STB-44: sbe24). Funnel plot asymmetry, an indicator of possible publication bias (16), was considered suggestive of study characteristics associated with variance and Z. When data were sufficient (Nstudies>=5), asymmetry was formally assessed using Begg and Mazumdar’s test (17) and Egger’s test (18) at α=0.10. Cochran’s Q two-sided homogeneity p-values (α=0.10 due to low power of the test) were used to assess overall heterogeneity in odds ratios (19).
Study characteristic analyses
Key study characteristics hypothesized to influence variation in the strength of control group SNP-smoking associations across studies were assessed using stratified meta-analysis and random-effects meta-regression, with the among-study variance estimated by restricted maximum likelihood (20). Stratified meta-analysis produces a summary ORz estimate for each stratum of a study characteristic. Meta-regression provides a formal comparison of the stratified estimates in the form of an estimated ratio of odds ratios.
Study characteristics were selected a priori. They included (1) study design (case-control, cohort, or convenience; patient-based control groups, healthy control groups), (2) continent, (3) ethnicity, (4) Hardy-Weinberg equilibrium p-value, (5) average age, (6) gender (% male), (7) study outcome (lung cancer, other cancer, non-cancer disease, non-disease), (8) minor allele frequency and (9) smoking prevalence. Study design was examined for all SNP-smoking combinations; additional study characteristics were examined for XRCC1 399, XPD 751 and XRCC3 241. Stratified random-effects meta-analyses were used when the overall SNP-smoking association had a Cochran’s Q p-value α<0.10, otherwise fixed effects meta-analysis was used, regardless of the homogeneity p-values of individual strata. To reduce the possibility of results being confounded by ethnicity (population stratification) in overall analyses and when examining the study characteristics likely to vary strongly by ethnicity (Hardy Weinberg equilibrium p-values and minor allele frequency) studies were stratified by ethnicity and treated as separate studies if possible. Sample size was not formally examined as a study characteristic because that would have essentially reproduced funnel plot analyses of variance. Variance (or precision) is the more important measure here, and sample size is not the sole determinant of precision. However, because the independence assumption is a large sample approximation, a sensitivity analysis was conducted in which large (N>=1000) studies were examined separately. Stata 9.2 was used for all analyses. Results for study characteristics were assessed for consistency across smoking categories and across SNPs.
Results
Eligible studies
The literature searches identified 228 articles for evaluation. Of these, 55 articles were eligible for inclusion. The primary reason for exclusion was that an article did not present the genotype-smoking distribution in controls (N=98, 57% of exclusions). Exclusion reasons for the remainder included: review article or abstract only (13%), did not assess any relevant SNPs (9%), and did not have any non-cases (10%). Finally, of the 55 studies eligible for inclusion, five were not included in final summary estimates because no data were presented using the dominant genetic model (21–22), there was no measure of adult smoking behavior (23), former smokers were excluded (24), or never smokers were included in lowest PY category (25). Fifty articles representing 46 distinct study populations were included in the final meta-analyses (brief study descriptions in Supplementary Table 1). The number of study populations included for each polymorphism was: XRCC1 Arg399Gln (N=32), XRCC1 Arg194Trp (N=16), XRCC1 Arg280His (N=8), XPD Lys751Gln (N=16), XPD Asp312Asn (N=9), and XRCC3 Arg241Gln (N=13). Thirty-seven studies presented the control distribution of genotype and ever/never smoking, 16 for current/ not current smoking and 14 for PY. Far fewer presented duration (N=4) and/or intensity (N=4). Case-control studies predominated with 12 population-based (26–37) and 23 hospital-based (38–60), four studies nested within cohorts (61–64) and two other case-control studies (65–66). Most control groups were from cancer case-control studies (N=39), one was from a case-control study of rheumatoid arthritis. Nine cohort or convenience sample studies examined non-cancer outcomes, predominantly measures of DNA damage (67–75).
Association between DNA repair gene variants and smoking behavior
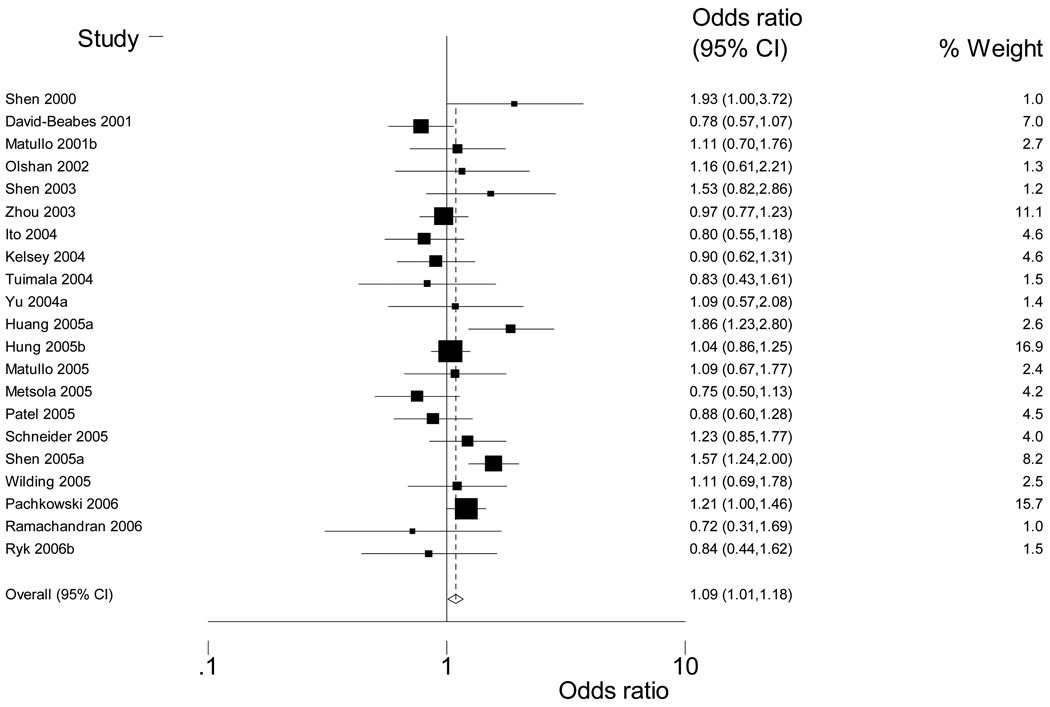
Across SNPs there was more variation in ORs assessing control-only G-E associations (ORzs) for measures of smoking amount (PY, duration, intensity) than for measures of smoking status (ever-never, current-not current) (Table 2). Ten of 11 summary estimates of smoking status fell between 0.9–1.1. Summary estimates for smoking amounts were distributed more broadly, with only five of 10 summary estimates between 0.9–1.1; the most extreme measures were found for duration and intensity. Although only two of 18 genotype-smoking groups were too heterogeneous for a fixed effects summary estimate, based on Cochran’s Q at alpha=0.10, nearly all groups had study estimates above and below the null. Individual study ORz (95% CI) are presented in Figure 1 and Supplementary Tables 3–8.
Table 2.
DNA repair gene variation and smoking summary estimates
| Ever-never | Current-Not current | Pack-years | Intensity | Duration | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q p- value |
ORz (95% CI)* |
N | Q p- value |
ORz (95% CI)† |
N | Q p-value |
ORz (95% CI)‡ |
N | Q p- value |
ORz (95% CI)§ |
N | Q p-value |
ORz (95% CI)□ |
|
| Gene and SNP | |||||||||||||||
| XRCC1 | |||||||||||||||
| Arg399Gln** | 21 | 0.01 | --- ††† | 11 | 0.40 | 1.0 (0.9, 1.1) | 9 | 0.30 | 1.2 (1.0, 1.5) | 4 | 0.49 | 1.5 (1.2, 1.9) | 2 | 0.03 | --- ††† |
| Arg194Trp†† | 12 | 0.62 | 1.0 (0.9, 1.1) | 6 | 0.68 | 1.1 (0.9, 1.3) | 2 | 0.73 | 1.1 (0.7, 1.6) | 2 | 0.89 | 1.1 (0.8, 1.6) | 3 | 0.47 | 0.7 (0.5, 0.9) |
| Arg280His‡‡ | 5 | 0.47 | 1.0 (0.8, 1.2) | 4 | 0.51 | 0.7 (0.5, 1.1) | 3 | 0.32 | 1.0 (0.6, 1.5) | 1 | -- | 0.9 (0.5, 1.8) | 1 | -- | 1.2 (0.6, 2.3) |
| XPD | |||||||||||||||
| Lys751Gln§§ | 12 | 0.46 | 0.9 (0.8, 1.1) | 6 | 0.25 | 1.1 (0.9, 1.3) | 7 | 0.02 | --- ††† | 0 | 0 | ||||
| Asp312Asn□□ | 9 | 0.79 | 1.1 (1.0, 1.2) | 1 | --- | 1.1 (0.7, 1.9) | 4 | 0.11 | 1.1 (0.8, 1.5) | 0 | 0 | ||||
| XRCC3 | |||||||||||||||
| Thr241Met*** | 9 | 0.52 | 1.0 (0.9, 1.2) | 7 | 0.73 | 0.9 (0.8, 1.1) | 4 | 0.67 | 0.8 (0.6, 1.2) | 0 | 0 | ||||
Abbreviations: CI=Confidence interval, na=not applicable, PY=pack-years, ORz=control-only genotype-smoking odds ratio, Q=Cochran’s test of heterogeneity, N=number of studies, G+=gene is positive for any variant allele G−=negative for variant allele (referent), E+=positive for smoking measures, E−=negative for smoking measure (referent), N=number of studies, SNP=single nucleotide polymorphism, Arg=Arginine, Gln=Glutamine, Trp=Tryptophan, His=Histidine, Met=methionine, Asp=Aspartic acid, Asn=Asparagine
Fixed effects summary estimates for G+/G− vs. E+/E−. G− is homozygous for the common allele (ref), G+ is the genotype with 1 or 2 variant alleles, E− (ref) is never smoker and E+ is ever smoking
Fixed effects summary estimates for G+/G− vs. E+/E−. G− is homozygous for the common allele (ref), G+ is the genotype with 1 or 2 variant alleles, E− (ref) is not current smoker and E+ is current smoking
Fixed effects summary estimates for G+/G− vs. E+/E−. G− is homozygous for the common allele (ref), G+ is the genotype with 1 or 2 variant alleles, E− (ref) is lowest non-zero PY category and E+ is highest category of PY
Fixed effects summary estimates for G+/G− vs. E+/E−. G− is homozygous for the common allele (ref), G+ is the genotype with 1 or 2 variant alleles, E− (ref) is lowest non-zero category of intensity (cig/day) and E+ is highest category of smoking intensity.
Fixed effects summary estimates for G+/G− vs. E+/E−. G− is homozygous for the common allele (ref), G+ is the genotype with 1 or 2 variant alleles, E− (ref) is lowest non-zero category of duration(yrs) and E+ is highest category of smoking duration.
Arg/Arg v. any Gln
Arg/Arg v. any Trp
Arg/Arg v. any His
Lys/lys v. any Gln
Asp/Asp v. any Asn
Thr/Thr v. any Met
Studies too heterogeneous for fixed effects summary estimate
Figure 1.
Weighted Forest Plot for XRCC1 399 and ever-never smoking
For XRCC1 399 any Gln and ever-never smoking, ORzs ranged from 0.7 (95% CI: 0.3, 1.7) (49) to 1.9 (95% CI: 1.0, 3.7) (34) (See Figure 1); three other measures of smoking behavior were homogeneous enough for a summary estimate of ORz: current smoker/not current (N=11), PY (N=9) and intensity (N=4) (Table 2). Higher PY and heavier smoking intensity, but not current vs. not current smoking, were associated with XRCC1 Arg399Gln (any Gln) [OR (95%CI): 1.2 (1.0, 1.5) and 1.5(1.2, 1.9), respectively]. For XRCC1 194 and 280, having the variant allele was associated with smoking duration [XRCC1 194: 0.7 (0.5, 0.9), XRCC1 280: 1.2 (0.6, 2.3)] and current smoking [XRCC1 280: 1.2 (0.6, 2.3)] though the number of studies was small. For the two XPD SNPs (751, 312) there was considerable variation in the association between XPD 751 variant allele and higher PY. Study estimates ranged from 1.4 (0.8, 2.6) (41) to 0.5 (0.3, 1.0) (53) (Table 2). Higher PY were associated with the variant allele for XRCC3 241 although the number of studies was small (N=4).
Sensitivity analyses
Among the studies that were assessed for current-not current smoking, a subset could also be assessed for never, former or current smoking (Supplementary Table 2). No consistent pattern emerged for comparisons of never smoking with former or current smoking. Absolute measures of PY, intensity and duration were calculated and compared to relative measures for consistency. Genotype-PY estimates for absolute cutpoints were comparable to estimates using relative categories although strata were sparse (Supplementary Table 2). Additionally, when studies with only smokers were dropped and never smoking was used as the reference category, results were essentially the same for relative and absolute measures of PY.
Genotype-smoking association between XRCC1 Arg399Gln and smoking intensity (cigarettes/day) could be estimated in four studies. There was an association between XRCC1 399 any Gln and greater smoking intensity, which was consistent across methods of smoking intensity categorization. Results did not change appreciably when studies without smoking amount were excluded from ever-never analyses, indicating that articles that presented dose were not driving estimates of smoking status. Results did not change appreciably when studies without smoking amount were excluded from ever-never analyses, indicating that articles that presented dose were not driving estimates of smoking status (data not shown).
There were six large (N>=1000) study populations, four each with data for XRCC1 399 & 194 ever-never smoking. ORzs for XRCC1 399 ever-never smoking showed evidence of heterogeneity [range of ORz(95% CI): 1.0(0.8, 1.2) – 1.6(1.2, 2.0)] (Supplementary Table 3)(32, 35, 42, 60). However, ORzs for XRCC1 194 were consistently null across studies (Supplementary Table 4)(32, 37, 42, 61). In the three large study populations with the relevant measures of smoking,(32, 42, 60) the magnitude of ORz was consistently different for status and amount for XRCC1 399 ever-never smoking; the one study population with smoking status and amount for XRCC1 194 had ORzs at the null.(42) Data were too sparse for further evaluation across large studies.
Funnel plot asymmetry
There was no evidence of funnel plot asymmetry for overall genotype-smoking associations (data not shown). In formal testing, the majority of p-values (75%) were >=0.3; the lowest p-value was p=0.14.
Study characteristics
Stratified associations and univariate meta-regression were evaluated across SNPs and smoking categories on the basis of consistency and direction. Study design was examined for all six SNPs for ever/never, current/not current smoking and PY. For smoking status, genotype-smoking associations for XRCC1 399 and 194 and XPD 751 and 312 were generally stronger for population-based case-control studies than for hospital-based or patient-based control groups, although the magnitude of the differences was small; the range of RORs was 0.7 to 0.9 for hospital/patient-based compared to population-based controls (referent) (Table 3). However, for smoking amount as measured by PY (2 evaluable SNPS, XRCC1 399 and XPD 751) the hospital-based/patient-based control groups showed stronger genotype-smoking associations than population-based control groups (range of RORs: 1.2–1.5). When examining PY, for all SNPs, the genotype-smoking association for population-based control groups was below the null. The remaining study characteristics were examined only for XRCC1 399, XPD 751 and XRCC3 241 (Tables 4, 5, and 6 respectively) due to sparse data for the other SNPs.
Table 3.
Genotype-smoking associations stratified by study design
| XRCC1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arg399Gln | Arg194Trp | Arg280His | ||||||||
| N | ORz (95% CI) |
Ratio of ORs (95%CI) † |
N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
N | ORz (95% CI)* |
Ratio of ORs (95%CI) † |
||
| Ever-never smoking | ||||||||||
| Case-based categories | ||||||||||
| Case-control | ||||||||||
| Population-based | 8 | 1.1 (0.9, 1.5) | Ref | 4 | 1.0 (0.9, 1.3) | Ref | 2 | 0.9 (0.7, 1.3) | Ref | |
| Hospital-based | 9 | 1.0 (0.9, 1.2) | 0.9 (0.7, 1.2) | 5 | 0.9 (0.8, 1.1) | 0.9 (0.7, 1.2) | 2 | 1.1 (0.8, 1.4) | 1.2 (0.8, 1.8) | |
| Other | 1 | 0.9 (0.6, 1.3) | 0.8 (0.4, 1.3) | 2 | 1.1 (0.9, 1.5) | 1.1 (0.8, 1.5) | 0 | |||
| Other | 3 | 1.0 (0.8, 1.4) | 0.9 (0.6, 1.4) | 1 | 0.7 (0.3, 1.4) | 0.7 (0.3, 1.4) | 1 | 0.5 (0.2, 1.1) | 0.5 (0.2, 1.3) | |
| Non-case-based categories | ||||||||||
| Case-control | ||||||||||
| Population controls | 8 | 1.1 (0.9, 1.5) | Ref | 4 | 1.0 (0.9, 1.3) | Ref | 2 | 0.9 (0.7, 1.3) | Ref | |
| Patient controls ‡ | 6 | 1.1 (0.9, 1.2) | 0.9 (0.7, 1.3) | 5 | 0.9 (0.8, 1.1) | 0.9 (0.7, 1.2) | 2 | 1.1 (0.8, 1.4) | 1.2 (0.8, 1.8) | |
| Non-patient controls§ | 4 | 0.9 (0.8, 1.1) | 0.8 (0.6, 1.1) | 2 | 1.1 (0.9, 1.5) | 1.1 (0.8, 1.5) | 0 | |||
| Other | 3 | 1.0 (0.8, 1.4) | 0.9 (0.6, 1.4) | 1 | 0.7 (0.3, 1.4) | 0.7 (0.3, 1.4) | 1 | 0.5 (0.2, 1.1) | 0.5 (0.2, 1.3) | |
| XRCC1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Arg399Gln | Arg194Trp | Arg280His | |||||||
| N | ORz (95% CI) |
Ratio of ORs (95%CI) † |
N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
|
| Current-not current smoking | |||||||||
| Case-based categories | |||||||||
| Case-control | |||||||||
| Population-based | 1 | 1.2 (1.0, 1.5) | Ref | 1 | 1.1 (0.8, 1.6) | NA | 1 | 0.9 (0.6, 1.4) | NA |
| Hospital-based | 3 | 0.9 (0.7, 1.2) | 0.7 (0.5, 1.0) | 2 | 0.9 (0.7, 1.3) | 0 | |||
| Unknown | 1 | 1.6 (0.7, 3.7) | 1.3 (0.5, 3.2) | 1 | 0.9 (0.4, 2.2) | 1 | 1.1 (0.2, 5.8) | ||
| Other | 6 | 0.8 (0.6, 1.1) | 0.7 (0.5, 1.0) | 2 | 1.5 (0.8, 2.7) | 2 | 0.5 (0.2, 1.0) | ||
| Non-case-based categories | |||||||||
| Case-control | |||||||||
| Population controls | 1 | 1.2 (1.0, 1.5) | Ref | 1 | 1.1 (0.8, 1.6) | NA | 1 | 0.9 (0.6, 1.4) | NA |
| Patient controls ‡ | 2 | 0.8 (0.6, 1.2) | 0.7 (0.5, 1.0) | 1 | 1.1 (0.6, 2.1) | 0 | |||
| Non-patient controls § | 1 | 1.0 (0.7, 1.5) | 0.8 (0.5, 1.3) | 1 | 0.8 (0.6, 1.2) | 0 | |||
| Unknown | 1 | 1.6 (0.7, 3.7) | 1.3 (0.5, 3.2) | 1 | 0.9 (0.4, 2.2) | 1 | 1.1 (0.2, 5.8) | ||
| Other | 6 | 0.8 (0.6, 1.1) | 0.7 (0.5, 1.0) | 2 | 1.5 (0.8, 2.7) | 2 | 0.5 (0.2, 1.0) | ||
| XRCC1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Arg399Gln | Arg194Trp | Arg280His | |||||||
| N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
N | ORz (95% CI) * |
Ratio of ORs (95%CI) † |
|
| Pack-years | |||||||||
| Case-based categories | |||||||||
| Case-control | |||||||||
| Population-based | 2 | 0.9 (0.6, 1.4) | Ref | 0 | NA | 1 | 0.5 (0.2, 1.5) | NA | |
| Hospital-based | 7 | 1.3 (1.1, 1.6) | 1.5 (0.9, 2.4) | 2 | 1.1 (0.7, 1.6) | 2 | 1.1 (0.7, 1.8) | ||
| Non-case-based categories | |||||||||
| Case-contrl | |||||||||
| Population controls | 2 | 0.9 (0.6, 1.4) | Ref | 0 | NA | 1 | 0.5 (0.2, 1.5) | NA | |
| Patient controls ‡ | 4 | 1.4 (1.1, 1.7) | 1.5 (0.9, 2.5) | 2 | 1.1 (0.7, 1.6) | 2 | 1.1 (0.7, 1.8) | ||
| Non-patient controls § | 3 | 1.3 (0.9, 1.8) | 1.4 (0.8, 2.5) | 0 | 0 | ||||
| XPD | XRCC3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lys751Gln | Asp312Asn | Thr241Met | |||||||
| N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
N | ORz (95% CI)* | Ratio of ORs (95%CI) † |
N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
|
| Ever-never smoking | |||||||||
| Case-based categories | |||||||||
| Case-control | |||||||||
| Population-based | 4 | 1.0 (0.8, 1.1) | Ref | 2 | 1.3 (1.0, 1.8) | Ref | 2 | 1.0 (0.8, 1.3) | Ref |
| Hospital-based | 7 | 0.9 (0.8, 1.1) | 0.9 (0.7, 1.2) | 7 | 1.0 (0.9, 1.2) | 0.8 (0.6, 1.1) | 4 | 1.0 (0.7, 1.3) | 1.0 (0.6, 1.4) |
| Other | 0 | 0 | 0 | ||||||
| Other | 1 | 1.0 (0.6, 1.6) | 1.0 (0.6, 1.7) | 0 | 3 | 1.2 (0.9, 1.6) | 1.2 (0.8, 1.8) | ||
| Non-case-based categories | |||||||||
| Case-control | |||||||||
| Population controls | 4 | 1.0 (0.8, 1.1) | Ref | 2 | 1.3 (1.0, 1.8) | Ref | 2 | 1.0 (0.8, 1.3) | Ref |
| Patient controls ‡ | 3 | 0.8 (0.6, 1.1) | 0.8 (0.5, 1.2) | 3 | 1.1 (0.9, 1.3) | 0.8 (0.6, 1.1) | 4 | 1.0 (0.7, 1.3) | 1.0 (0.6, 1.4) |
| Non-patient controls § | 4 | 1.0 (0.8, 1.2) | 1.0 (0.7, 1.4) | 4 | 1.0 (0.8, 1.2) | 0.7 (0.5, 1.1) | 0 | ||
| Other | 1 | 1.0 (0.6, 1.6) | 1.0 (0.5, 1.7) | 0 | 3 | 1.2 (0.9, 1.6) | 1.2 (0.8, 1.8) | ||
| XPD | XRCC3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lys751Gln | Asp312Asn | Thr241Met | |||||||
| N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
N | ORz (95% CI)* | Ratio of ORs (95%CI) † |
N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
|
| Current-not current smoking | |||||||||
| Case-based categories | |||||||||
| Case-control | |||||||||
| Population-based | 1 | 1.0 (0.7, 1.3) | NA | 0 | 1 | 1.0 (0.7, 1.5) | Ref | ||
| Hospital-based | 1 | 0.7 (0.5, 1.2) | 1 | 1.1 (0.7, 1.9) | NA | 2 | 0.9 (0.6, 1.2) | 0.8 (0.5, 1.4) | |
| Other | 0 | 0 | 1 | 2.0 (0.6, 6.6) | 2.0 (0.6, 6.8) | ||||
| Other | 4 | 1.4 (1.0, 1.8) | 0 | 3 | 0.9 (0.6, 1.2) | 0.8 (0.5, 1.4) | |||
| Non-case-based categories | |||||||||
| Case-control | |||||||||
| Population controls | 1 | 1.0 (0.7, 1.3) | NA | 0 | 1 | 1.0 (0.7, 1.5) | Ref | ||
| Patient controls ‡ | 1 | 0.7 (0.5, 1.2) | 1 | 1.1 (0.7, 1.9) | NA | 2 | 0.9 (0.6, 1.2) | 0.8 (0.5, 1.4) | |
| Non-patient controls § | 0 | 0 | 1 | 2.0 (0.6, 6.6) | 2.0 (0.6, 6.8) | ||||
| Other | 4 | 1.4 (1.0, 1.8) | 0 | 3 | 0.9 (0.6, 1.2) | 0.8 (0.5, 1.4) | |||
| XPD | XRCC3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lys751Gln | Asp312Asn | Thr241Met | |||||||
| N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
N | ORz (95% CI)* | Ratio of ORs (95%CI) † |
N | ORz (95% CI) * | Ratio of ORs (95%CI) † |
|
| Pack-years | |||||||||
| Case-based categories | |||||||||
| Case-control | |||||||||
| Population-based | 2 | 0.8 (0.5, 1.3) | Ref | 0 | 1 | 0.8 (0.4, 1.4) | NA | ||
| Hospital-based | 5 | 1.3 (0.8, 2.1) | 1.5 (0.7, 3.5) | 4 | 1.1 (0.8, 1.5) | NA | 3 | 0.8 (0.5, 1.3) | |
| Other | 0 | 0 | 0 | ||||||
| Non-case-based categories | |||||||||
| Case-control | |||||||||
| Population controls | 2 | 0.8 (0.5, 1.3) | Ref | 0 | 1 | 0.8 (0.4, 1.4) | NA | ||
| Patient controls ‡ | 2 | 1.0 (0.6, 1.6) | 1.2 (0.5, 2.6) | 1 | 0.9 (0.5, 1.4) | NA | 2 | 0.8 (0.5, 1.3) | |
| Non-patient controls § | 3 | 1.6 (0.8, 3.1) | 2.0 (0.9, 4.3) | 3 | 1.3 (0.9, 1.8) | 1 | 1.1 (0.4, 2.7) | ||
Abbreviations: CI=Confidence interval, HWE = Hardy Weinberg equilibrium, MAF=minor allele frequency, na=not applicable, ORz=control-only genotype-smoking odds ratio, PY=pack-years, N=number of studies, Ref=referent, Q=Cochran’s test of homogeneity, Arg=Arginine, Gln=Glutamine, Trp=Tryptophan, His=Histidine, Met=methionine, Asp=Aspartic acid, Asn=Asparagine
Unadjusted OR (95% CI): Genotype contrast for all SNPs is A/A (ref) vs. any a, where A is the more common allele and a is the less common; random effects for XRCC1 Arg399Gln ever-never & XPD Lys751Gln, fixed effects for others
Ratio of Odds Ratios: Meta-regression used to compare odds ratios in given study design stratum to the odds ratio in the designated reference stratum
Patient controls: controls are persons attending a hospital or disease clinic for treatment or diagnosis, does not include patients at wellness or check-up clinics
Non-patient controls: Case-control study participants who are not patients (i.e. not treated at hospital or disease clinic); they may be friend and family controls, cohort members in a nested case-control etc.; also excludes population-based controls
Table 4.
XRCC1 Arg399Gln and Smoking: Overall and by study characteristics
| Ever-never * | Current-Not current † | PY ‡,§ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) □ |
N | Q p-value |
ORz (95% CI) |
Ratio of ORs (95% CI) |
N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) |
|
| Overall | ||||||||||||
| Not stratified by ethnicity w/in study | 21 | 0.01 | 11 | 0.4 | 9 | 0.3 | ||||||
| Stratified by ethnicity within study ** | 23 | 0.02 | 12 | 0.3 | na | |||||||
| By Study Characteristic | ||||||||||||
| Continent | ||||||||||||
| North America | 7 | 0.01 | 1.1 (0.9, 1.3) | Ref | 2 | 0.6 | 1.2 (1.0, 1.5) | Ref | 2 | 0.1 | 1.2 (0.8, 1.7) | 1.1 (0.7, 1.7) |
| Europe | 11 | 0.2 | 1.1 (0.9, 1.3) | 1.0 (0.8, 1.3) | 5 | 0.5 | 0.8 (0.6, 1.0) | 0.7 (0.5, 0.9) | 5 | 0.8 | 1.1 (0.9, 1.4) | Ref |
| Asia | 3 | 0.1 | 1.1 (0.7, 1.9) | 1.0 (0.7, 1.6) | 4 | 0.7 | 1.0 (0.8, 1.3) | 0.8 (0.6, 1.1) | 2 | 0.6 | 1.9 (1.2, 2.9) | 1.7 (1.0, 2.7) |
| Ethnicity/nationality ** | ||||||||||||
| Single-ethnicity studies **,†† | 14 | 0.3 | 1.0 (0.9, 1.2) | Ref | 4 | 0.1 | 1.0 (0.8, 1.2) | Ref | 5 | 0.4 | 1.1 (0.9, 1.5) | Ref |
| Multi-ethnic studies †† | 2 | 0.01 | 1.2 (0.7, 2.1) | 1.2 (0.9, 1.7) | 1 | na | 0.9 (0.2, 3.4) | 0.9 (0.2, 3.5) | 0 | |||
| Unknown ethnicity | 7 | 0.1 | 1.1 (0.9, 1.3) | 1.1 (0.8, 1.4) | 7 | 0.6 | 0.9 (0.7, 1.1) | 0.9 (0.6, 1.3) | 4 | 0.2 | 1.3 (1.1, 1.7) | 1.2 (0.8, 1.8) |
| White >=99% ‡‡ | 10 | 0.6 | 1.0 (0.9, 1.1) | Ref | 2 | 0.2 | 0.9 (0.6, 1.2) | na | 5 | 0.4 | 1.1 (0.9, 1.5) | na |
| African American >=99% | 2 | 0.03 | 1.1 (0.5, 2.5) | 1.1 (0.7, 1.8) | 1 | na | 2.1 (1.1, 3.9) | 0 | ||||
| Han >=99% | 2 | 0.2 | 1.4 (0.8, 2.5) | 1.5 (0.8, 2.5) | 1 | na | 1.0 (0.7, 1.4) | 0 | ||||
| Multi-ethnic studies | 2 | 0.01 | 1.2 (0.7, 2.1) | 1.3 (0.9, 1.8) | 1 | na | 0.9 (0.2, 3.4) | 0 | ||||
| HWE p-value ** | ||||||||||||
| Single-ethnicity (continuous) | 13 | 0.2 | 1.0 (0.9, 1.2) | 0.8 (0.5, 1.3) | 4 | 0.1 | 1.0 (0.8, 1.2) | 1.3 (0.2, 7.2) | 5 | 0.4 | 1.1 (0.9, 1.5) | 16.9 (0.4, 713) |
| p <0.05 | 0 | |||||||||||
| p< 0.10 | 1 | --- | 0.9 (0.6, 1.3) | na | 0 | 0 | ||||||
| p>=0.10 | 21 | 0.02 | 1.1 (1.0, 1.2) | 12 | 9 | |||||||
| Age | ||||||||||||
| Age non-missing | 20 | 0.01 | 1.1 (1.0, 1.2) | 1.0 (1.0, 1.0) | 10 | 0.3 | 1.0 (0.9, 1.1) | 1.0 (1.0, 1.0) | 9 | 0.3 | 1.2 (1.0, 1.5) | 1.0 (1.0, 1.1) |
| <= 47.9y §§ | 1 | --- | 0.8 (0.4, 1.6) | na | 5 | 0.3 | 0.9 (0.7, 1.1) | Ref | 0 | na | ||
| > 47.9 y | 19 | 0.01 | 1.1 (1.0, 1.2) | 5 | 0.4 | 1.1 (0.9, 1.3) | 1.2 (0.9, 1.7) | 9 | 0.3 | 1.2 (1.0, 1.5) | ||
| <=59y ** | 8 | 0.1 | 1.1 (0.9, 1.3) | Ref | 8 | 0.6 | 0.9 (0.7, 1.1) | Ref | 3 | 0.1 | 1.1 (0.8, 1.6) | Ref |
| >59y | 12 | 0.02 | 1.1 (0.9, 1.3) | 1.0 (0.8, 1.3) | 2 | 0.2 | 1.1 (0.9, 1.4) | 1.3 (1.0, 1.7) | 6 | 0.4 | 1.3 (1.0, 1.6) | 1.2 (0.8, 1.8) |
| At or below median | 12 | 0.1 | 1 (0.9, 1.2) | Ref | 5 | 0.3 | 0.9 (0.7, 1.1) | Ref | 5 | 0.4 | 1.2 (1.0, 1.5) | Ref |
| Above median | 8 | 0.02 | 1.2 (0.9, 1.4) | 1.1 (0.9, 1.4) | 5 | 0.4 | 1.1 (0.9, 1.3) | 1.2 (0.9, 1.7) | 4 | 0.2 | 1.3 (1.0, 1.8) | 1.1 (0.7, 1.6) |
| Gender | ||||||||||||
| Percent male (mixed gender only) | 13 | 0.1 | 1.0 (0.9, 1.2) | 1.4 (0.6, 3.5) | 4 | 0.7 | 1.0 (0.8, 1.3) | 0.9 (0.1, 8) | 6 | 0.1 | 1.3 (1.0, 1.5) | 1 (0.1, 11.4) |
| All female | 5 | 0.01 | 1.1 (0.8, 1.4) | Ref | 2 | 0.3 | 1.2 (0.9, 1.5) | Ref | 1 | -- | 0.9 (0.4, 1.8) | Ref |
| Mixed gender | 13 | 0.1 | 1.0 (0.9, 1.2) | 1.0 (0.7, 1.3) | 4 | 0.7 | 1.0 (0.8, 1.3) | 0.9 (0.6, 1.2) | 6 | 0.1 | 1.3 (1.0, 1.5) | 1.4 (0.7, 2.9) |
| All male | 3 | 0.7 | 1.2 (0.9, 1.6) | 1.1 (0.7, 1.7) | 4 | 0.7 | 0.7 (0.6, 1.0) | 0.6 (0.4, 0.9) | 2 | 0.7 | 1.3 (0.8, 2.2) | 1.5 (0.6, 3.6) |
| Study outcome | ||||||||||||
| Lung cancer | 6 | 0.4 | 1.0 (0.9, 1.1) | Ref | 1 | --- | 0.9 (0.6, 1.3) | Ref | 6 | 0.2 | 1.3 (1.1, 1.6) | Ref |
| Other cancer | 12 | 0.02 | 1.2 (1.0, 1.4) | 1.2 (1.0, 1.6) | 3 | 0.3 | 1.1 (0.9, 1.3) | 1.2 (0.8, 2) | 3 | 0.7 | 1.0 (0.7, 1.5) | 0.8 (0.5, 1.2) |
| Non-cancer disease | 0 | 1 | --- | 1.6 (0.6, 3.7) | 1.8 (0.7, 4.7) | 0 | ||||||
| Non-disease | 3 | 0.7 | 1.0 (0.8, 1.4) | 1.1 (0.7, 1.6) | 6 | 0.6 | 0.8 (0.6, 1.1) | 0.9 (0.6, 1.5) | 0 | |||
| Lung cancer | 6 | 0.4 | 1.0 (0.9, 1.1) | Ref | 1 | --- | 0.9 (0.6, 1.3) | na | 6 | 0.2 | 1.3 (1.1, 1.6) | Ref |
| All other | 15 | 0.04 | 1.2 (1.0, 1.3) | 1.2 (1.0, 1.5) | 10 | 0.3 | 1 (0.9, 1.2) | 3 | 0.7 | 1 (0.7, 1.5) | 0.8 (0.5, 1.2) | |
| MAF † | ||||||||||||
| MAF (cutpoints assigned by tertiles across SNP) | ||||||||||||
| 0.10–0.27 | 6 | 0.1 | 1.1 (0.7, 1.5) | Ref | 4 | 0.1 | 1.1 (0.9, 1.4) | Ref | 2 | 0.6 | 1.9 (1.2, 2.9) | Ref |
| >0.27–0.36 | 10 | 0.01 | 1.1 (1.0, 1.3) | 1.1 (0.8, 1.5) | 4 | 0.4 | 0.9 (0.6, 1.4) | 0.8 (0.5, 1.4) | 7 | 0.5 | 1.1 (0.9, 1.4) | 0.6 (0.4, 1.0) |
| >0.36–0.50 | 6 | 0.9 | 1 (0.8, 1.2) | 1 (0.7, 1.4) | 4 | 0.7 | 0.8 (0.7, 1.1) | 0.8 (0.5, 1.1) | 0 | |||
| MAF-assigned ethnicity | ||||||||||||
| White | 15 | 0.3 | 1.1 (0.9, 1.2) | Ref | 6 | 0.5 | 0.9 (0.7, 1.1) | Ref | 7 | 0.5 | 1.1 (0.9, 1.4) | Ref |
| African American ‡‡‡ | 3 | 0.1 | 1 (0.5, 1.8) | 0.9 (0.6, 1.5) | 1 | 2.1 (1.1, 3.9) | 2.4 (1.2, 4.7) | 0 | ||||
| Han | 3 | 0.1 | 1.1 (0.7, 1.9) | 1.0 (0.7, 1.5) | 4 | 0.7 | 1.0 0.8, 1.3) | 1.1 (0.8, 1.6) | 2 | 0.6 | 1.9 (1.2, 2.9) | 1.6 (1.0, 2.6) |
| Multi-ethnic studies | 2 | 0.01 | 1.2 (0.7, 2.1) | 1.2 (0.8, 1.7) | 1 | -- | 0.9 0.2, 3.4) | 1 (0.3, 4.1) | 0 | |||
| Smoking prevalence §§§ | ||||||||||||
| Continuous | 21 | 0.01 | 1.7 (0.6, 4.7) | 11 | 0.4 | 1.0 (0.9, 1.1) | 0.2 (0.1, 0.7) | 9 | 0.3 | 1.2 (1.0, 1.5) | 0.7 (0.2, 2.6) | |
| >0–0.507 | 5 | 0.2 | 1 (0.8, 1.2) | Ref | 9 | 0.5 | 1.0 (0.9, 1.2) | Ref | 6 | 0.4 | 1.3 (1.1, 1.6) | Ref |
| >0.507–1 | 16 | 0.01 | 1.1 (1.0, 1.3) | 1.1 (0.9, 1.5) | 2 | 0.8 | 0.7 (0.5, 1.1) | 0.7 (0.5, 1.1) | 3 | 0.3 | 0.9 (0.6, 1.4) | 0.7 (0.4, 1.1) |
Abbreviations: CI=Confidence interval, na=not applicable, HWE = Hardy Weinberg equilibrium PY=pack-years, ORz=control-only genotype-smoking odds ratio (bolded), N=number of studies, Ref=referent, Q=Cochran’s test of homogeneity, SNP=single nucleotide polymorphism, MAF=minor allele frequency, Arg=Arginine, Gln=Glutamine, Trp=Tryptophan, His=Histidine, Met=methionine, Asp=Aspartic acid, Asn=Asparagine
OR=Unadjusted odds ratio for XRCC1 Arg399Gln: XRCC1 Arg/Arg (ref) vs. any Gln, never moking (ref) vs. ever smoking, random effects estimates
OR=Unadjusted odds ratio for XRCC1 Arg399Gln: XRCC1 Arg/Arg (ref) vs. any Gln, not current smoker (ref) vs. current smoker, fixed effects estimates
OR=Unadjusted odds ratio for XRCC1 Arg399Gln: XRCC1 Arg/Arg (ref) vs. any Gln, lightest smokers (ref) vs. heaviest smokers [lightest excludes never smokers], fixed effects
PY contrast is between lightest non-zero category of pack-years (ref) vs. heaviest category of PY
Ratio of Odds Ratios: Compares odds ratio in given study characteristic stratum to the odds ratio in the designated reference stratum for that study characteristic by meta-regression
Studies that can be stratified by ethnicity are included as separate single-ethnicity studies; studies w 99%–100% of 1 ethnicity are classified as single-ethnicity
Only includes studies with explicitly stated ethnic makeup
White = Caucasian, white or non-Hispanic white; African American = African American or black; Han = Han, Han Chinese or ethnic Chinese; Japan, Korea and China may include ethnic minorities.
Median of studies included in XRCC1 399 current/not current smoker analyses.
Categories based on thirds from studies included in XRCC1 399 PY analyses (<=59y, >59y–63y, >63y)
Median of all studies w age info (all SNPs) w age info [range:23.6–69y, mean: 56.51y SD: 9.84y]
Median proportion male in all studies (all SNPs, all smoking exposures): 0.69
For 399 ever-never 1 study from Hungary is included using MAF as proxy for ethnicity
Smoking prevalence is contrast-specific (defined as "ever", "current" or "heavier PY" as appropriate)
Table 5.
XPD Lys751Gln and Smoking: Overall and by study characteristics
| Ever-never * | Current-Not current † | PY ‡,§ | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) □ |
N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) |
N | Q p-value |
ORz (95% CI) |
Ratio of ORs (95% CI) |
||
| Overall | |||||||||||||
| 12 | 0.5 | 0.9 (0.8, 1.1) | 6 | 0.2 | 1.1 (0.9, 1.3) | 7 | 0.019 | 1.2 (1.0, 1.5) | |||||
| By Study Characteristic | |||||||||||||
| Continent | |||||||||||||
| North America | 4 | 0.4 | 0.9 (0.8, 1) | Ref | 1 | --- | 0.9 (0.7, 1.3) | Ref | 3 | 0.006 | 1.5 (0.7, 3.2) | Ref | |
| Europe | 6 | 0.3 | 1.0 (0.8, 1.3) | 1.2 (0.9, 1.5) | 5 | 0.2 | 1.2 (0.9, 1.5) | 1.2 (0.8, 1.8) | 3 | 0.503 | 1.0 (0.7, 1.4) | 0.7 (0.3, 1.5) | |
| Asia | 2 | 0.5 | 1.0 (0.6, 1.7) | 1.1 (0.7, 1.9) | 0 | 1 | na | 0.8 (0.3, 1.7) | 0.5 (0.1, 1.9) | ||||
| Ethnicity/nationality | |||||||||||||
| Single-ethnicity studies ** | 6 | 0.3 | 1.0 (0.8, 1.2) | Ref | 1 | --- | 1.2 (0.8, 1.8) | na | 5 | 0.097 | 1.4 (0.9, 2.1) | na | |
| Multi-ethnic studies †† | 2 | 0.6 | 0.8 (0.7, 1.0) | 0.8 (0.6, 1.1) | 1 | --- | 0.9 (0.7, 1.3) | 1 | --- | 0.8 (0.5, 1.3) | |||
| Unknown ethnicity | 4 | 0.7 | 1.0 (0.8, 1.3) | 1.1 (0.8, 1.4) | 4 | 0.1 | 1.1 (0.8, 1.5) | 1 | --- | 0.8 (0.4, 1.4) | |||
| White >=99% ‡‡ | 4 | 0.2 | 1.0 (0.8, 1.2) | Ref | 1 | --- | 1.2 (0.8, 1.8) | na | 4 | 0.181 | 1.6 (1.0, 2.4) | na | |
| African American >=99% | 0 | 1.0 (0.6, 1.8) | 0 | 0 | |||||||||
| Han >=99% | 2 | 0.5 | 1.0 (0.6, 1.7) | 0.8 (0.6, 1.1) | 0 | 1 | --- | 0.8 (0.3, 1.7) | |||||
| HWE p-value | |||||||||||||
| Continuous (single ethnicity) | 6 | 0.3 | 1.0 (0.8, 1.2) | 0.8 (0.5, 1.2) | 1 | --- | 1.2 (0.8, 1.8) | na | 5 | 0.097 | 1.4 (0.9, 2.1) | 2.2 (0, 93.7) | |
| HWE p <0.05 | 1 | --- | 1.1 (0.8, 1.7) | Ref | 2 | 0.6 | 1.3 (0.9, 1.8) | Ref | 0 | na | |||
| HWE p <0.10 | 2 | 0.9 | 1.1 (0.8, 1.5) | Ref | 2 | 0.6 | 1.3 (0.9, 1.8) | Ref | 1 | --- | 0.8 (0.4, 1.4) | na | |
| HWE p >=0.10 | 10 | 0.4 | 0.9 (0.8, 1.0) | 0.8 (0.6, 1.1) | 4 | 0.1 | 1.0 (0.8, 1.3) | 0.8 (0.5, 1.2) | 6 | 0.022 | 1.2 (0.8, 1.9) | ||
| Age | |||||||||||||
| Age non-missing (continuous) | 12 | 0.5 | 0.9 (0.8, 1.1) | 1.0 (1.0, 1.0) | 6 | 0.2 | 1.1 (0.9, 1.3) | 1.0 (1.0, 1.0) | 7 | 0.019 | 1.1 (0.8, 1.7) | 1.0 (0.9, 1.1) | |
| <= 47.9y §§ | 0 | na | 3 | 0.5 | 1.4 (1.0, 1.9) | Ref | 0 | na | |||||
| > 47.9 y | 12 | 0.5 | 0.9 (0.8, 1.1) | 0 (0, 0) | 3 | 0.4 | 0.9 (0.7, 1.2) | 0.7 (0.5, 1.0) | 7 | 0.019 | 1.1 (0.8, 1.7) | ||
| <=59y *** | 7 | 0.6 | 0.9 (0.8, 1.0) | Ref | 6 | 0.2 | 1.1 (0.9, 1.3) | na | 4 | 0.054 | 1.4 (0.8, 2.4) | Ref | |
| >59y | 5 | 0.2 | 1.0 (0.8, 1.2) | 1.1 (0.9, 1.4) | 0 | 3 | 0.362 | 0.9 (0.6, 1.2) | 0.6 (0.3, 1.2) | ||||
| At or below median | 6 | 0.5 | 0.9 (0.8, 1.1) | Ref | 3 | 0.5 | 1.4 (1.0, 1.9) | Ref | 4 | 0.054 | 1.4 (0.8, 2.4) | Ref | |
| Above median | 6 | 0.3 | 1.0 (0.8, 1.1) | 1.1 (0.9, 1.4) | 3 | 0.4 | 0.9 (0.7, 1.2) | 0.7 (0.5, 1.0) | 3 | 0.362 | 0.9 (0.6, 1.2) | 0.6 (0.3, 1.2) | |
| Gender | |||||||||||||
| Percent male, mixed gender only | 8 | 0.9 | 1.0 (0.9, 1.2) | 0.8 (0.3, 2.4) | 2 | 0.3 | 1.5 (1.0, 2.3) | na | 5 | 0.006 | 1.2 (0.7, 2) | 0 (0, 0.1) | |
| All female | 2 | 0.1 | 0.9 (0.7, 1.1) | Ref | 1 | --- | 0.9 (0.7, 1.3) | Ref | 1 | na | 0.9 (0.4, 1.9) | na | |
| Mixed gender | 8 | 0.9 | 1.0 (0.9, 1.2) | 1.1 (0.9, 1.5) | 2 | 0.3 | 1.5 (1.0, 2.3) | 1.6 (0.9, 2.7) | 5 | 0.006 | 1.2 (0.7, 2) | ||
| All male | 2 | 0.2 | 0.7 (0.5, 1.1) | 0.8 (0.5, 1.3) | 3 | 0.2 | 1.0 (0.8, 1.4) | 1.1 (0.7, 1.7) | 1 | na | 1.3 (0.7, 2.5) | ||
| Study outcome | |||||||||||||
| Lung cancer | 3 | 0.5 | 1.0 (0.8, 1.2) | Ref | 0 | 3 | 0.080 | 1.6 (0.8, 3.1) | Ref | ||||
| Other cancer | 8 | 0.2 | 0.9 (0.8, 1.1) | 0.9 (0.7, 1.2) | 2 | 0.4 | 0.9 (0.7, 1.2) | Ref | 4 | 0.565 | 0.9 (0.7, 1.2) | 0.5 (0.3, 0.9) | |
| Non-cancer disease | 0 | 0 | 0 | ||||||||||
| Non-disease | 1 | --- | 1.0 (0.6, 1.6) | 1.0 (0.6, 1.7) | 4 | 0.7 | 1.3 (1.0, 1.8) | 1.5 (1.0, 2.2) | 0 | ||||
| Lung cancer | 3 | 0.5 | 1.0 (0.8, 1.2) | Ref | 0 | na | 3 | 0.080 | 1.6 (0.8, 3.1) | Ref | |||
| All other | 9 | 0.3 | 0.9 (0.8, 1.1) | 0.9 (0.7, 1.2) | 6 | 0.2 | 1.1 (0.9, 1.3) | 4 | 0.565 | 0.9 (0.7, 1.2) | 0.5 (0.3, 0.9) | ||
| MAF | |||||||||||||
| MAF non-missing | 12 | 0.5 | 0.9 (0.8, 1.1) | 1.3 (0.3, 6.2) | 6 | 0.2 | 1.1 (0.9, 1.3) | 0 (0, 3772.6) | 7 | 1.2 (0, 45.7) | |||
| MAF (cutpoints assigned by median across SNP) | |||||||||||||
| 0.01–0.37 | 7 | 0.5 | 0.9 (0.8, 1.0) | Ref | 2 | 0.3 | 1.0 (0.7, 1.3) | Ref | 5 | 0.016 | 1.3 (0.8, 2.1) | Ref | |
| >0.37–0.50 | 5 | 0.7 | 1.1 (0.9, 1.4) | 1.2 (1.0, 1.6) | 4 | 0.2 | 1.1 (0.9, 1.5) | 1.1 (0.7, 1.8) | 2 | 0.768 | 0.8 (0.5, 1.3) | 0.7 (0.3, 1.5) | |
| MAF-assigned ethnicity | |||||||||||||
| White | 8 | 0.4 | 1.0 (0.9, 1.2) | Ref | 5 | 0.2 | 1.2 (0.9, 1.5) | na | 5 | 0.052 | 1.3 (0.9, 2.1) | na | |
| African American | 0 | 0 | 0 | ||||||||||
| Han | 2 | 0.5 | 1.0 (0.6, 1.7) | 1.0 (0.6, 1.7) | 0 | na | 1 | --- | 0.8 (0.3, 1.7) | na | |||
| Multi-ethnic studies | 2 | 0.6 | 0.8 (0.7, 1.0) | 0.8 (0.6, 1.0) | 1 | --- | 0.9 (0.7, 1.3) | 1 | --- | 0.8 (0.5, 1.3) | |||
| Smoking prevalence ‡‡‡ | |||||||||||||
| Continuous | 12 | 0.5 | 0.9 (0.8, 1.1) | 0.3 (0.1, 1.1) | 6 | 0.2 | 1.1 (0.9, 1.3) | 2 (0.5, 8.6) | 7 | 0.3 (0, 7.8) | |||
| >0–0.507 | 3 | 0.9 | 1.2 (0.9, 1.6) | Ref | 5 | 0.2 | 1.1 (0.9, 1.3) | na | 5 | 0.010 | 1.0 (0.6, 1.6) | Ref | |
| >0.507–1 | 9 | 0.5 | 0.9 (0.8, 1.0) | 0.7 (0.5, 1.0) | 1 | --- | 1.2 (0.8, 1.8) | 2 | 0.359 | 1.5 (0.9, 2.6) | 1.6 (0.7, 3.9) | ||
Abbreviations: CI=Confidence interval, na=not applicable, HWE = Hardy Weinberg equilibrium, PY=pack-years, ORz=control-only genotype-smoking odds ratio,), N=number of studies, Ref=referent, Q=Cochran’s test of homogeneity, SNP=single nucleotide polymorphism, Arg=Arginine, Gln=Glutamine, Trp=Tryptophan, His=Histidine, Met=methionine, Asp=Aspartic acid, Asn=Asparagine
OR=Unadjusted odds ratio for XPD Lys751Gln: Lys/Lys (ref) vs. any Gln, never smoking (ref) vs. ever smoking, fixed effects
OR=Unadjusted odds ratio for XPD Lys751Gln: Lys/Lys (ref) vs. any Gln, not current smoker (ref) vs. current smoker, fixed effects
OR=Unadjusted odds ratio for XPD Lys751Gln: Lys/Lys (ref) vs. any Gln, lightest non-zero smokers (ref) vs. heaviest smokers; stratified random effects
PY contrast is between lightest non-zero category of pack-years (ref) vs. heaviest category of PY
Ratio of Odds Ratios: Compares OR in given study characteristic stratum to the OR in the designated reference stratum for that study characteristic by meta-regression
Studies w 99%–100% of 1 ethnicity are classified as single-ethnicity
Only includes studies with explicitly stated ethnic makeup
White = Caucasian, white or non-Hispanic white; African American = African American or black; Han = Han, Han Chinese or ethnic Chinese
Median of studies included in XRCC1 751 current/not current smoker analyses.
Categories based on thirds from studies included in XRCC1 399 PY analyses (<=59y, >59y–63y, >63y)
Median of all studies w age info (all SNPs) w age info [range:23.6–69y, mean: 56.7y SD: 9.8y]
Median proportion male in all studies (all SNPs, all smoking exposures): 0.69
Smoking prevalence is contrast-specific (defined as "ever", "current" or "heavier PY" as appropriate)
Table 6.
XRCC3 Thr241Met and Smoking: Overall and by study characteristics
| Ever-never * | Current-Not current † | PY ‡,§ | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) □ |
N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) □ |
N | Q p- value |
ORz (95% CI) |
Ratio of ORs (95% CI) □ |
||
| Overall | |||||||||||||
| 9 | 0.5 | 1.0 (0.9, 1.2) | 7 | 0.7 | 0.9 (0.8, 1.1) | 4 | 0.7 | 0.8 (0.6, 1.2) | |||||
| By Study Characteristic | |||||||||||||
| Continent | |||||||||||||
| North America | 2 | 0.1 | 0.9 (0.6, 1.3) | Ref | 1 | --- | 0.8 (0.5, 1.2) | na | 2 | 0.3 | 0.7 (0.4, 1.3) | Ref | |
| Europe | 7 | 0.7 | 1.1 (0.9, 1.3) | 1.1 (0.8, 1.7) | 5 | 0.9 | 0.9 (0.7, 1.1) | 2 | 0.6 | 0.9 (0.6, 1.4) | 1.2 (0.6, 2.5) | ||
| Asia | 0 | 1 | --- | 2.0 (0.6, 6.6) | 0 | ||||||||
| Ethnicity/nationality | |||||||||||||
| Single-ethnicity studies ** | 3 | 0.7 | 0.8 (0.6, 1.1) | Ref | 3 | 0.3 | 0.8 (0.6, 1.1) | Ref | 2 | 0.9 | 1.0 (0.6, 1.7) | na | |
| Multi-ethnic studies †† | 2 | 0.2 | 1.0 (0.7, 1.3) | 1.2 (0.8, 1.9) | 1 | --- | 1.0 (0.7, 1.5) | 1.2 (0.7, 2) | 1 | --- | 0.6 (0.3, 1.2) | ||
| Unknown ethnicity | 4 | 0.9 | 1.2 (1.0, 1.5) | 1.5 (1.0, 2.2) | 3 | 0.7 | 0.9 (0.7, 1.3) | 1.1 (0.7, 1.7) | 1 | --- | 0.8 (0.4, 1.4) | ||
| White >=99% ‡‡ | 3 | 0.7 | 0.8 (0.6, 1.1) | Ref | 2 | 0.9 | 0.8 (0.6, 1.1) | Ref | 2 | 0.9 | 1.0 (0.6, 1.7) | na | |
| Han >=99% | 0 | 1 | --- | 2.0 (0.6, 6.6) | 0 | ||||||||
| HWE p-value | |||||||||||||
| Continuous (single ethnicity) | 3 | 0.7 | 0.8 (0.6, 1.1) | 1.0 (0.3, 3.4) | 3 | 0.3 | 0.8 (0.6, 1.1) | 0.4 (0.1, 1.6) | 2 | 0.9 | 1.0 (0.6, 1.7) | ||
| HWE p <0.05 | 0 | 1 | --- | 2.0 (0.6, 6.6) | na | 0 | na | ||||||
| HWE p <0.10 | 0 | na | 1 | --- | 2.0 (0.6, 6.6) | na | 0 | na | |||||
| HWE p >=0.10 | 9 | 0.5 | 1.0 (0.9, 1.2) | 6 | 0.9 | 0.9 (0.7, 1.1) | 4 | 0.7 | 0.8 (0.6, 1.2) | ||||
| Age | |||||||||||||
| Age non-missing | 9 | 0.5 | 1.0 (0.9, 1.2) | 1.0 (1.0, 1.0) | 7 | 0.7 | 0.9 (0.8, 1.1) | 0.9 (0.8, 1.1) | 4 | 0.7 | 0.8 (0.6, 1.2) | 0.9 (0.8, 1.1) | |
| <= 47.9y §§ | 1 | --- | na | 2 | 0.6 | 0.8 (0.6, 1.1) | Ref | 0 | na | ||||
| > 47.9 y | 8 | 0.4 | 1.0 (0.9, 1.2) | 5 | 0.6 | 1.0 (0.8, 1.2) | 1.2 (0.8, 1.9) | 4 | 0.7 | 0.8 (0.6, 1.2) | |||
| <=59y *** | 5 | 0.3 | 1 (0.8, 1.2) | Ref | 6 | 0.9 | 0.9 (0.7, 1.1) | na | 1 | --- | 1.1 (0.4, 2.7) | na | |
| >59y | 4 | 0.7 | 1.1 (0.9, 1.5) | 1.2 (0.8, 1.6) | 1 | --- | 2.0 (0.6, 6.6) | 3 | 0.5 | 0.8 (0.5, 1.1) | |||
| At or below median | 5 | 0.3 | 1.0 (0.8, 1.2) | Ref | 4 | 0.8 | 0.8 (0.7, 1.1) | Ref | 3 | 0.8 | 0.9 (0.6, 1.4) | na | |
| Above median | 4 | 0.7 | 1.1 (0.9, 1.5) | 1.2 (0.8, 1.6) | 3 | 0.5 | 1.0 (0.8, 1.4) | 1.2 (0.8, 1.8) | 1 | --- | 0.6 (0.3, 1.2) | ||
| Gender | |||||||||||||
| Percent male, mixed gender only | 6 | 0.3 | 1 (0.9, 1.3) | 1.1 (0.1, 12.6) | 4 | 0.5 | 1.0 (0.8, 1.3) | 0.1 (0, 3.4) | |||||
| All female | 0 | 0 | 0 | ||||||||||
| Mixed gender | 6 | 0.3 | 1.0 (0.9, 1.3) | Ref | 4 | 0.5 | 1.0 (0.8, 1.3) | Ref | 3 | 0.6 | 0.8 (0.5, 1.1) | na | |
| All male | 3 | 0.8 | 1.0 (0.8, 1.4) | 1.0 (0.7, 1.4) | 3 | 0.8 | 0.8 (0.6, 1.1) | 0.9 (0.6, 1.3) | 1 | --- | 1.0 (0.5, 1.9) | ||
| Study outcome | |||||||||||||
| Lung cancer | 0 | 0 | 1 | --- | 1.1 (0.4, 2.7) | na | |||||||
| Other cancer | 5 | 0.4 | 1 (0.8, 1.2) | Ref | 4 | 0.5 | 1.0 (0.7, 1.2) | Ref | 3 | 0.5 | 0.8 (0.5, 1.1) | ||
| Non-cancer disease | 0 | 0 | 0 | ||||||||||
| Non-disease | 4 | 0.7 | 1.2 (0.9, 1.6) | 1.2 (0.8, 1.7) | 3 | 0.7 | 0.9 (0.6, 1.2) | 0.9 (0.6, 1.3) | 0 | ||||
| Lung cancer | 0 | 0 | na | 1 | --- | 1.1 (0.4, 2.7) | na | ||||||
| All other | 9 | 0.5 | 1.0 (0.9, 1.2) | na | 6 | 0.7 | 0.9 (0.8, 1.1) | 3 | 0.5 | 0.8 (0.5, 1.1) | |||
| MAF | |||||||||||||
| MAF non-missing | 9 | 0.5 | 1.0 (0.9, 1.2) | 0.9 (0, 235.8) | 7 | 0.7 | 0.9 (0.8, 1.1) | 0.2 (0, 4.6) | 4 | 0.7 | 0.8 (0.6, 1.2) | 0 (0, 45050) | |
| MAF (cutpoints assigned by median across SNP) | |||||||||||||
| 0.01–0.37 | 5 | 0.3 | 1.0 (0.8, 1.3) | Ref | 2 | 0.1 | 0.9 (0.6, 1.4) | Ref | 4 | 0.7 | 0.8 (0.6, 1.2) | na | |
| >0.37–0.50 | 4 | 0.5 | 1.0 (0.8, 1.3) | 1.0 (0.7, 1.4) | 5 | 0.9 | 0.9 (0.7, 1.1) | 1.0 (0.6, 1.7) | 0 | ||||
| MAF-assigned ethnicity | |||||||||||||
| White | 7 | 0.5 | 1.1 (0.9, 1.3) | Ref | 5 | 0.9 | 0.9 (0.7, 1.1) | na | 3 | 0.8 | 0.9 (0.6, 1.4) | na | |
| Han | 0 | 1 | --- | 2.0 (0.6, 6.6) | 0 | ||||||||
| Multi-ethnic studies | 2 | 0.2 | 1.0 (0.7, 1.3) | 0.9 (0.7, 1.3) | 1 | --- | 1.0 (0.7, 1.5) | 1 | --- | 0.6 (0.3, 1.2) | |||
| Smoking prevalence 13 | |||||||||||||
| Continuous | 9 | 0.5 | 1.0 (0.9, 1.2) | 0.1 (0, 1.8) | 7 | 0.7 | 0.9 (0.8, 1.1) | 0.5 (0.1, 3.1) | 4 | 0.7 | 0.8 (0.6, 1.2) | 2.4 (0.1, 56.6) | |
| >0–0.507 | 0 | na | 6 | 0.7 | 0.9 (0.8, 1.2) | na | 2 | 0.5 | 0.7 (0.4, 1.1) | na | |||
| >0.507–1 | 9 | 0.5 | 1.0 (0.9, 1.2) | 1 | --- | 0.8 (0.6, 1.2) | 2 | 0.9 | 1.0 (0.6, 1.7) | ||||
Abbreviations: CI=Confidence interval, na=not applicable, HWE = Hardy Weinberg equilibrium, PY=pack-years, ORz=control-only genotype-smoking odds ratio,), N=number of studies, Ref=referent, Q=Cochran’s test of homogeneity, SNP=single nucleotide polymorphism, Arg=Arginine, Gln=Glutamine, Trp=Tryptophan, His=Histidine, Met=methionine, Asp=Aspartic acid, Asn=Asparagine
OR=Unadjusted odds ratio for XRCC3 Thr241Met: Thr/Thr (ref) vs. any Met, never smoking (ref) vs. ever smoking, fixed effects
OR=Unadjusted odds ratio for XRCC3 Thr241Met: Thr/Thr (ref) vs. any Met, not current smoker (ref) vs. current smoker, fixed effects
OR=Unadjusted odds ratio for XRCC3 Thr241Met: Thr/Thr (ref) vs. any Met, lightest non-zero smokers (ref) vs. heaviest smokers; stratified random effects
PY contrast is between lightest non-zero category of pack-years (ref) vs. heaviest category of PY
Ratio of Odds Ratios: Compares odds ratio in given study characteristic stratum to the odds ratio in the designated reference stratum for that study characteristic by meta-regression
Studies w 99%–100% of 1 ethnicity are classified as single-ethnicity
Only includes studies with explicitly stated ethnic makeup
White = Caucasian, white or non-Hispanic white; African American = African American or black; Han = Han, Han Chinese or ethnic Chinese
Median of studies included in XRCC1 751 current/not current smoker analyses.
Categories based on thirds from studies included in XRCC1 399 PY analyses (<=59y, >59y–63y, >63y)
Median of all studies w age info (all SNPs) w age info [range:23.6–69y, mean: 56.7y SD: 9.8y]
Median proportion male in all studies (all SNPs, all smoking exposures): 0.69
Smoking prevalence is contrast-specific (defined as "ever", "current" or "heavier PY" as appropriate)
For PY, lung cancer studies were above the null for all three SNPs. When compared to studies of other cancers the genotype-smoking association was stronger for lung cancer studies (referent) compared to other cancer studies [ROR= 0.8(0.5, 1.2) and 0.5(0.3, 0.9) for XRCC1 399 and XPD 751, respectively]. All studies with PY were cancer studies. Older average age of study participants weakly but consistently showed stronger associations between ever smoking and variant allele for XRCC1 399, XPD 751 and XRCC3 241 than did younger age. For XRCC1 399 only, this was evident across all three smoking categories. Also, for XRCC1 399 current-not current smokers and PY only, studies with lower minor allele frequencies (N=3) showed stronger associations (~2.0) than those with higher MAF. These three studies had only African-American or Asian participants. No strong and/or consistent patterns emerged for other study characteristics examined.
Discussion
This systematic review and meta-analysis of DNA repair genotypes and smoking behavior in control data was conducted with the goal of examining the independence assumption of case-only studies of gene-environment interaction. There was considerable variation in estimates of Z for XRCC1 399 ever-never smoking and XPD 751 PY of smoking. Point estimates of ORz varied as much as 5-fold, even when studies were homogeneous enough for a summary estimate. Summary estimates for individual SNPs varied across smoking categorizations, with larger magnitudes of association generally found for measures of smoking dose (PY, intensity, duration) than for smoking status (ever-never, current-not current). There was a weak association between XRCC1 399 and higher smoking dose (PY, intensity). No study characteristics examined strongly predicted the magnitude of association although study outcome (lung cancer vs. other cancer for PY), study design (population-based vs. hospital/patient-based), and age warrant further investigation.
Although the validity of case-only estimates rests on the independence assumption (2, 65), literature on independence assumption verification is limited. Data simulations have demonstrated that small violations of the independence assumption can strongly bias the case-only interaction parameter (7). Even an ORz of 1.2 biased the COR by nearly 30%. Further, when Z ≠ 1 in population subgroups, the COR for those subgroups will be biased as well.
However, little empirical work has been conducted to quantitatively assess the magnitude of control-only associations (ORz) between DNA repair gene variations and smoking. A population-based study (N=339) of Japanese males assessed association between ‘habitual smoking’ (ever/never) and a panel of 153 SNPs in 40 candidate genes, including the DNA repair genes OGG1 and NUDT1(MTH1) (66). Association was found between smoking and 3 of 4 of the SNPs in OGG1 (0.4–0.6, borderline statistical significance).
Smoking amount (PY and/or intensity) may be causally associated with variation in XRCC1 399, or with a polymorphism in linkage disequilibrium with XRCC1 399. There is evidence that the XRCC1 399 and XPD 751 variants are functional (67–69). Different aspects of smoking behavior (smoking initiation, smoking cessation, intensity etc.) operate through multiple overlapping pathways (70) therefore would not be expected to be identically affected by DNA repair variation. This is supported by the differing results for smoking status and amount for several SNPs (XRCC1 399, XRCC1 280, XPD 751, XRCC3 241). There is some evidence that variation in DNA repair activity may affect neurological and/or respiratory outcomes, which could in turn affect smoking behavior (71–75). If the variants are functional, or linked to functional variants, heterogeneity could be due to gene-environment interaction in specific populations.
There are also several possible non-causal explanations for these finding. Although publication bias is a concern with meta-analyses, visual inspection of funnel plots and formal tests of asymmetry argue against this. Spurious results for XRCC1 399 and smoking amount could be caused by selection bias in a subsample of studies. Just over half of the studies with smoking amount information for XRCC1 399 were lung cancer studies (8 of 14) and lung cancer studies had on average higher ORzs than other cancer studies for all PY analyses. The connection between smoking and lung cancer is well known, possibly leading to more variation in response rates or recall by smoking history and/or family history of cancer, but the direction of possible bias is unpredictable. The ORz for the one XRCC1 399 study that explicitly excluded participants with smoking-related diseases was essentially the same as the summary estimate (42).
Population stratification could have contributed to the heterogeneity in XRCC1 399 ever-never and XPD 751 PY estimates since the variant alleles are found at different frequencies in different ethnic groups within the same study, and smoking behavior may also differ by ethnicity. Although this cannot be rigorously assessed without individual level data, there were no clear patterns in ORz for any SNP for study-level ethnicity, either by stated ethnicity, when stratified by single-ethnicity vs. multi-ethnicity studies, or when MAF was used as a crude proxy to assign ethnicity for studies with unknown ethnic makeup. Finally, chance could play a role, particularly given the large number of associations examined and sparse data for many analyses. However, in the four studies with large sample sizes (N>=1000) for XRCC1 399 ever-never smoking, ORzs ranged from 1.0 (0.8, 1.2) to 1.6 (1.2, 2.0), while ORz was essentially null across the four large studies that examined XPD 194 ever-never smoking [ORzs=0.9, 1.0, 1.1 and 1.1]. Further, the magnitudes of ORz differed across smoking status and amount for XRCC1 399 (3 populations) but not for XRCC1 194 (1 population). This large-sample sensitivity analysis is consistent with the overall interpretation of ORz’s population-specificity for each SNP, rather than chance alone driving the heterogeneity among studies.
Implications for stand-alone case-only studies
Z is a measure of the magnitude of bias in the COR. If Z=1, the case-only estimate of interaction is not biased by genotype-environment association in the underlying population (65). Commonly, this assumption is assessed in control data from a small number of outside studies, using significance testing. Significance testing alone is not sufficient for assessment of potential bias (76). Rarely is Z estimated and/or adjusted for, analogous to other forms of bias such as confounding.
Results from this project illustrate some of the pitfalls of this approach. For instance, for XRCC1 399 ever-never smoking, 18 of the 21 included studies have estimates that are not statistically significantly different than the null value of 1.0. Considering any of these in a statistical significance testing framework would lead to the conclusion that the independence assumption was valid; therefore a case-only study estimate of interaction would not be biased, at least from independence assumption violation. However, the range of ORzs for these 18 studies is 0.7–1.6, many with wide CIs, indicating the substantial range of potential bias of the effect estimate. Given that different conclusions can be drawn from subsets of smoking behavior and that less than half of the studies that collect control genotype and smoking information present it in publications, this ever-never approach seems inappropriate.
In the estimation framework, results from this project demonstrate the difficulty of using ancillary data to assess the independence assumption. Even when the Cochran’s Q p-value is high, such as for XRCC1 399 current-not current smoking (p=0.4), point estimates of ORz can vary as much as 5-fold [2.1(1.1, 3.9) for African Americans (27) to 0.4(0.1, 1.2) (77)]. Without further information that certain study characteristics might be influential, there is no good way to decide which of the available ancillary control groups might best represent the underlying (unmeasured) population for a proposed case-only study. Further, it is necessary to do a broad literature search to even to be aware of the possible values of ORz and range of bias in the COR. Additionally, since both summary estimates and individual study estimates vary across smoking categories, it is important that the independence assumption be evaluated for all smoking categories that will be used in the case-only analyses. For investigations of smoking amount, it will be difficult for many SNPs to locate enough published control group data to even assess the possible range of the magnitude of bias.
This study has several strengths. Using a comprehensive search strategy in collaboration with information specialists increased power to detect and investigate heterogeneity between studies. Sample size was large for smoking status analyses and relatively large for XRCC1 399 and XPD 751 PY analyses. There were sufficient data for many studies to compare ORz for smoking status and amount within studies, and by smoking category across multiple SNPs. However, of the searched studies that collected the appropriate information only about 1/3 presented it such that it could be abstracted for meta-analysis, limiting sample size, especially for measures of smoking amount.
Only unadjusted odds ratios could be calculated so study estimates may have been confounded. Although some study characteristics could be determined accurately from articles, others were more likely to be misclassified. In particular, average age of study participants was difficult to determine. However, the fact that age was not a central study feature for any of the studies makes it likely that misclassification is non-differential with respect to smoking and genotype. Several potentially informative study characteristics could not be examined because too few articles presented the relevant information using the same metric. In particular, response rates, which may vary by smoking behavior and family/personal history of cancer (78–81), and control group exclusion criteria, were presented very differently. Only two of the 12 articles with multi-ethnic study populations presented data stratified by ethnicity, complicating interpretation of HWE p-value, ethnicity and MAF as study characteristics. Few studies presented enough control group information to examine multiple measures of smoking in the same study population.
This systematic review of control-group associations between smoking and widely studied polymorphisms in DNA repair genes was conducted to accomplish several objectives. The overarching goal was to enable investigators to make more effective use of ancillary data to evaluate the independence assumption prior to launching a stand-alone case-only study. Results from this study suggest that the independence assumption is frequently violated and caution is warranted before proceeding with any case-only interaction analysis. At a minimum, the independence assumption should be more rigorously evaluated than is often done. For a case-only analysis of a case-control study, separate ORzs should be calculated for each anticipated COR in the relevant subgroup before proceeding. Evaluation of the independence assumption for a proposed stand-alone case-only study should include, whenever possible, results from studies similar to the current study, relevant literature reviews, and a thorough search for individual studies with control or cohort data to ascertain at least the range of ORzs, both overall and in relevant subgroups. Finally, it serves as a reminder that in the traditional case-control study, interaction is a contrast between control-only association and case-only association, and interaction can be driven by unanticipated associations in controls.
Evaluation of the independence assumption for case-only interaction studies would be greatly improved with more transparency and finer detail in published articles. This could perhaps be accomplished by expanding supplementary online tables to include selected joint genotype-smoking distributions in non-case groups. If it could reliably be shown that Z=1 across individual studies, better use could be made of data pooling from control groups and cohorts for selected SNPs and exposures, especially where individual level data on potential confounders can be provided. However, despite the current emphasis on pooling controls, our results indicate that investigators should not proceed with case-only studies without rigorously evaluating the independence assumption in individual studies.
Supplementary Material
References
- 1.Prentice RL, Vollmer WM, Kalbfleisch JD. On the use of case series to identify disease risk factors. Biometrics. 1984;40:445–458. [PubMed] [Google Scholar]
- 2.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 3.Yang Q, Khoury MJ, Flanders WD. Sample size requirements in case-only designs to detect gene-environment interaction. Am J Epidemiol. 1997;146:713–720. doi: 10.1093/oxfordjournals.aje.a009346. [DOI] [PubMed] [Google Scholar]
- 4.Infante-Rivard C, Labuda D, Krajinovic M, Sinnett D. Risk of childhood leukemia associated with exposure to pesticides and with gene polymorphisms. Epidemiology. 1999;10:481–487. [PubMed] [Google Scholar]
- 5.Greenland S, Poole C. Invariants and noninvariants in the concept of interdependent effects. Scand J Work Environ Health. 1988;14:125–129. doi: 10.5271/sjweh.1945. [DOI] [PubMed] [Google Scholar]
- 6.Rothman KJ. Modern Epidemiology. 3rd edition. Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 7.Albert PS, Ratnasinghe D, Tangrea J, Wacholder S. Limitations of the case-only design for identifying gene-environment interactions. American Journal of Epidemiology. 2001;154:687–693. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- 8.Tan Q, Yashin AI, Bladbjerg EM, et al. A case-only approach for assessing gene by sex interaction in human longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B129–B133. doi: 10.1093/gerona/57.4.b129. [DOI] [PubMed] [Google Scholar]
- 9.Weinberg CR, Umbach DM. Choosing a retrospective design to assess joint genetic and environmental contributions to risk. American Journal of Epidemiology. 2000;152:197–203. doi: 10.1093/aje/152.3.197. [see comments.]. [Review] [32 refs] [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Fallin MD, Kao WH. Genetic dissection methods: designs used for tests of gene-environment interaction. Curr Opin Genet Dev. 2004;14:241–245. doi: 10.1016/j.gde.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Manuguerra M, Saletta F, Karagas MR, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 12.Wu XF, Zhao H, Suk R, Christiani DC. Genetic susceptibility to tobacco-related cancer. Oncogene. 2004;23:6500–6523. doi: 10.1038/sj.onc.1207811. [DOI] [PubMed] [Google Scholar]
- 13.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007;4:59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54:267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Qu T, Morimoto K. X-ray repair cross-complementing group 1 polymorphisms and cancer risks in Asian populations: a mini review. Cancer Detect Prev. 2005;29:215–220. doi: 10.1016/j.cdp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Sterne JA, Gavaghan D, Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. J Clin Epidemiol. 2000;53:1119–1129. doi: 10.1016/s0895-4356(00)00242-0. [DOI] [PubMed] [Google Scholar]
- 17.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 18.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy RJ, Thompson SG. Detecting and describing heterogeneity in meta-analysis. Stat Med. 1998;17:841–856. doi: 10.1002/(sici)1097-0258(19980430)17:8<841::aid-sim781>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Thompson SG, Sharp SJ. Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med. 1999;18:2693–2708. doi: 10.1002/(sici)1097-0258(19991030)18:20<2693::aid-sim235>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Stern MC, Johnson LR, Bell DA, Taylor JA. XPD codon 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2002;11:1004–1011. [PubMed] [Google Scholar]
- 22.Stern MC, Siegmund KD, Conti DV, Corral R, Haile RW. XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev. 2006;15:2384–2390. doi: 10.1158/1055-9965.EPI-06-0381. [DOI] [PubMed] [Google Scholar]
- 23.Figueiredo JC, Knight JA, Briollais L, Andrulis IL, Ozcelik H. Polymorphisms XRCC1-R399Q and XRCC3-T241M and the risk of breast cancer at the Ontario Site of the Breast Cancer Family Registry. Cancer Epidemiology Biomarkers & Prevention. 2004;13:583–591. [PubMed] [Google Scholar]
- 24.Affatato AA, Wolfe KJ, Lopez MS, Hallberg C, Ammenheuser MM, Abdel-Rahman SZ. Effect of XPD/ERCC2 polymorphisms on chromosome aberration frequencies in smokers and on sensitivity to the mutagenic tobacco-specific nitrosamine NNK. Environmental and Molecular Mutagenesis. 2004;44:65–73. doi: 10.1002/em.20032. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Liang D, Spitz MR, et al. XRCC3 genetic polymorphism, smoking, and lung carcinoma risk in minority populations. Cancer. 2003;98:1701–1706. doi: 10.1002/cncr.11692. [DOI] [PubMed] [Google Scholar]
- 26.David-Beabes GL, London SJ. Genetic polymorphism of XRCC1 and lung cancer risk among African-Americans and Caucasians. Lung Cancer. 2001;34:333–339. doi: 10.1016/s0169-5002(01)00256-2. [DOI] [PubMed] [Google Scholar]
- 27.Duell EJ, Millikan RC, Pittman GS, et al. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:217–222. [PubMed] [Google Scholar]
- 28.Duell EJ, Holly EA, Bracci PM, Wiencke JK, Kelsey KT. A population-based study of the Arg399Gln polymorphism in X-ray repair cross- complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62:4630–4636. [PubMed] [Google Scholar]
- 29.Huang WY, Chow WH, Rothman N, et al. Selected DNA repair polymorphisms and gastric cancer in Poland. Carcinogenesis. 2005;26:1354–1359. doi: 10.1093/carcin/bgi084. [DOI] [PubMed] [Google Scholar]
- 30.Justenhoven C, Hamann U, Pesch B, et al. ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiology Biomarkers & Prevention. 2004;13:2059–2064. [PubMed] [Google Scholar]
- 31.Kelsey KT, Park S, Nelson HH, Karagas MR. A population-based case-control study of the XRCC1 Arg399Gln polymorphism and susceptibility to bladder cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1337–1341. [PubMed] [Google Scholar]
- 32.Pachkowski BF, Winkel S, Kubota Y, Swenberg JA, Millikan RC, Nakamura J. XRCC1 genotype and breast cancer: functional studies and epidemiologic data show interactions between XRCC1 codon 280 His and smoking. Cancer Res. 2006;66:2860–2868. doi: 10.1158/0008-5472.CAN-05-3388. [DOI] [PubMed] [Google Scholar]
- 33.Ryk C, Kumar R, Thirumaran RK, Hou SM. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54:285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Shen HB, Xu YC, Qian Y, et al. Polymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese population. International Journal of Cancer. 2000;88:601–606. doi: 10.1002/1097-0215(20001115)88:4<601::aid-ijc13>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, Gammon MD, Terry MB, et al. Polymorphisms in XRCC1 modify the association between polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, dietary antioxidants, and breast cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2005;14:336–342. doi: 10.1158/1055-9965.EPI-04-0414. [DOI] [PubMed] [Google Scholar]
- 36.Smedby KE, Lindgren CM, Hjalgrim H, et al. Variation in DNA repair genes ERCC2, XRCC1, and XRCC3 and risk of follicular lymphoma. Cancer Epidemiol Biomarkers Prev. 2006;15:258–265. doi: 10.1158/1055-9965.EPI-05-0583. [DOI] [PubMed] [Google Scholar]
- 37.Terry MB, Gammon MD, Zhang FF, et al. Polymorphism in the DNA repair gene XPD, polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, and breast cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2004;13:2053–2058. [PubMed] [Google Scholar]
- 38.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22:593–597. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Closas M, Malats N, Real FX, et al. Genetic variation in the nucleoticle excision repair pathway and bladder cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2006;15:536–542. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 40.Harms C, Salama SA, Sierra-Torres CH, Cajas-Salazar N, Au WW. Polymorphisms in DNA repair genes, chromosome aberrations, and lung cancer. Environ Mol Mutagen. 2004;44:74–82. doi: 10.1002/em.20031. [DOI] [PubMed] [Google Scholar]
- 41.Hou SM, Falt S, Angelini S, et al. The XPD variant alleles are associated with increased aromatic DNA adduct level and lung cancer risk. Carcinogenesis. 2002;23:599–603. doi: 10.1093/carcin/23.4.599. [DOI] [PubMed] [Google Scholar]
- 42.Hung RJ, Brennan P, Canzian F, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. Journal of the National Cancer Institute. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 43.Ito H, Matsuo K, Hamajima N, et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25:1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- 44.Jiao L, Hassan MM, Bondy ML, Abbruzzese JL, Evans DB, Li D. The XPD Asp312Asn and Lys751Gln polymorphisms, corresponding haplotype, and pancreatic cancer risk. Cancer Lett. 2007;245:61–68. doi: 10.1016/j.canlet.2005.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matullo G, Guarrera S, Sacerdote C, et al. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 46.Metsola K, Kataja V, Sillanpaa P, et al. XRCC1 and XPD genetic polymorphisms, smoking and breast cancer risk in a Finnish case-control study. Breast Cancer Res. 2005;7:R987–R997. doi: 10.1186/bcr1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olshan AF, Watson MA, Weissler MC, Bell DA. XRCC1 polymorphisms and head and neck cancer. Cancer Lett. 2002;178:181–186. doi: 10.1016/s0304-3835(01)00822-9. [DOI] [PubMed] [Google Scholar]
- 48.Park JY, Lee SY, Jeon HS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- 49.Ramachandran S, Ramadas K, Hariharan R, Kumar RR, Pillai MR. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncology. 2006;42:350–362. doi: 10.1016/j.oraloncology.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Schabath MB, Delclos GL, Grossman HB, et al. Polymorphisms in XPD Exons 10 and 23 and bladder cancer risk. Cancer Epidemiology Biomarkers & Prevention. 2005;14:878–884. doi: 10.1158/1055-9965.EPI-04-0235. [DOI] [PubMed] [Google Scholar]
- 51.Schneider J, Classen V, Bernges U, Philipp M. XRCC1 polymorphism and lung cancer risk in relation to tobacco smoking. Int J Mol Med. 2005;16:709–716. [PubMed] [Google Scholar]
- 52.Shen H, Sturgis EM, Dahlstrom KR, Zheng Y, Spitz MR, Wei Q. A variant of the DNA repair gene XRCC3 and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Int J Cancer. 2002;99:869–872. doi: 10.1002/ijc.10413. [DOI] [PubMed] [Google Scholar]
- 53.Shen M, Hung RJ, Brennan P, et al. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in northern Italy. Cancer Epidemiol Biomarkers Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- 54.Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:125–131. [PubMed] [Google Scholar]
- 55.Stern MC, Umbach DM, Lunn RM, Taylor JA. DNA repair gene XRCC3 codon 241 polymorphism, its interaction with smoking and XRCC1 polymorphisms, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:939–943. [PubMed] [Google Scholar]
- 56.Xing D, Tan W, Wei Q, Lin D. Polymorphisms of the DNA repair gene XPD and risk of lung cancer in a Chinese population. Lung Cancer. 2002;38:123–129. doi: 10.1016/s0169-5002(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 57.Yu HP, Wang XL, Sun X, et al. Polymorphisms in the DNA repair gene XPD and susceptibility to esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:10–15. doi: 10.1016/j.cancergencyto.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 58.Yu HP, Zhang XY, Wang XL, et al. DNA repair gene XRCC1 polymorphisms, smoking, and esophageal cancer risk. Cancer Detect Prev. 2004;28:194–199. doi: 10.1016/j.cdp.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 59.Zhou W, Liu G, Miller DP, et al. Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res. 2002;62:1377–1381. [PubMed] [Google Scholar]
- 60.Zhou W, Liu G, Miller DP, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:359–365. [PubMed] [Google Scholar]
- 61.Han JL, Hankinson SE, De Vivo I, et al. A prospective study of XRCC1 haplotypes and their interaction with plasma carotenoids on breast cancer risk. Cancer Research. 2003;63:8536–8541. [PubMed] [Google Scholar]
- 62.Jin MJ, Chen K, Song L, et al. The association of the DNA repair gene XRCC3 Thr241Met polymorphism with susceptibility to colorectal cancer in a Chinese population. Cancer Genet Cytogenet. 2005;163:38–43. doi: 10.1016/j.cancergencyto.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Misra RR, Ratnasinghe D, Tangrea JA, et al. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003;191:171–178. doi: 10.1016/s0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- 64.Patel AV, Calle EE, Pavluck AL, Feigelson HS, Thun MJ, Rodriguez C. A prospective study of XRCC1 (X-ray cross-complementing group 1) polymorphisms and breast cancer risk. Breast Cancer Res. 2005;7:R1168–R1173. doi: 10.1186/bcr1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao Y, Miao XP, Huang MY, et al. Polymorphisms of XRCC1 genes and risk of nasopharyngeal carcinoma in the Cantonese population. BMC Cancer. 2006;6:167. doi: 10.1186/1471-2407-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koyama A, Kubota Y, Shimamura T, Horiuchi S. Possible association of the X-ray cross complementing gene 1 (XRCC1) Arg280His polymorphism as a risk for rheumatoid arthritis. Rheumatol Int. 2006;26:749–751. doi: 10.1007/s00296-005-0066-3. [DOI] [PubMed] [Google Scholar]
- 67.Kocabas NA, Karahalil B. XRCC1 Arg399Gln genetic polymorphism in a Turkish population. Int J Toxicol. 2006;25:419–422. doi: 10.1080/10915810600870567. [DOI] [PubMed] [Google Scholar]
- 68.Lei YC, Hwang SJ, Chang CC, et al. Effects on sister chromatid exchange frequency of polymorphisms in DNA repair gene XRCC1 in smokers. Mutat Res. 2002;519:93–101. doi: 10.1016/s1383-5718(02)00127-4. [DOI] [PubMed] [Google Scholar]
- 69.Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: Effects on aflatoxin B-1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59:2557–2561. [PubMed] [Google Scholar]
- 70.Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 71.Skjelbred CF, Svendsen M, Haugan V, et al. Influence of DNA repair gene polymorphisms of hOGG1, XRCC1, XRCC3, ERCC2 and the folate metabolism gene MTHFR on chromosomal aberration frequencies. Mutat Res. 2006;602:151–162. doi: 10.1016/j.mrfmmm.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Tuimala J, Szekely G, Wikman H, et al. Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: effects on levels of sister chromatid exchanges and chromosomal aberrations. Mutat Res. 2004;554:319–333. doi: 10.1016/j.mrfmmm.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Wilding CS, Relton CL, Rees GS, Tarone RE, Whitehouse CA, Tawn EJ. DNA repair gene polymorphisms in relation to chromosome aberration frequencies in retired radiation workers. Mutat Res. 2005;570:137–145. doi: 10.1016/j.mrfmmm.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Zijno A, Verdina A, Galati R, et al. Influence of DNA repair polymorphisms on biomarkers of genotoxic damage in peripheral lymphocytes of healthy subjects. Mutat Res. 2006;600:184–192. doi: 10.1016/j.mrfmmm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 75.Hoffmann H, Isner C, Hogel J, Speit G. Genetic polymorphisms and the effect of cigarette smoking in the comet assay. Mutagenesis. 2005;20:359–364. doi: 10.1093/mutage/gei049. [DOI] [PubMed] [Google Scholar]
- 76.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144:207–213. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Yoshimura K, Hanaoka T, et al. Association of habitual smoking and drinking with single nucleotide polymorphism (SNP) in 40 candidate genes: data from random population-based Japanese samples. J Hum Genet. 2005 doi: 10.1007/s10038-004-0221-9. [DOI] [PubMed] [Google Scholar]
- 78.Naccarati A, Soucek P, Stetina R, et al. Genetic polymorphisms and possible gene-gene interactions in metabolic and DNA repair genes: Effects on DNA damage. Mutat Res. 2006;593:22–31. doi: 10.1016/j.mrfmmm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 79.Vodicka P, Kumar R, Stetina R, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25:757–763. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]
- 80.Vodicka P, Stetina R, Polakova V, et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28:657–664. doi: 10.1093/carcin/bgl187. [DOI] [PubMed] [Google Scholar]
- 81.Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann Med. 2003;35:94–121. doi: 10.1080/07853890310010014. [DOI] [PubMed] [Google Scholar]
- 82.Andrews AD, Barrett SF, Robbins JH. Xeroderma pigmentosum neurological abnormalities correlate with colony-forming ability after ultraviolet radiation. Proc Natl Acad Sci USA. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inoshima I, Kuwano K, Hamada N, et al. Induction of CDK inhibitor p21 gene as a new therapeutic strategy against pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2004;286:L727–L733. doi: 10.1152/ajplung.00209.2003. [DOI] [PubMed] [Google Scholar]
- 84.Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 85.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shackelford RE, Manuszak RP, Heard SC, Link CJ, Wang S. Pharmacological manipulation of ataxia-telangiectasia kinase activity as a treatment for Parkinson's disease. Med Hypotheses. 2005;64:736–741. doi: 10.1016/j.mehy.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 87.Rothman KJ, Greenland S. Modern Epidemiology. Philadelphia: Lippincott-Raven Publishers; 1998. [Google Scholar]
- 88.Hoffmann H, Isner C, Hogel J, Speit G. Genetic polymorphisms and the effect of cigarette smoking in the comet assay. Mutagenesis. 2005;20:359–364. doi: 10.1093/mutage/gei049. [DOI] [PubMed] [Google Scholar]
- 89.Morabia A, Stellman SD, Wynder EL. Smoking prevalence in neighborhood and hospital controls: implications for hospital-based case-control studies. J Clin Epidemiol. 1996;49:885–889. doi: 10.1016/0895-4356(96)00026-1. [DOI] [PubMed] [Google Scholar]
- 90.Ramos E, Lopes C, Barros H. Investigating the effect of nonparticipation using a population-based case-control study on myocardial infarction. Ann Epidemiol. 2004;14:437–441. doi: 10.1016/j.annepidem.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 91.Holt VL, Martin DP, LoGerfo JP. Correlates and effect of non-response in a postpartum survey of obstetrical care quality. J Clin Epidemiol. 1997;50:1117–1122. doi: 10.1016/s0895-4356(97)00096-6. [DOI] [PubMed] [Google Scholar]
- 92.Heilbrun LK, Nomura A, Stemmermann GN. The effects of nonresponse in a prospective study of cancer. Am J Epidemiol. 1982;116:353–363. doi: 10.1093/oxfordjournals.aje.a113419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.