Abstract
Heterochronous data sets comprise molecular sequences sampled at different points in time. If the temporal range of the sampled sequences is large relative to the rate of mutation, the sampling times can directly calibrate evolutionary rates to calendar time. Here, we extend this calibration process to provide a full probabilistic method that utilizes temporal information in heterochronous data sets to estimate sampling times (leaf-ages) for sequenced for which this information unavailable. Our method is similar to relaxing the constraints of the molecular clock on specific lineages within a phylogenetic tree. Using a combination of synthetic and empirical data sets, we demonstrate that the method estimates leaf-ages reliably and accurately. Potential applications of our approach include incorporating samples of uncertain or radiocarbon-infinite age into ancient DNA analyses, evaluating the temporal signal in a particular sequence or data set, and exploring the reliability of sequence ages that are somehow contentious.
Keywords: heterochronous sequences, ancient DNA, molecular clock, viral evolution, measurably evolving populations
Introduction
The incorporation of temporal information into molecular phylogenetic and genealogic analyses means that evolutionary processes can be investigated on a natural timescale of years, centuries, or millennia. This is commonly achieved by one of two methods. If the sequences of interest are sampled at effectively the same time (“isochronous” data) then an evolutionary timescale can be calibrated by assigning a date, or date range, to one or more divergence events in the tree. If data sets comprise sequences sampled at different time points (“heterochronous” data), these can be calibrated by fixing the age of each sequence (the leaves of the tree) to the known age of the specimen from which the sequence was amplified (e.g., Rambaut 2000). Both approaches result in an estimated rate of evolution and a corresponding phylogenetic timescale, which can then be used to test hypotheses about the timing and nature of evolutionary and demographic events, such as dates of divergence among lineages or changes in population size or structure (Drummond, Pybus, Rambaut, Forsberg, and Rodrigo 2003).
The two major sources of heterochronous sequence data are rapidly evolving RNA and DNA viruses, whose high mutation rates enable the generation of phylogenetically informative sequence diversity within historical time frames (Drummond, Pybus, and Rambaut 2003), and ancient DNA data isolated from preserved remains (Hofreiter, Serre, et al. 2001) that may be up to several hundred thousands of years old (Willerslev et al. 2007). In both cases, the period over which sequences are isolated is sufficiently long relative to the mutation rate to allow estimation of the evolutionary rate (Drummond, Pybus, Rambaut, Forsberg, and Rodrigo 2003).
The temporal information associated with RNA virus sequences typically represents the day, month, or year of sample isolation and/or storage (Taubenberger et al. 1997). Heterochronous viral data sets have been used to estimate rates of mutation for specific viruses (Jenkins et al. 2002), to investigate rates of evolution and adaptation within hosts (e.g., Lemey et al. 2006, 2007), and to infer epidemic dynamics within and between populations of susceptible hosts (Rambaut 2000; Pybus and Rambaut 2009).
For ancient DNA data sets, ages of the genetic sequences are most often approximated using radiocarbon dates that are estimated from the same specimens from which the DNA sequences are amplified (e.g., Shapiro et al. 2004; Bunce et al. 2009; Campos et al. 2010). Dates from material associated with stratigraphic context have also been used as calibrating information, however (e.g., Lambert et al. 2002; Valdiosera et al. 2008). For ancient DNA sequences, leaf-ages are normally assigned some number of thousands of years before the present (ka BP). Molecular clock analyses of these data have been used to identify periods of population turnover (e.g., Hadly et al. 1998; Barnes et al. 2002; Hofreiter et al. 2004) to estimate divergence times within species (e.g., Shapiro et al. 2004; Debruyne et al. 2008; Stiller et al. 2010) and to correlate population-level changes in genetic diversity with external events, such as climate change (e.g., Hadly et al. 2004; Chan et al. 2006; Barnett et al. 2009; Campos et al. 2010). For example, there has been considerable debate about the relative roles of climate change associated with the last glacial maximum (LGM; ca 21 ka BP) and the appearance and increase in human populations (ca 14 ka BP) in the recent disappearance of the North American megafauna (Alroy 2001; Barnosky et al. 2004; Stuart et al. 2004). Heterochronous data sets comprising sequences directly dated to before, during, and after these events allow explicit tests of these alternative hypotheses.
Despite significant technical and chemical pretreatment advances in Accelerator Mass Spectrometry radiocarbon dating (Bronk Ramsey et al. 2004), the oldest samples for which finite radiocarbon dates can be routinely generated are around 50-–55 ka BP (e.g., Barnett et al. 2009). The period 0–55 ka BP includes several specific large-scale environmental events that are likely to have affected the distribution and abundance of plant and animal species. However, there are a number of circumstances under which the leaf-ages of ancient DNA or viral sequences may be unknown, or at best, highly uncertain. First, ancient mitochondrial DNA (mtDNA) sequences are routinely amplified from permafrost-preserved specimens older than the 50–55 ka BP radiocarbon limit. For example, nearly 100 of the bison sequences reported in Shapiro et al (2004) were too old to be assigned finite radiocarbon ages, and ancient DNA sequences have been reported that are perhaps as old as 800 ka BP (Willerslev et al. 2007). In this case, only censored temporal information is often available (i.e., age > 55 ka BP). Second, ancient DNA samples are routinely recovered from situations in which the stratigraphic context provides calibrating information (e.g., Lambert et al. 2002; Coolen and Overmann 2007; Valdiosera et al. 2008) but only within a wide range of uncertainty, such that assigning a specific mean or median date to such sequences is statistically inappropriate (Ho and Phillips 2009). Third, for rapidly evolving viral sequences, the date of sampling may simply be unknown due to the loss or absence of accurate archival information. Even if the viral sampling date is known to the nearest year, it may be important to know the isolation date more accurately. Fourth, it may also be important to independently verify posited sampling dates due to their extreme age (Zhu et al. 1998; Taubenberger et al. 2005; Worobey et al. 2008) or because they are in some way contentious (Sonoda et al. 2000; Coolen and Overmann 2007). Because frozen viral isolates do not accumulate mutations while in storage, a leaf-dating method also has the potential to identify transmitting viruses that, after a period of storage, have been released into the environment (Worobey 2008).
Despite these obvious problems, few studies have attempted to estimate the unknown age or sampling date of heterochronous sequences using molecular clock methods, and none have investigated the statistical reliability of such methods. Perhaps the most similar analysis was that undertaken by Korber et al. (1998), who validated their molecular clock of HIV-1 by testing whether it could accurately predict the date of an “old” isolate sampled in 1959. However, in that case, the approach consists of the visual inspection of the fit to a linear regression of viral sampling date against genetic distance rather than a statistical analysis or estimation procedure.
Here, we investigate a statistical framework for the estimation of leaf-dates using molecular clock models when the sample age or isolation date is either unknown or highly uncertain. Following Drummond (2002), we estimate the age of individual DNA sequences using the temporal calibrating information from other sequences in the data set. Methodologically, the leaf-dating method is similar to relaxing the constraints of the molecular clock on specific lineages within a phylogenetic tree. Using a combination of simulations and empirical analyses, we show that leaf-ages can be estimated reliably and accurately using our approach. Although the analyses presented here only perform leaf-dating on one sequence within any given data set, the method can be readily extended to multiple sequences within the same analysis.
Materials and Methods
We developed and implemented a leaf-dating method that estimates the age or date of isolation of individual sequences within the Bayesian Markov chain Monte Carlo (MCMC) framework provided by the software package BEAST (Drummond and Rambaut 2007). BEAST allows dates/times to be specified for each sequence in a sample alignment and uses this information to estimate a timescale for the evolutionary history of the sample. The models implemented in BEAST accomplish this by fixing the external nodes of the tree (the leaves) to the specified dates and then sampling the times of the internal nodes of the tree from their posterior probability distribution using MCMC. The length of each branch in units of time is mapped to an expected number of substitutions per site using a vector of molecular evolutionary rates. The simplest model assigns the same single rate to every branch (the strict molecular clock model). BEAST also implements methods that allow the evolutionary rate to vary among branches (relaxed molecular clock models) such that the vector of rates follows a specified parametric distribution (Drummond et al. 2006). Under these models, BEAST can simultaneously infer the tree topology, the times of the internal nodes, the rate of evolution and any parameters of the associated coalescent, and substitution models (Drummond et al. 2002). As is required in Bayesian inference, all of these parameters are assigned one of a wide variety of possible prior probability distributions. MCMC sampling is then used to obtain estimates of marginal posterior probability distributions for any parameters of interest.
In previous molecular clock implementations, all nodes in the tree are given dates/ages, that is, internal nodes are treated as unknown parameters, although the dates/ages of the tree external nodes (leaves) are assumed to be known. To estimate the age of an individual sequence, we extend the framework introduced above to estimate the time associated with the sequence, that is, the sequence’s leaf-age is treated as a random variable. Thus, an additional parameter for the age of the external node is introduced and is treated identically to the internal node age parameters in terms of proposals made by the MCMC kernel. See Drummond et al. (2006) for further specifications and parameterizations of the molecular clock models used here.
Synthetic Data Sets
To explore the ability of our leaf-dating model to recover sample ages, we first estimated the dates of randomly chosen sequences within synthetic heterochronous data sets. We generated sequence alignments of 1,000 nt in length, each including 50 taxa sampled at different times, by simulating sequence evolution down 1,000 random trees. We simulated sequences using a Jukes–Cantor model of nucleotide substitution under a strict molecular clock with a rate of 2.5 × 10−7 subs/site/year (Rambaut and Grassly 1997). Each tree represents a random sample from the constant size serial-sample coalescent model (Rodrigo et al. 1999), with a population size equal to 105 haploid individuals. The ages of the 50 individuals were 0, 0, 2,000, 2,000, … , 48,000, 48,000 years. The evolutionary rate and sampling times used in these simulations are representative of those found in typical analyses of ancient mtDNA data sets.
For each of the 1,000 simulated data sets, a single leaf was chosen at random and its known date was removed. Each data set was analyzed separately in BEAST using the method outlined above and the age of undated leaf was estimated. This procedure represents a “leave-one-out cross-validation” design and is an effective approach to examining estimator performance. BEAST analyses were performed under the true model, that is, a strict clock, a constant size coalescent model, and the Jukes–Cantor model of nucleotide substitution. For each analysis, we ran a single MCMC chain for 5 million generations, with samples drawn from the chain every 5,000 generations, of which the first 10% was discarded as burn-in.
Empirical Data Sets
We selected two empirical heterochronous data sets for further validation of the leaf-dating method, enabling us to test our approach when the true evolutionary model is unknown: 1) a data set of partial (1,404 nt) E gene sequences of Dengue-2 (DEN-2) virus subtype II (Carrington et al. 2005) comprising 89 samples isolated between 1981 and 2002 and 2) a data set representing 166 ancient and modern bison (Bison priscus) for which 602 nt of mitochondrial control region sequences are available (Shapiro et al. 2004) with radiocarbon ages calibrated using CalPal_2007_HULU (http://www.calpal-online.de/). Sample ages within the bison data set ranged from 0 to 55,000 years old.
For both empirical data sets, we estimated the leaf-age of each sequence in the alignment in a separate BEAST analysis. Because the youngest sampling date places a hard upper bound on the evolutionary timescale, it is impossible to overestimate (estimated age older than the true age) the age of the youngest sequences in each data set. We therefore excluded the youngest sampled sequences from both data sets from the verification procedure. However, requiring strictly positive leaf-ages is not warranted for all problems. For example, measurable evolving viral populations may contain unknown leaf-ages that are more recent than the youngest known age that is assumed to represent time 0 in the tree.
For the DEN-2 analyses, we assumed a strict molecular clock, the GTR + G model of nucleotide substitution and a constant coalescent model with a diffuse, log-normally distributed effective population size, as was selected as the best fitting coalescent model in the initial publication (Carrington et al. 2005). A single MCMC chain of 10 million generations was run with samples recorded from the chain every 1,000 generations, discarding the first 10% as burn-in. For the bison analyses, we assumed a strict molecular clock and the HKY + G model of nucleotide substitution. Previous work has shown that simple coalescent models are insufficient to explain the complex demographic history of bison (Shapiro et al. 2004; Drummond et al. 2005). Therefore, we performed each bison analysis using two different flexible coalescent models: a piece-wise constant, multiple change-point (MCP) process, often referred to as the Skyline (Drummond et al. 2005), with 12 groups, and a Gaussian Markov random field (GMRF) process, the Skyride (Minin et al. 2008). For each bison analysis, we ran a single MCMC chain for 50 million generations, with samples recorded from the chain every 5,000 generations, again discarding the first 10% as burn-in. Thus, in total, we performed 85 DEN-2 analyses (one for every leaf except the youngest one) and 250 bison analyses (one for every nonyoungest leaf for each of two coalescent models). For all analyses, we evaluated parameter mixing and convergence to the stationary distribution using Tracer v 1.4 (available from: http://tree.bio.ed.ac.uk/software/tracer/).
Results
Simulated Data Sets
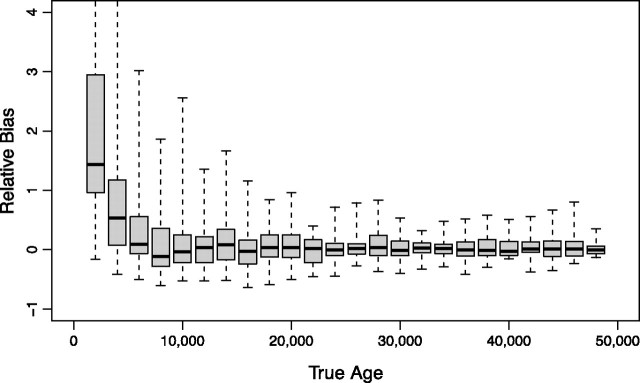
In the simulated data sets, the true ages of the leaves ranged from 0 to 48,000 years. In each of our 1,000 analyses, one leaf was selected at random and its known age was removed. In 40 cases, a leaf with a sample age of zero was chosen; these were excluded from the statistical verification analysis (see above for explanation). Of the 960 remaining analyses, the true sequence age was recovered within the 95% highest posterior density (HPD) interval of the leaf-age estimates 95% of the time, demonstrating that our leaf-dating method performs well and has favorable statistical coverage properties. More importantly, comparison of the posterior mean estimated leaf-age with the true age revealed no significant bias in the estimates for true ages greater than or equal to 6,000 years old (fig. 1), indicating that the posterior mean leaf-age estimates are equally likely to overestimate as to underestimate the true age. Mean square error in these estimates decreases with increasing true age. Here, the difference in expected number of substitutions between leaf-ages at zero and those more distant in the tree increases, yielding on average more informative data for leaf-age estimation. Modest bias is observed for true ages very close to or equal zero, which is wholly expected because, for this example, the estimated dates cannot be smaller than zero, as the youngest sequence in the data set if from a modern bison. The lower bound on unbiased estimates naturally depends both on real times and the mutation rate.
FIG. 1.
Relative bias in posterior mean leaf-age estimates from 960 leave-one-out analyses of synthetic data sets with varying true leaf-ages. For true ages sufficiently distinct from zero, the posterior mean estimator is unbiased.
Empirical Data Sets
Dengue-2
Of the 85 sequences in this data set that were older than the youngest sampled sequence, the true age was contained within the 95% HPD intervals of the leaf-age estimate for 79 leaves (93%; supplementary table S1, Supplementary Material online). Of the six sequences for which the real age was not included in the HPD, only one (D2DR_1984) resulted in a significant difference in the marginal likelihood, assessed by calculation of the Bayes factor comparing the strict clock model and the leaf-age model (BF; the ratio of the marginal likelihoods with respect to the prior of the two models) (Suchard et al. 2001). The BF strongly favors relaxing the strict clock assumption on the branch leading to D2DR_1984 (log10 BF = 3.99), suggesting that either the age assigned to the sequence or the sequence itself is incorrect or that this particular sequence is evolving at a markedly different rate than the other sequences in the DEN-2 data set.
Bison
Leaf-ages for 125 bison mtDNA control region sequences (leaf-ages were not estimated for 41 modern bison) were estimated under two different coalescent models (the MCP and GMRF models). Both coalescent models gave consistent results: the true age fell outside the 95% HPD interval of the estimated leaf-age for 18 (14.4%) and 19 (15.2%) bison sequences when assuming the GMRF and MCP coalescent models, respectively. Of these, 17 were estimated incorrectly in both analyses (table 1). No significant correlation was observed between the calibration age of the sequence and failure of the method to recover the true age (Wilcoxon rank sum test with continuity correction: W = 751, P = 0.14).
Table 1.
Twenty Bison for Which the Leave-One-Out Analysis Failed to Recover the True Age Within the 95% HPDs of the Estimated Leaf-Age.
| Strict Molecular Clock |
Relaxed Clock |
|||
| Sample ID | Calibrated Age (BP) | GMRF Mean (95% HPD) | MCP Mean (95% HPD) | MCP Mean (95% HPD) |
| BS111 | 25,920 ± 503 | 16,566 (8,826–25,075) | 16,373(8,823–25,628) | 16,170 (9,072–23,940) |
| BS146 | 13,662 ± 58 | 60,806 (28,641–89,428) | 65,235 (20,633–106,620) | 55,930 (22,450–84,100) |
| BS148 | 7,326 ± 56 | 14,091 (8,284–20,545) | 13,965 (8,364–19,833) | 14,220 (9,087–20,490) |
| BS161 | 25,181 ± 176 | 70,688 (36,052–101,030) | 80,242 (30,538–125,386) | 65,010 (27,900–94,470) |
| BS176 | 14,232 ± 129 | 46,145 (23,488–68,808) | 52,947 (21,632–83,714) | 43,690 (19,410–66,600) |
| BS196 | 23,123 ± 196 | 64,575 (30,175–94,040) | 66,860 (28,657–102,425) | 57,670 (23,410–86,460) |
| BS202 | 12,381 ± 120 | 80,369 (47,807–106,660) | 82,771 (19,936–119,486) | 69,190 (18,720–94,030) |
| BS253 | 14,753 ± 112 | 8,530(1,769–14,766) | 8,266 (2,058–14,140) | 8,749(1,948–15,660) |
| BS258 | 26,606 ± 184 | 31,004(8,346–59,351) | 206,542 (105,637–333,502) | 37,130(11,720-69,840) |
| BS286 | 54,134 ± 2,800 | 83,049 (64,269–101,972) | 90,412 (66,629–116,325) | 73,570(52,480–96,780) |
| BS292 | 40,991 ± 686 | 67,453 (43,506–91,413) | 70,453 (42,618–94,697) | 62,680(35,570–86,290) |
| BS297 | 12,867 ± 53 | 62,262 (41,667–79,291) | 64,290 (42,197–83,495) | 58,980 (35,120–78,390) |
| BS329 | 32,370 ± 256 | 67,021 (40,663–91,504) | 70,817 (43,050–97,395) | 60,880(24,900–87,330) |
| BS365 | 51,433 ± 4,004 | 29,187 (11,700–46,697) | 28,916 (11,986–47,484) | 33,140(12,550–52,400) |
| BS388 | 32,933 ± 336 | 85,768 (49,700–113,587) | 117,733 (52,790–178,806) | 78,570 (38,190–106,500) |
| BS389 | 20,337 ± 87 | 52,175 (34,999–72,762) | 52,238 (33,254–75,360) | 53,000 (30,760–79,110) |
| BS398 | 32,732 ± 317 | 86,951 (63,479–108,490) | 99,544 (60,419–142,781) | 79,330 (55,530–100,000) |
| BS400 | 50,204 ± 3,427 | 21,796 (12,297–33,451) | 21,807 (11,094–33,915) | 23,270 (10,210–43,990) |
| BS405 | 27,653 ± 195 | 78,249 (41,825–111,633) | 124,010 (48,004–207,590) | 68,910(25,820–99,180) |
| BS478 | 39,836 ± 261 | 17,416 (2,988–32,063) | 18,784 (3,171–36,834) | 18,050 (3,332–37,250) |
Note.—Bold, italicized values are those for which the true age is recovered by the analysis.
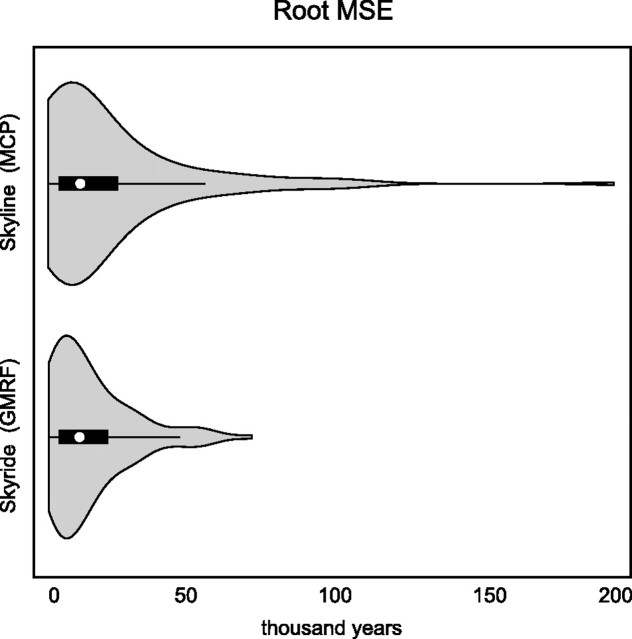
To examine differences in results between the two coalescent models, we calculated the square root of the mean squared error (rMSE) of the leaf-age estimate for each analysis (supplementary table S2, Supplementary Material online). The rMSE integrates both the variance and bias of an estimator and is an effective tool with which to evaluate statistical performance when variances may differ and there remains potential for bias. Of the sequences, whose true age was contained within the 95% HPD interval of the estimated leaf-age, the posterior average rMSE for the analyses assuming a MCP process was 16,292 years, whereas the same statistic for the GMRF analyses was 12,013 years. To understand this difference, we examined the distribution of rMSE estimates (fig. 2). From the figure, average behavior is similar across the flexible coalescent models; however, rMSE estimates under the MCP model can result in a very long-tailed distribution, with a small number of leave-one-out analyses generating highly skewed leaf-age estimates.
FIG. 2.
Empirical distributions of rMSE when estimating leaf-dates under flexible coalescent models based on a multiple change-point (MCP) process and GMRF.
The molecular clock is a likely source of error in estimating leaf-ages (see expanded discussion below). To explore this further, we reran each of the 20 bison leaf-dating analyses for which the strict clock model failed to recover the true date, this time allowing the evolutionary rate to be relaxed across the entire genealogy. The analyses were performed as described above, assuming the GMRF model and the uncorrelated lognormal relaxed clock (Drummond et al. 2006). Very similar results were obtained using the relaxed and strict clock models (table 1). However, when the molecular clock is relaxed across the entire tree rather than just on the single branch, the true age is contained within the 95% HPD intervals of the estimated leaf-age for seven of the bison sequences that previously failed.
Discussion
Our results indicate that the leaf-age model is powerful enough to recover temporal signal from sequence data provided that the calibrating information within other portions of the data set is sufficient. Analysis of simulated data sets demonstrates convincingly that, when all other sources of error are accounted for (i.e., the analysis is performed under the true model), the leaf-age estimator is an unbiased predictor of the true age of a sequence and has correct properties of statistical coverage. Figure 1 shows that bias is greater when the estimated leaf-age is close to its boundary (in this case, 0). Such behavior is expected because the variable being estimated (the sequence age) is a strictly nonnegative quantity and as the value approaches the boundary only ages that are older than the true age can be estimated. Implementing an estimator from a model that allows for negative leaf-ages may circumvent this but would be biologically meaningless when the youngest sample included in the data set was sampled, effectively, today (such a negative leaf-age would become the youngest sample, suggesting that all the other samples are older than ascribed).
The results of the empirical data analyses were mixed. Our leaf-dating method performed very well when applied to the DEN-2 data set, as the known dates of sampling fell within the 95% HPD intervals of our estimates 93% of the time. This is extremely encouraging, as it is almost certainly true that the evolutionary and coalescent models used in our analyses are a gross simplification of natural processes; yet, we were able to estimate leaf-dates reliably and accurately. In addition to demonstrating the effectiveness of our leaf-dating approach, our DEN-2 virus result also serve to validate the molecular clock methods used, for if terminal nodes throughout the tree are estimated with accuracy then it logically follows the internal nodes, including the tree root, are similarly dated correctly.
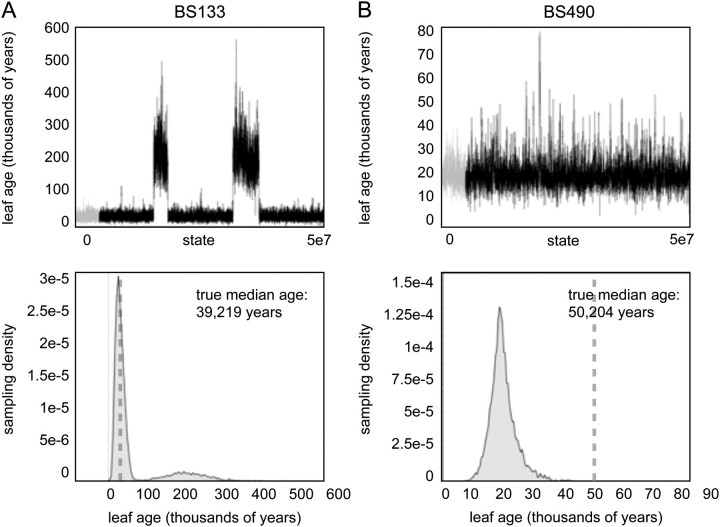
For the bison data set, the leaf-ages (as determined by radiocarbon dating) were estimated incorrectly approximately 15% of the time (table 1, supplementary table S2, Supplementary Material online). There was no noticeable pattern in the age of the tips for which the leaf-dating method fails. However, in general, the same leaf-ages, which differ significantly from their priors in all cases, are incorrectly estimated under both coalescent models. This suggests two things: first, that the incorrect dates likely arise from some heterogeneity in the evolutionary process that is not accounted for in the models and second, that the leaf-date estimates depend more on the data than on the prior distributions assumed. The MCP process assumes that effective population sizes change according to an exponential Markovian process (Drummond et al. 2005). In rare cases, with limited information about the root age that occurs when the unknown dated leaf attaches near the root, this nonstationary process can introduce excess variability (fig. 3A); the GMRF, on the other hand, is stationary (Minin et al. 2008) and modestly outperforms the MCP in these situations.
FIG. 3.
Examples of two different leave-one-out analyses for which the true age was not recovered within the 95% HPDs of the leaf-age estimates. In (A), the analysis identifies two similarly likely leaf-ages, whereas in B the analysis identifies a single, precise estimate that does not coincide with the radiocarbon date of the specimen from which the sequence was isolated. By evaluating the trace files from each leaf-age estimate, it is possible to identify potentially erroneous sequences and to devise an appropriate strategy to authenticate these sequences.
Although the vast majority of bison sequence ages were estimated correctly, it is unsatisfactory that some were not and important to consider the possible causes of such error. First, and most obviously, the “true” ages of the specimens may have been incorrectly reported or estimated. For example, the radiocarbon ages we used in our analysis may be incorrect or subject to error, in particular as the age of the specimen approaches the limits of this technique.
Second, the sequences themselves may be incorrect or be erroneous in some way. This is particularly problematic for ancient DNA data, where the degraded nature of the samples results in predictable patterns of DNA damage and base misincorporation by the polymerases used in polymerase chain reaction (Hofreiter, Jaenicke, et al. 2001; Gilbert et al. 2003, 2005; Binladen et al. 2006). Previously, an analysis of the bison data set incorporating the postmortem damage (PMD) model showed that this particular aDNA data set contained very little damage (Rambaut et al. 2009). The PMD model assumes that the probability that any given nucleotide remained undamaged decays exponentially with age; the oldest sequences in the bison data set demonstrated, on average, only 0.74 damaged sites, a level that produced no qualitative change in the demographic reconstructions. In its current implementation, the PMD model assumes a common decay rate for every site of every sequence (such that the probability of a site being undamaged decays exponentially with age). A useful extension of the PMD model may be to provide a probabilistic expectation that each individual sequence is damaged, thereby making it possible to identify problematic sequences for additional assessment. Although incorporating both PMD models and leaf-dating in a single analysis is possible, the common decay rate PMD model and leaf-dating are expected to be only weakly identifiable in the sampling density of the observed sequence data, serving primarily to increase variance on the leaf-age estimate without extracting much additional information from the data. Furthermore, the individual decay rate PMD model and a random leaf-age are not identifiable. Consequently, prior assumptions will dominate inference as to whether a sequence is damaged or incorrectly dated.
Third, some aspect of the evolutionary models used may be unrealistic and a possible source of error. We consider each model component in turn:
(i) The phylogenetic model assumes that the sequences do not undergo recombination. Recombination is very unlikely to be present in our ancient mtDNA bison data set but may be a potential cause of error in the analysis of some viruses that do recombine readily, such as HIV-1 (in which case the method introduced here may be used to “detect” putatively recombinant sequences).
(ii) The coalescent prior distribution may too constraining or unrepresentative of the tree shapes supported by the data. However, similar results were obtained when the bison data set was estimated under two different coalescent models, each of which is highly flexible, this is unlikely to be a significant cause of estimation error.
(iii) The molecular clock model did not accurately model temporal sequence evolution. The molecular clock determines the estimated evolutionary timescale and is therefore, a priori, the most likely source of error in estimating leaf-dates. The strict molecular clock used in our analysis does not incorporate rate heterogeneity among lineages, which is common in many heterochronous data sets (Korsten et al. 2009; Magiorkinis et al. 2009). If this variation is ignored then leaves attached to terminal branches that evolve unusually rapidly or slowly will have their ages poorly estimated.
Note that the discussion above does not directly address the biological assumptions underlying model components, such as the coalescent and the molecular clock. This is deliberate, as we wish to highlight a common misconception. In population-level analyses, it is often assumed that it is necessary to assume neutral evolution in order to accurately estimate divergence times using a molecular clock. If divergence times are the primary parameters of interest, then the clock model used can be thought of as a statistical or phenomenological description of the relationship between genetic distance and time rather than an explicit model of a biological process. Viewed this way, all that is important is that the model “fits” the data well, statistically speaking. The same argument can be applied to the coalescent model, which, when being treated as a nuisance parameter, need only represent a suitable range of tree shapes and sizes. In such cases, the assumptions of the coalescent model (e.g., random sampling or panmixis) are not necessary conditions for accurate date estimation. As a result, estimation of leaf-dates is likely to be highly robust provided that the clock model used incorporates sufficient rate heterogeneity.
Although it is often not feasible to discriminate among the sources of error outlined above, it may be possible to identify problematic sequences by evaluating the output of the leaf-dating analysis. For example, a common problem among the small number of analyses that fail to recover the true leaf-age is that the sequences appear to be equally or nearly equally likely to fall in two locations in the genealogy, with each of these resulting in very different leaf-age estimates. This pattern is seen clearly by plotting the estimated marginal posterior distribution leaf-ages, which has an unusually high variance (fig. 3A). This result may be due to errors within the sequence itself, which could potentially be resolved by resequencing. Alternately, very precise leaf-age estimates not containing the true age may indicate a problem with the true age (fig. 3B). For ancient DNA data, recovering an additional radiocarbon date or confirming information about the stratigraphic context of the sample can be useful to rule out this potential source of error. For viral sequences, a reexamination of the documentation associated with the isolate may reveal an annotation or transcription error.
Although the estimated ages recovered by the leaf-dating method are often associated with wide credible intervals, our leaf-dating method provides a means to include in molecular clock analyses data for which little or no temporal information is known. Incorporating additional sequence data can improve the resolution of the phylogenetic, demographic, and geographic history of the sampled sequences and can extend significantly the temporal range of the analysis. Additionally, estimating leaf-ages can provide an independent means of assessing both the authenticity of heterochronous sequences and the ages to which the sequences have been ascribed, which is often a significant concern in ancient DNA research.
The possibility of treating leaf-ages as random variables also allows uncertainty to be modeled explicitly. This enables the incorporation of the uncertainty associated with layer dating, for example, in the form of a uniform prior across the age range of the source stratum. In addition, the error in isotopic dating can be reflected by choosing an appropriate prior distribution for the corresponding leaf-age (Ho and Phillips 2009).
We hope that further uses for the leaf-dating method may be found. As one potential application, consider the forensic or archaeological examination of biological tissue from which rapidly evolving viral sequences (e.g., influenza) are recoverable—by estimating the date of such sequences using our method, it will be possible to posit a time of death.
Supplementary Material
Supplementary tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was partially supported by the National Institute of Health (R01 GM083603, R01 GM083983, and R01 GM086887), National Science Foundation (ARC 0909456), and the National Evolutionary Synthesis Center (NESCent), NSF #EF-0423641.
References
- Alroy J. A multispecies overkill simulation of the end-Pleistocene megafaunal mass extinction. Science. 2001;292:1893–1896. doi: 10.1126/science.1059342. [DOI] [PubMed] [Google Scholar]
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- Barnett R, Shapiro B, Barnes I, et al. (17 co-authors) Phylogeography of lions (Panthera leo ssp.) reveals three distinct taxa and a late Pleistocene reduction in genetic diversity. Mol Ecol. 2009;18:1668–1677. doi: 10.1111/j.1365-294X.2009.04134.x. [DOI] [PubMed] [Google Scholar]
- Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. Assessing the causes of late Pleistocene extinctions on the continents. Science. 2004;306:70–75. doi: 10.1126/science.1101476. [DOI] [PubMed] [Google Scholar]
- Binladen J, Wiuf C, Gilbert MTP, et al. (11 co-authors) Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronk Ramsey C, Higham TFG, Bowles A, Hedges R. Improvements to the pretreatment of bone at Oxford. Radiocarbon. 2004;46:155–163. [Google Scholar]
- Bunce M, Worthy TH, Phillips MJ, et al. (11 co-authors) The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc Natl Acad Sci U S A. 2009;106:20646–20651. doi: 10.1073/pnas.0906660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos PF, Willerslev E, Sher A, et al. (20 co-authors) Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. Proc Natl Acad Sci U S A. 2010;107:5675–5680. doi: 10.1073/pnas.0907189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Anderson CNK, Hadly EA. Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. PLoS Genet. 2006;2:451–460. doi: 10.1371/journal.pgen.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MJ, Overmann J. 217,000-year-old DNA sequences of green sulfur bacteria in Mediterranean sapropels and their implications for the reconstruction of the paleoenvironment. Environ Microbiol. 2007;9:238–249. doi: 10.1111/j.1462-2920.2006.01134.x. [DOI] [PubMed] [Google Scholar]
- Debruyne R, Chu G, King CE, et al. (21 co-authors) Out of America: ancient DNA evidence for a new world origin of late quaternary woolly mammoths. Curr Biol. 2008;18:1320–1326. doi: 10.1016/j.cub.2008.07.061. [DOI] [PubMed] [Google Scholar]
- Drummond A, Pybus OG, Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv Parasitol. 2003;54:331–358. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- Drummond AJ. Computational and statistical inference for molecular evolution and population genetics. Biological sciences. Auckland (New Zealand): University of Auckland; 2002. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. Measurably evolving populations. Trends Ecol Evol. 2003;18:481–488. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Bandelt HJ, Hofreiter M, Barnes I. Assessing ancient DNA studies. Trends Ecol Evol. 2005;20:541–544. doi: 10.1016/j.tree.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Characterization of genetic miscoding lesions caused by postmortem damage. Am J Hum Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadly EA, Kohn MH, Leonard JA, Wayne RK. A genetic record of population isolation in pocket gophers during Holocene climatic change. Proc Natl Acad Sci U S A. 1998;95:6893–6896. doi: 10.1073/pnas.95.12.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadly EA, Ramakrishnan U, Chan YL, van Tuinen M, O'Keefe K, Spaeth PA, Conroy CJ. Genetic response to climatic change: insights from ancient DNA and phylochronology. PLoS Biol. 2004;2:1600–1609. doi: 10.1371/journal.pbio.0020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Paabo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Serre D, Poinar HN, Kuch M, Paabo S. Ancient DNA. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Serre D, Rohland N, Rabeder G, Nagel D, Conard N, Munzel S, Paabo S. Lack of phylogeography in European mammals before the last glaciation. Proc Natl Acad Sci U S A. 2004;101:12963–12968. doi: 10.1073/pnas.0403618101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Korber B, Theiler J, Wolinsky S. Limitations of a molecular clock applied to considerations of the origin of HIV-1. Science. 1998;280:1868–1871. doi: 10.1126/science.280.5371.1868. [DOI] [PubMed] [Google Scholar]
- Korsten M, Ho SYW, Davison J, et al. (24 co-authors) Sudden expansion of a single brown bear maternal lineage across northern continental Eurasia after the last ice age: a general demographic model for mammals? Mol Ecol. 2009;18:1963–1969. doi: 10.1111/j.1365-294x.2009.04163.x. [DOI] [PubMed] [Google Scholar]
- Lambert DM, Ritchie PA, Millar CD, Holland B, Drummond AJ, Baroni C. Rates of evolution in ancient DNA from Adelie penguins. Science. 2002;295:2270–2273. doi: 10.1126/science.1068105. [DOI] [PubMed] [Google Scholar]
- Lemey P, Pond SLK, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3:282–292. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SYW, Shapiro B, Pybus O, Allain JP, Hatzakis A. The global spread of Hepatitis C Virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 2009;6:e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Ho SYW, Drummond AJ, Shapiro B. Accommodating the effect of ancient DNA damage on inferences of demographic histories. Mol Biol Evol. 2009;26:245–248. doi: 10.1093/molbev/msn256. [DOI] [PubMed] [Google Scholar]
- Rodrigo AG, Shpaer EG, Delwart EL, Iversen AK, Gallo MV, Brojatsch J, Hirsch MS, Walker BD, Mullins JI. Coalescent estimates of HIV-1 generation time in vivo. Proc Natl Acad Sci U S A. 1999;96:2187–2191. doi: 10.1073/pnas.96.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Drummond AJ, Rambaut A, et al. (27 co-authors) Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Li HC, Cartier L, Nunez L, Tajima K. Ancient HTLV type 1 provirus DNA of Andean mummy. AIDS Res Hum Retrovir. 2000;16:1753–1756. doi: 10.1089/08892220050193263. [DOI] [PubMed] [Google Scholar]
- Stiller M, Baryshnikov G, Bocherens H, et al. (12 co-authors) Withering away—25,000 years of genetic decline preceded cave bear extinction. Mol Biol Evol. 2010;27:975–978. doi: 10.1093/molbev/msq083. [DOI] [PubMed] [Google Scholar]
- Stuart AJ, Kosintsev PA, Higham TF, Lister AM. Pleistocene to Holocene extinction dynamics in giant deer and woolly mammoth. Nature. 2004;431:684–689. doi: 10.1038/nature02890. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Krafft AE, Bijwaard KE, Fanning TG. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Valdiosera CE, Garcia-Garitagoitia JL, Garcia N, et al. (11 co-authors) Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos) Proc Natl Acad Sci U S A. 2008;105:5123–5128. doi: 10.1073/pnas.0712223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, Cappellini E, Boomsma W, et al. (29 co-authors) Ancient biomolecules from deep ice cores reveal a forested Southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus. J Virol. 2008;82:3769–3774. doi: 10.1128/JVI.02207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Gemmel M, Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.