SUMMARY
Fas-Associated protein with Death Domain (FADD) was originally reported as a pro-apoptotic adaptor molecule that mediates receptor induced apoptosis. Recent studies have revealed a potential role of FADD in NF-κB activation, embryogenesis, and cell cycle regulation and proliferation. Over-expression of FADD and its phosphorylation have been associated with the transformed phenotype in many cancers and is therefore a potential target for therapeutic intervention. In an effort to delineate signaling events that lead to FADD phosphorylation and to identify novel compounds that impinge on this pathway, we developed a cell based reporter for FADD kinase activity. The reporter assay, optimized for a high throughput screen (HTS), measures bioluminescence in response to modulation of FADD kinase activity in live cells. In addition, the potential use of the reporter cell line in the rapid evaluation of pharmacologic properties of HTS hits in mouse models has been demonstrated.
Keywords: FADD, phosphorylation, non-invasive molecular imaging, bioluminescence, kinase activity
INTRODUCTION
FADD (FAS-ASSOCIATED DEATH DOMAIN CONTAINING PROTEIN) was originally identified as a critical adapter for formation of the death-inducing signaling complex (DISC),1–3 which plays a key role in relaying death signals originating from death receptors (such as Fas, DR4, and DR5). Subsequent studies revealed a potential role of FADD in NF-κB (nuclear factor–kappaB) activation, embryogenesis, cell cycle progression, and proliferation.4 Recent studies have led to a better understanding of the FADD gene and its location on chromosome 11q13.3, which is a hot spot for chromosomal amplification in a number of human cancers such as carcinomas of breast, bladder, esophagus, lung, and head and neck.5,6 Our recent studies provide evidence for overexpression of FADD mRNA and protein in human lung adenocarcinoma compared to normal lung.7 In addition, FADD overexpression in lung tumor biopsies was shown to correlate with poor clinical outcome.7 We have also shown that high levels of phosphorylated FADD (p-FADD), which is predominantly localized to the nucleus in lung tumor tissues, is a biomarker for aggressive disease and poor clinical outcome.5 The basis for this correlation is that p-FADD is a potent mediator of NF-κB activation. NF-κB is a heterodimeric protein composed of different combinations of members of the Rel family of transcription factors.8 A role of NF-κB as a regulator of cell fate decisions, such as resistance to programmed cell death and lack of proliferation control, and therefore in tumorigenesis has been well established.
Phosphorylation of FADD at serine 194 has been shown to be mediated by casein kinase Iα (CK1α)9 by FIST (FADD interacting serine threonine kinase),10 as well as by PKC-zeta.11 CK1α and FIST are the FADD kinases commonly identified in cancer cells. FIST activity can be downregulated in response to JNK inhibition,10 resulting in decreased FADD phosphorylation. In addition, CK1α inhibitors (CKI-7) also inhibit FADD phosphorylation. However, the regulation and role of FADD kinases in cancer is not well understood. To identify lead compounds that modulate FADD phosphorylation as potential therapeutic agents and to better define molecular events that lead to FADD phosphorylation, we describe here the development of a molecular imaging-based reporter for FADD kinase activity (FADD kinase reporter, FKR) and the optimization of the cell-based assay for high-throughput screening (HTS).
EXPERIMENTAL PROCEDURES
Plasmid construction
FKR was generated in the mammalian expression vector pEF. N-terminal firefly luciferase gene (NLuc) was amplified by PCR,12 using primers that generated a product comprising a SalI restriction site followed by a Kozak consensus sequence, with a NotI restriction site at the 3′ end. FHA2 was amplified from the Rad53p FHA2 domain with a sense primer containing a NotI site and a reverse primer containing an XbaI site. C-terminal firefly luciferase gene (C-Luc) was amplified using primers that produce a 5′ XbaI followed by the FADD kinase substrate sequence, 142–208 amino acids harboring serine 194, flanked by a linker (GGSGG) at each side, with a 3′ EcoRI restriction site after the terminating codon. FKR mutant was constructed by changing the phosphorylation site, serine 194, to alanine using the Quick Change kit (Stratagene, La Jolla, CA). All plasmids were verified by automatic DNA sequencing.
Cell culture and transfection
The human head and neck squamous carcinoma (UMSCC-1) cells were provided by Thomas Carey, Head and Neck Oncology Program, University of Michigan. Human lung epithelial carcinoma (A549) cells were purchased from the American Type Culture Collection. UMSCC1 and A549 (lung cancer) cells were grown in RPMI-1640 (Invitrogen, Carlsbad, CA) and Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen), respectively. Complete medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS; GIBCO, Carlsbad, CA) and 100 U/mL penicillin/streptomycin. Cell cultures were maintained in a humi-dified incubator at 37°C and 5% CO2.
To construct stable cell lines, the FKR plasmids (wild type and mutant) were stably transfected into UMSCC1 and A549 cells using Fugene (Roche Diagnostics, Indianapolis, IN), and stable clones were selected with 500 μg/mL G418 (Invitrogen). Resulting clones were isolated and cultured for further analysis by Western blot for determination of expression levels of the recombinant protein.
Antibodies and reagents
Rabbit polyclonal antibodies to phospho-FADD and c-Jun were purchased from Cell Signaling Technology (Danvers, MA). Goat polyclonal antibodies to CK1α and firefly luciferase were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Chemicon (Millipore, Billerica, MA), respectively. Mouse monoclonal antibodies to FADD were purchased from BD Pharmingen (San Diego, CA). Actin antibody was purchased from Sigma Aldrich (St. Louis, MO).
JNK Inhibitor (SP600125) was obtained from Calbiochem (EMD Chemicals, San Diego, CA) and CKI-7 from Toronto Chemical Research (New York, Ontario, Canada). Luciferin was obtained from Biosynth (Naperville, IL).
Small Interfering RNA (siRNA) transfection analysis
The siRNA sequences for sense strand CK1α and FIST/HIPK3 are 5′-CCAGGCAUCCCCAGUUG CUTT-3′, 5′-AAU ACU UAC GAA GUC CUU CAU-3′ respectively, and were synthesized from Invitrogen. FADD siGENOME Smart Pool and non-silencing siRNA (NSS) were synthesized by Dharmacon Research. siRNA transfection was carried out using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cells were analyzed 48–72 hours later for bioluminescence imaging followed by western blot.
Western blotting and immunoprecipitation
UMSCC1 FKR cells in culture dishes were collected and centrifuged at 1,800 × g for 5 min at 4°C. Cell pellets were washed twice with cold PBS and then lysed with a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 50 mM NaF, and 1 mM Na3VO4 and supplemented with complete protease inhibitors mixture (Roche Diagnostics). Cells in lysis buffer were rocked at 4°C for 30 min. The lysates were then cleared by centrifugation. The supernatants collected were estimated for protein content by a detergent-compatible protein assay kit from Bio-Rad (Hercules, CA). Lysates with equal amounts of protein were separated by Laemmli SDS/PAGE, and the levels of protein expression were detected by western blot analysis with anti-FADD (1:2000), p-FADD (1:1000), CK1α (1:200), HIPK3 (1:500), luciferase (1:5000) and actin (1:1000). Specific signals were visualized by using the Enhanced Chemiluminescence (ECL) Western Blotting System (GE Healthcare, Piscataway, NJ).
For immunoprecipitation, cell extracts (2000μg) were incubated with the luciferase antibody for 1 hour. Immune complexes were captured using protein G–Sepharose (GE Healthcare, Piscataway, NJ) and washed three times using RIPA buffer. The resulting pellet was boiled for five minutes in sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, 0.125M tris-HCL, pH 6.8) and resolved by SDS/PAGE.
96-well assay
A549 FKR cells were seeded in 96-well plates, using a Titertek Multidrop Microplate Dispenser (Thermo Fisher Scientific, Waltham, MA) and incubated 24 h prior to compound addition. Compound stocks were prepared in DMSO and diluted 1:100 in PBS. Then, 10 μL of the intermediate stock was added to each assay plate to achieve final concentrations from 3 nM to 50 μM using the Beckman Biomek NXP Laboratory Automation Workstation (Beckman Coulter, Fullerton, CA). The cells were incubated with test compounds at 37°C, 5% CO2 for 6 h. The final DMSO concentration was 0.1%.
Variability tests were conducted on 3 types of signals. Maximum and minimum signals were obtained by treating plates with 30 μM JNK inhibitor and DMSO, respectively. To estimate signal variability at points between the maximum and minimum signals, JNK inhibitor was used at a concentration of 15 μM. Results were analyzed by plotting the signal from each well against the well number in which the wells were ordered by row and then by column.
In vivo studies
A549 FKR (1.0 × 106) cells were transplanted into the flanks of nu/nu CD-1 nude mice (Charles River Laboratories, Wilmington, MA). When tumors reached approximately 100 mm3 in volume, the mice were randomized into 3 groups, and treatment was initiated. A CK1α inhibitor in DMSO was given via intraperitoneal injection at doses of 0.2 and 0.5 mg/kg. DMSO was used as control.
Bioluminescence imaging
Live-cell luminescent imaging was achieved by adding D-luciferin (200 μg/mL final concentration) to the assay medium following compound incubation. Photon counts were acquired 5 to 10 min after 37°C incubation with D-luciferin using the IVIS imaging system (Caliper Life Sciences, Hopkinton, MA). An exposure time of 1 min was used for acquisition. Living Image 3.0 software was used for data analysis (Caliper Life Sciences). For in vivo bioluminescence, mice were anesthetized using a 2% isofluorane/air mixture and injected with a single dose of 150 mg/kg D-luciferin in PBS intraperitoneally. Image acquisition was initiated 5 min following injection of luciferin. Consecutive bioluminescence images were acquired before treatment and then every 3 h for 9 h.
Data analysis
Data were collected from at least two independent experiments with three or more replicates per experiment. Graph Pad Prism v.5 nonlinear regression analysis (GraphPad Software, San Diego, CA) was used to generate the 50% inhibition concentration (IC50) values. Fold induction was calculated as signal to noise ratios. The Z-factor was calculated as previously described.13
RESULTS
Development and validation of bioluminescent FADD Kinase Reporter, FKR
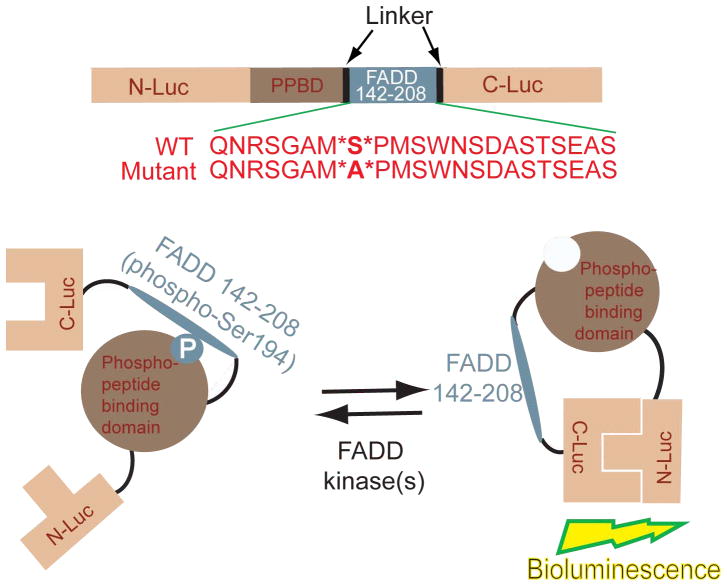
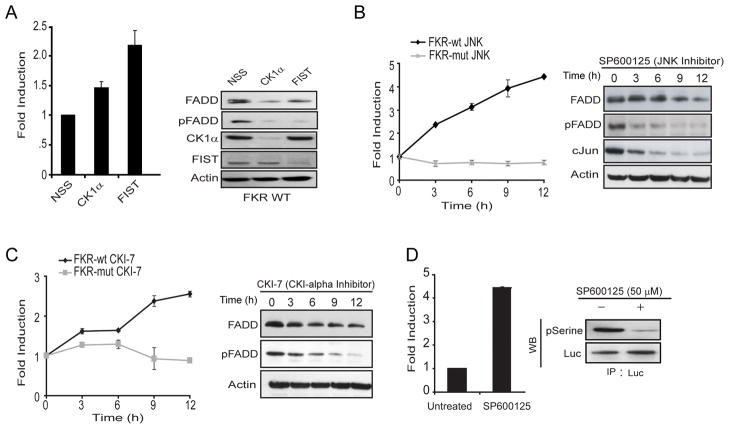
Rather than study each of the known FADD kinases and their regulation in normal versus tumor cells individually, we chose to develop a pan reporter for FADD kinase activity (FADD Kinase Reporter, FKR), that would non-invasively report on changes in FADD kinase activity in living cells. This reporter was based on a platform we recently described14 for imaging of Akt, except that the target peptide was substituted with residues 142–208 of FADD which encompasses Ser194, the primary site for phosphorylation in FADD (Fig. 1). The strategy for monitoring FADD kinases using FKR is based on the concept that in the presence of FADD kinase activity, phosphorylation of the target peptide at the serine residue (Ser194 of FADD) would prevent reconstitution of the two halves of the split luciferase due to steric constraints by virtue of the interaction between the phospho-serine and the FHA2 phospho-serine binding domain (Fig. 1). Inhibition of FADD kinase activity would result in loss of interaction between these domains allowing for reconstitution of luciferase activity which can then be imaged optically using luciferin as a substrate (Fig. 1). To validate that FKR was indeed a reporter for FADD kinase activity, we transfected FKR expressing UMSCC1 (UMSCC1 FKR-wt), head and neck squamous carcinoma cells with siRNA for FIST/HIPK3 or CK1α, two of the best studied FADD kinases in epithelial cells. As shown in Figure 2A, compared to cells transfected with a non-silencing siRNA, cells transfected with the FIST/HIPK3 or CK1α specific siRNA demonstrated a 2.2 and 1.5-fold increase in luciferase activity, respectively. Western blot analysis of these samples confirmed that transfection with siRNA for FIST/HIPK3 and CK1α resulted in decreased levels of the respective proteins. Inhibition of either of these two FADD kinases resulted in decreased phosphorylation of FADD, as evidenced by a decrease in phospho-FADD specific reactivity. The FADD specific antibody detected a doublet under control conditions but a single band when either of the FADD kinases were targeted using siRNA (Fig. 2A). To further validate that FKR was reporting on FADD kinase activity and that the reporter was quantitative, we treated UMSCC1 FKR-wt as well as UMSCC1 cells expressing mutant FKR (FKR-mut) with SP600125, a small molecule JNK inhibitor. A time dependent increase in bioluminescence activity, with a corresponding decrease in phospho-FADD and c-Jun levels was observed following treatment with SP600125 (Fig. 2B). Similarly, inhibition of the other FADD kinase, CK1α using a small molecule inhibitor (CKI-7) resulted in a time dependent increase of bioluminescence activity in UMSCC1 FKR-wt cells but not in UMSCC1 FKR-mut cells (Fig. 2C). Western blot analysis of these samples demonstrated that the increase in bioluminescence activity correlated with a decrease in FADD kinase activity as evidenced by a decrease in phospho-FADD levels (Fig. 2B and 2C). Lastly, to validate that FKR is a substrate for FADD kinases, we immunoprecipitated the FKR using a luciferase specific antibody from SP600125 treated and untreated cells. As with the previous experiment described above, a four-fold increase in bioluminescence activity (Fig. 2D) was monitored when UMSCC1 FKR-wt cells were treated with SP600125. A corresponding decrease in phosphorylation of FKR was also observed when the immunoprecipitated FKR was probed with phospho-serine antibody. This is consistent with the notion that FKR is a substrate for FADD kinase activity and that changes in its phosphorylation status are reflected by changes in its bioluminescence activity.
FIG. 1.
Construction of the FADD Kinase Reporter (FKR). The model shows a diagrammatic representation of the domain structure of FKR and the sequence of the C terminal domain which harbors serine 194, the target for FADD kinases. N-Luc and C-Luc are the N- and C-terminal domains of firefly luciferase that are fused to the FHA2 appropriate ends of the reporter. Two versions of the FKR were developed: the FKR (WT), which contains the wild type FADD peptide sequence and the FKR (Mutant), which contains a serine 194 substitution to alanine at the primary phosphorylation site. Phosphorylation of the FADD peptide at serine 194 results in interaction with FHA2 phosphopeptide binding domain (PPBD) causing steric constraints on C-luc and N-Luc. Inhibition of FADD kinase results in decreased binding of phospho-peptide and PPBD enabling N-Luc and C-Luc interaction to restore enzymatic activity.
FIG. 2.
Validation of the FADD Kinase Reporter (FKR). (A) UMSCC1 FKR stable cells were transfected with siRNA targeting CK1α, FIST/HIPK3 or non-silencing siRNA (NSS). Bioluminescence was measured after 48 hours of transfection. Cell lysates were prepared and analyzed by western blotting using antibodies specific for FADD, phospho-FADD, CK1α, FIST/HIPK3 or actin as a loading control. (B, C) UMSCC1 FKR (WT and Mutant) stable cells were treated with 50 μM JNK inhibitor (SP600125) and 250 μM CKI-7 inhibitor. Bioluminescence activity was measured at the indicated time points. Cell lysates were prepared and analyzed by western blotting using the indicated antibodies. (D) UMSCC1 FKR stable cells were treated with 50 μM JNK inhibitor (SP600125). Bioluminescence activity was measured after 12 hours of treatment. Cells were lysed and cell lysates were precipitated with luciferase (Luc) antibody. The products were analyzed by p-Serine and Luc immunoblot analysis.
HTS optimization and validation
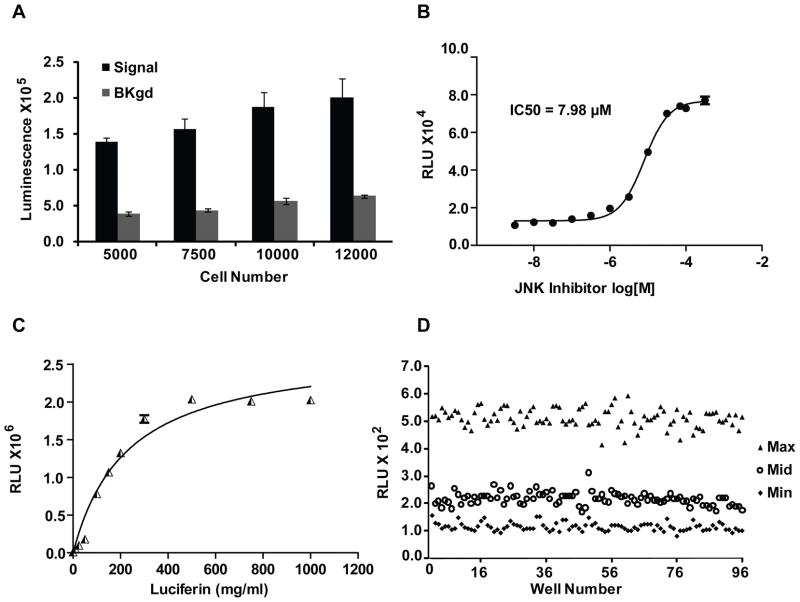
A549 FKR bioluminescence activity and background response was compared at seeding densities from 5000 to 12,000 cells per well in 96-well plates. Bioluminescence intensity was found to increase proportionally to cell number, indicating that signal intensity correlated directly with cell number per well (Fig. 3A). Comparison of signal-to-noise ratio at each density tested identified a proportional increase in background bio luminescence proportional to signal intensity. These data show that FKR signal-to-noise ratio is constant across a given range of cell densities and is minimally affected by cell number variation. Cell-based assays often have a narrow range of cell densities required to achieve an optimal response. Our data demonstrate that variability in seeding densities minimally affects A549 FKR signal-to-noise ratio, thereby enabling flexibility in cell plating. To validate the pharmacology of the miniaturized assay format and to define a parameter in which to rank order the efficacy of HTS hits based on potency, the IC50 for JNK inhibitor was determined. Data show a dose-dependent increase in bioluminescence activity in response to JNK inhibition (Fig. 3B). Based on previous studies demonstrating the role of P-glycoprotein (PGP) in the extrusion of luciferin from intracellular compartments,15 the effect of luciferin concentration on the bioluminescent response was evaluated. Cells were treated with increasing doses of D-luciferin to obtain saturating substrate conditions. The obtained results demonstrate measurable bioluminescence at 100 mg/mL luciferin, with saturating concentrations reached at 400 mg/mL (Fig. 3C). To assess the quality of the assay protocol in a 96-well plate format, a study designed with control plates expressing high, medium, and low responses was run to assess plate uniformity, signal-to-background ratio, and Z factor. The assay resulted in an average Z factor of 0.62, demonstrating the robustness of the A549 FKR cell line required for HTS. There was also minimal edge effects demonstrating that the assay is amenable to high-throughput cell plating instrumentation and liquid handling automation (Fig. 3D). The effect of DMSO on signal generation was examined. The data indicated that assay robustness was unaffected by DMSO concentrations ranging from 0% to 1% (data not shown).
FIG. 3.
HTS Optimization. (A) Effect of seeding density on signal and background in 96-well plates. Cells were seeded at 5000, 7500, 10000 and 12000 cells/well. Signal and background represent A549 FKR cells treated with 20 μM JNK inhibitor and DMSO respectively. Values represent the mean ± SEM of 2 separate experiments, each with 4 replicates. (B) 96-well dose dependent inhibition of FADD kinase activity in A549 FKR cells. Cells treated with JNK inhibitor (SP600125) as described in Materials and Methods. Three independent experiments were performed with 8 replicates for each dose. Data points represent the mean ± SEM for each condition. (C) Effect of luciferin concentration on cellular response. A549 FKR cells were treated with multiple doses of luciferin and imaged 5–10 minutes following substrate addition. (D) Representative results of 96-well assay performance. Cells were treated with 0, 15 and 30 μM JNK inhibitor for min, mid and max measurements respectively. The average Z-factor of 0.62 indicates assay suitability for high throughput screening.
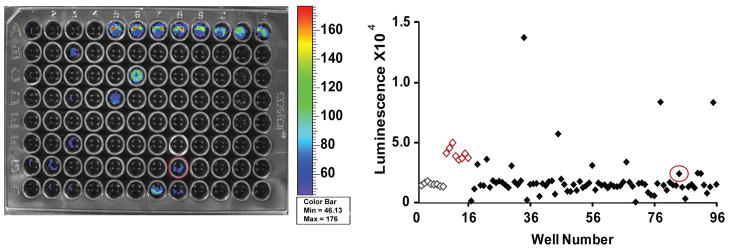
A validation screen, results of which are shown in Figure 4, was run to evaluate assay effectiveness in identifying FADD kinase inhibitors in the 96-well microplate format. The Kinase Inhibitor Library, KI-80 (TimTec LLC, Newark, DE), included 80 compounds with known kinase activities. The screen was run on 2 separate days and included intraplate controls as well as negative and positive control plates on each day to measure assay performance. The screen was run under optimized assay conditions and yielded an average Z factor of 0.60. The utility of the A549 FKR cell line to identify compounds that inhibit FADD phosphorylation was demonstrated by the validation screen in its ability to positively identify FADD kinase inhibitors as well as upstream modulators of FADD kinase activity. JNK inhibitor (SP600125) was present in the 80-compound library, and its identification as a positive hit demonstrated the fidelity of the A549 FKR cell line to identify FADD kinase inhibitors.
FIG. 4.
A549 FKR Validation Screen. The 80-compound screen was conducted under optimized assay conditions. Positive control JNK inhibitor (SP600125) was placed in wells A5-A12 and negative control, DMSO in wells A1-A4 and H9-H12 (controls identified on scatter plot by red and gray for positive and negative, respectively). Control compound JNK inhibitor, located in well G8 (circled), was part of the compound set and demonstrated bioluminescence activity in the validation screen thereby demonstrating assay utility in identifying modulators of FADD kinase activity.
A validation screen, results of which are shown in figure 4, was run to evaluate assay effectiveness in identifying FADD kinase inhibitors in the 96-well microplate format. The Kinase Inhibitor Library, KI-80 (TimTec LLC, Newark, DE) included 80 compounds with known kinase activities. The screen was run on two separate days and included intraplate controls as well as negative and positive control plates on each day to measure assay performance. The screen was run under optimized assay conditions and yielded an average Z-factor of 0.60. The utility of the A549 FKR cell line to identify compounds that inhibit FADD phosphorylation was demonstrated by the validation screen in its ability to positively identify FADD kinase inhbitors as well as upstream modulators of FADD kinase activity. JNK inhibitor (SP600125) was present in the 80-compound library and its identification as a positive hit demonstrated the fidelity of the A549 FKR cell line to identify FADD kinase inhibitors.
A549 FKR in vivo imaging
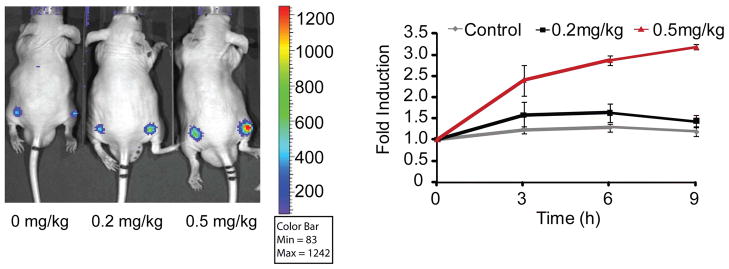
To demonstrate the utility of the reporter beyond HTS, A549 FKR cells were implanted subcutaneously as a xenograft model to assess the in vivo bioavailability and efficacy of FADD kinase inhibitors identified by the assay. When the A549 FKR tumors reached an approximate volume of 100 mm3, mice were treated with a CK1α inhibitor or with vehicle control. Bioluminescence activity was monitored over time. A time dependent increase in bioluminescence was observed in mice treated with 0.5 mg/kg, whereas animals treated with 0.2 mg/kg did not show an increase over the 9-hour measurement period (Fig. 5). These results suggest a steep dose threshold at which FADD kinase inhibition occurs. These data demonstrate that inhibition of FADD phosphorylation by a small molecule a CK1α inhibitor exhibit a time- and dose-dependent increase in bioluminescence activity, thereby demonstrating that the A549 FKR xenograft model can be used to validate drug target interactions in a biological context and to rapidly attrite HTS hits that exhibit poor bioavailability.
FIG. 5.
Time and Dose Imaging of FKR Activity. Representative image of CD-1 nude mice transplanted with A549 FKR cells and treated with 0.2 mg/kg and 0.5 mg/kg CK1α inhibitor. DMSO served as the control. Mice were imaged at 0, 3, 6 and 9 hours following compound injection. Graph represents induction of bioluminescence at each time point. Fold induction was calculated for each time point as the ratio of bioluminescence activity following treatment and the basal bioluminescence prior to treatment. Data shows a time and dose dependent increase in bioluminescence in vivo.
DISCUSSION
In an attempt to define the molecular basis of high phospho-FADD levels in tumor versus normal tissue (in HNSCC and lung cancer), we developed FKR, a reporter wherein FADD kinase activity, irrespective of the kinase involved, could be quantitatively and non-invasively monitored in living cells. This assay was adapted from our recently described reporter system for Akt also a Ser/Thr kinase,14 except that the Akt kinase substrate sequence was substituted with residues 142–208 of FADD which includes Ser194, the target for all known FADD kinases. We found that inhibition of FADD kinase activity through the use of siRNA or by small molecule resulted in a decrease in levels of phospho-FADD and a corresponding activation of the reporter’s bioluminescence activity in a dose and time dependent manner. The design of the reporter wherein inhibition of kinase activity would result in activation of the reporter (a positive signal) provides for numerous advantages. Firstly, the signal to noise is significantly improved and secondly, false positives are minimized (cytotoxic compounds would appear as false positives in an assay wherein inhibition of a kinase would result in loss of reporter activity). In contrast, cells expressing FKR-mut wherein a serine to alanine substitution was introduced (corresponding to Ser194 of FADD), inhibition of FADD kinase activity, did not result in a significant change in bioluminescence thereby demonstrating the specificity of the reporter.
The luciferase complementation reporter system described here possesses 1/100th the activity of wild-type luciferase in the absence of complementation and as much as 1/10th the activity of wild-type luciferase activity in the presence of complementation.16 Since the mutant reporter lacks the target phosphorylation site (Serine mutated to Alanine), it would be constitutively active and therefore possess a level of luciferase activity that is comparable to FKR in response to a FADD kinase inhibitor. In this regard, the mutant reporter possesses 1/10th the activity of wild-type luciferase. The presence of compounds in the library that stabilize the half life of luciferase and/or enhance expression of the reporter by up-regulating its transcription or translation would appear as positive hits. Since these compounds exert their FADD kinase independent activities equally on the mutant and the wild-type reporter, a secondary screen would identify them as false positives. This makes the mutant reporter a useful tool in the identification of false positives from a high throughput screen.
Further validation of the reporter was provided by studies demonstrating inhibition of JNK resulted in a four-fold increase in bioluminescence and a corresponding decrease in serine phosphorylation of FKR, demonstrating that the reporter itself was a substrate for FADD kinases. Recent studies in our lab have demonstrated that phosphorylation of FADD results in its stabilization while inhibition of FADD phosphorylation results in its ubiquitin dependent degradation. Our studies using MG132, a proteosome inhibitor, have confirmed that a decrease in total FADD levels in response to FADD dephosphorylation (i.e. in the presence of a FADD kinase inhibitor) can be reversed by inhibiting proteosomal degradation. (Supplementary Figure 2)
The FADD Kinase Reporter (FKR) allows for noninvasive monitoring of FADD-phosphorylation in live cells quantitatively and dynamically in real time. There are numerous advantages to screening compounds in a live cell assay as compared to a purified biochemical assay. These include the ability to evaluate drug-target interactions in a physiologically relevant context. For example, compounds unable to diffuse to the target site or are cytotoxic, go undetected, thereby leading to their immediate elimination from further investigation. In addition, the bioluminescence read out can be used quantitatively to determine optimal time points to measure the response of interest. While inherent toxicity is not detected in biochemical assays, live cell assays enable the measurement of short and long term responses, which helps rule out off-target effects such as toxicity to cells. Biochemical assays, however, are advantageous for high throughput screening due to parameters such as high sensitivity and the ability to define screening conditions.
Our data demonstrate that the A549 FKR signal to background ratio is independent of cell density within a given range as evidenced by the proportional increase in signal and background with increasing cell density. When optimizing an assay to a high throughput format, it is advantageous to incorporate flexibility into the screening protocol. Parameters including poor cell growth and differential seeding densities can affect assay results and reproducibility. However, our data demonstrates consistency in signal to background ratio independent of cell density and therefore allows for flexibility in cell plating conditions when conducting HTS. Treatment of cells with SP600125 (JNK inhibitor) results in a dose dependent increase in bioluminescence corresponding to a decrease in FADD kinase activity. The pharmacology of the assay can be used to measure day to day assay variability and to rank order compound hits according to potency. These optimization data were derived using the 96-well assay format; however optimization for the 384-well format has been conducted and can be found online as Supplementary Figure 1
The efficacy of FKR to selectively identify inhibitors of FADD kinase activity was demonstrated by the validation screen. The screen resulted in the positive identification of known FADD kinase modulators i.e. inhibitors of the MAPK pathway, previously shown to inhibit FADD phosphorylation in acute lymphocytic leukemia cells.17 Our positive control JNK inhibitor (SP600125), a known FADD kinase inhibitor, was also part of the compound set and demonstrated activity in the assay (Fig. 4). Notably, 10 μM JNK inhibitor as present within the compound collection demonstrated lower bioluminescent activity as compared to the 10 μM JNK inhibitor positive control wells. This is likely due to the compromised compound integrity as a result of multiple freeze thaw cycles. The ability of the assay to identify compounds that impinge on FADD phosphorylation from a diverse set of small molecule kinase inhibitors demonstrates the utility of the assay at selectively identifying compounds that modulate FADD kinase activity. However, the ability of FKR to identify FADD kinase inhibitors as well as compounds involved in upstream signaling events suggests the need for secondary screens.
Our current experience has identified two major sources of false positives against FKR. The first being compounds with inherent luminescence and the second source are compounds that modulate signaling events upstream of the target. The former can be eliminated with the FKR-mut reporter. Resolution of the latter, i.e., how to identify compounds specific to the inhibition of one of the known FADD kinases as opposed to upstream events, would require a purified system. However, the ability of the screen to identify upstream signaling events could be beneficial in identifying signaling hubs that impinge on the target kinase.
Identifying compound hits from a high throughput screen with desirable pharmacological properties requires tedious PK/PD and ADME/Tox studies and is both time consuming and labor intensive. The in vivo results presented here allows for the identification of compounds with desirable pharmacologic properties by rapidly evaluating drug target interactions in vivo thereby providing preliminary insight as to whether the drug is a potential therapeutic agent. The FKR cell-based assay platform will provide rapid identification of lead compounds from HTS, as well as providing in vivo validation of active compounds identified by HTS by rapidly validating drug target interaction and dose and schedule optimization.
Supplementary Material
Acknowledgments
This work was supported by the US National Institutes of Health research grants R01CA129623 (AR), R21CA131859 (AR), U24CA083099 (BDR) P50CA093990 (BDR) and by a developmental project to MSB by the National Institutes of Health through the University of Michigan’s Head and Neck SPORE Grant (5 P50 CADE97248).
References
- 1.Zhang J, Winoto A. A mouse Fas-associated protein with homology to the human Mort1/FADD protein is essential for Fas-induced apoptosis. Mol Cell Biol. 1996;16:2756–2763. doi: 10.1128/mcb.16.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 3.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 4.Hu WH, Johnson H, Shu HB. Activation of NF-kappaB by FADD, Casper, and caspase-8. J Biol Chem. 2000;275:10838–10844. doi: 10.1074/jbc.275.15.10838. [DOI] [PubMed] [Google Scholar]
- 5.Bhojani MS, Chen G, Ross BD, Beer DG, Rehemtulla A. Nuclear localized phosphorylated FADD induces cell proliferation and is associated with aggressive lung cancer. Cell Cycle. 2005;4:1478–1481. doi: 10.4161/cc.4.11.2188. [DOI] [PubMed] [Google Scholar]
- 6.Gibcus JH, Menkema L, Mastik MF, Hermsen MA, de Bock GH, van Velthuysen ML, Takes RP, Kok K, Alvarez Marcos CA, van der Laan BF, van den Brekel MW, Langendijk JA, Kluin PM, van der Wal JE, Schuuring E. Amplicon mapping and expression profiling identify the Fas-associated death domain gene as a new driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin Cancer Res. 2007;13:6257–6266. doi: 10.1158/1078-0432.CCR-07-1247. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Bhojani MS, Heaford AC, Chang DC, Laxman B, Thomas DG, Griffin LB, Yu J, Coppola JM, Giordano TJ, Lin L, Adams D, Orringer MB, Ross BD, Beer DG, Rehemtulla A. Phosphorylated FADD induces NF-kappaB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. Proc Natl Acad Sci U S A. 2005;102:12507–12512. doi: 10.1073/pnas.0500397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 9.Alappat EC, Feig C, Boyerinas B, Volkland J, Samuels M, Murmann AE, Thorburn A, Kidd VJ, Slaughter CA, Osborn SL, Winoto A, Tang WJ, Peter ME. Phosphorylation of FADD at serine 194 by CKIalpha regulates its nonapoptotic activities. Mol Cell. 2005;19:321–332. doi: 10.1016/j.molcel.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Rochat-Steiner V, Becker K, Micheau O, Schneider P, Burns K, Tschopp J. FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J Exp Med. 2000;192:1165–1174. doi: 10.1084/jem.192.8.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Thonel A, Bettaieb A, Jean C, Laurent G, Quillet-Mary A. Role of protein kinase C zeta isoform in Fas resistance of immature myeloid KG1a leukemic cells. Blood. 2001;98:3770–3777. doi: 10.1182/blood.v98.13.3770. [DOI] [PubMed] [Google Scholar]
- 12.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, Holland EC, Ross BD, Rehemtulla A. Molecular imaging of Akt kinase activity. Nat Med. 2007;13:1114–1119. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Bressler JP, Neal J, Lal B, Bhang HE, Laterra J, Pomper MG. ABCG2/BCRP expression modulates D-Luciferin based bioluminescence imaging. Cancer Res. 2007;67:9389–9397. doi: 10.1158/0008-5472.CAN-07-0944. [DOI] [PubMed] [Google Scholar]
- 16.Paulmurugan R, Gambhir SS. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem. 2005;77:1295–1302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng XW, Chandra J, Loegering D, Van Becelaere K, Kottke TJ, Gore SD, Karp JE, Sebolt-Leopold J, Kaufmann SH. Central role of Fas-associated death domain protein in apoptosis induction by the mitogen-activated protein kinase kinase inhibitor CI-1040 (PD184352) in acute lymphocytic leukemia cells in vitro. J Biol Chem. 2003;278:47326–47339. doi: 10.1074/jbc.M304793200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.