Abstract
A rapid, high throughput readout for single-nucleotide polymorphism (SNP) analysis was developed employing single base chain extension and cytometric analysis of an array of fluorescent microspheres. An array of fluorescent microspheres was coupled with uniquely identifying sequences, termed complementary ZipCodes (cZipCodes), which allowed for multiplexing possibilities. For a given assay, querying a polymorphic base involved extending an oligonucleotide containing both a ZipCode and a SNP-specific sequence with a DNA polymerase and a pair of fluoresceinated dideoxynucleotides. To capture the reaction products for analysis, the ZipCode portion of the oligonucleotide was hybridized with its cZipCodes on the microsphere. Flow cytometry was used for microsphere decoding and SNP typing by detecting the fluorescein label captured on the microspheres. In addition to multiplexing capability, the ZipCode system allows multiple sets of SNPs to be analyzed by a limited set of cZipCode-attached microspheres. A standard set of non-cross reactive ZipCodes was established experimentally and the accuracy of the system was validated by comparison with genotypes determined by other technologies. From a total of 58 SNPs, 55 SNPs were successfully analyzed in the first pass using this assay format and all 181 genotypes across the 55 SNPs were correct. These data demonstrate that the microsphere-based single base chain extension (SBCE) method is a sensitive and reliable assay. It can be readily adapted to an automated, high-throughput genotyping system.
[Primer sequences used in this study are available as online supplementary materials at www.genome.org.]
Analysis of DNA sequence variation has led to advances in the mapping of human disease genes (Sheffield et al. 1995). Recently, identification of single nucleotide polymorphisms (SNPs) and the application of SNP data have been the focus for human genetics research and genomic drug discovery. SNPs, appearing at an estimated one in one thousand base pairs and totaling >3 million in the human genome (Cooper et al. 1985), are the prevalent genetic variations. These biallelic markers offer the potential for identification of disease-causing genes and drug targets, development and redefining of diagnostics, and the establishment of markers for individualized medicines. With this goal in mind, ten of the world's largest pharmaceutical companies and five leading academic laboratories formed The SNP Consortium (TSC), to discover and map hundreds of thousands of SNPs in the human genome and to develop a map of high density SNP markers (Marshall 1999). Extremely efficient and cost effective technology will be required to utilize information from mapped SNPs for genotype profiling in thousands of patient and control DNA samples.
A number of different techniques have been reported in the literature for analyzing single-nucleotide polymorphisms. Conventional methods include single strand conformation polymorphism analysis (Orita et al. 1989), gel-based restriction fragment length polymorphism (RFLP), allele-specific oligonucleotide (ASO) hybridization (Saiki et al. 1989), oligonucleotide ligation assay (OLA) (Landegren et al. 1988), and primer extension assay (Syvanen et al. 1990). New technologies (e.g., chips, mass spectrometry, slides) have been developed and incorporated into the detection and readout of allele signals based on either hybridization or enzymatic discrimination (Livak et al. 1995; Chen et al. 1997; Fu et al. 1998; Tyagi et al. 1998; Chen et al. 1999; Gilles et al. 1999).
Microspheres have been used as solid support for biological reaction assays (McHugh 1994). An array of fluorescent polystyrene microspheres (Kettman et al. 1998) offer a novel technology platform capable of multiplexed assaying of numerous SNPs with increased flexibility over traditional assays. The standard set of 64 multiplexed microspheres is identified individually by red and orange fluorescence using a flow cytometer; signals are assayed by a green fluorochrome (Fulton et al. 1997; McDade and Fulton 1997; Kettman et al. 1998). We have developed a microsphere-based SNP assay that utilizes a DNA polymerase for single base chain extension (SBCE) for allele detection. This genotyping method has been used widely in other formats and has been proven to be highly specific and reliable (Nikiforov et al. 1994; Chen et al. 1999; Syvanen 1999). In our system, a DNA sequence (termed ZipCode) at the 5′ end of the capture oligonucleotide probe allows the resulting enzymatic reaction product to be captured by its complementary sequence (cZipCode), which has been coupled to a specific fluorescent microsphere. In this study, we demonstrate that microsphere-based SBCE is a flexible and reliable technology that can be adapted to high-throughput genotyping of DNA samples.
RESULTS
Analysis of SNPs in Multiplexed Reactions
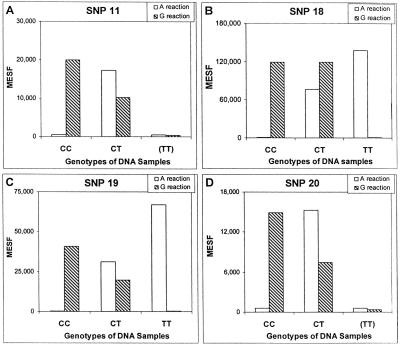
A primary advantage of the Luminex fluorescent microsphere technology is the capacity for conducting multiple biological reactions simultaneously in a single reaction vessel (i.e., well). By synthesizing stocks of unique pairings between microspheres and cZipCodes (DNA sequences), each fluorescent microsphere becomes the address (hybridization target) for a single SNP. Each SNP then simply requires an assigned ZipCode encoded in the same capture oligonucleotide to permit multiplexing (see details in Fig. 1). Each SNP is assayed for both alleles in two separate wells and the pair of values is used for determining the genotype. To test this, four polymorphisms with T and C alleles were assayed in multiplex reactions as described below. The four SNPs were amplified individually from either homozygous (CC or TT) or heterozygous (CT) genomic DNAs, and the PCR products were pooled separately according to their known genotypes [e.g., one pool consists of the products (CC) from the four SNPs]. SBCE reactions were performed with the four anti-sense capture oligonucleotides as primers to incorporate either A or G dideoxynucleotides. All of the four SNPs were genotyped correctly based on signal strength as measured by molecules of equivalent soluble fluorochrome (MESF values). The background MESF values were in the hundreds and represent only a few percent of the specific signals (Fig. 2). It is interesting to note that the signals for both the A and the G reactions were close to the background in the absence of specific PCR template (TT) for SNP11 and SNP20 (Fig. 2A,D). This indicates the absence of hybridization of those capture oligonucleotides and the other unrelated DNA templates. The results were nearly identical for the T and C reactions using the capture oligos for the opposite strand.
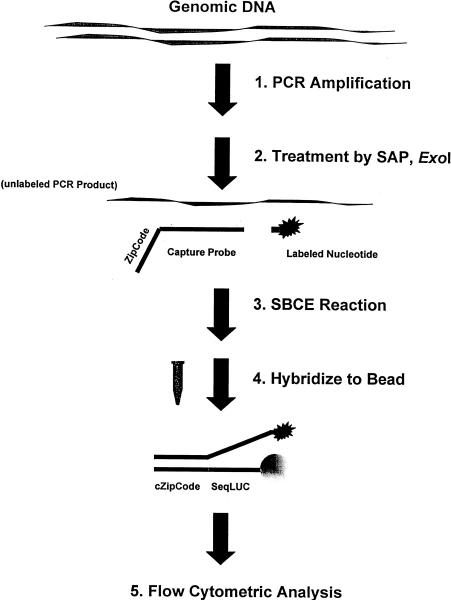
Figure 1.
Schematic presentation of the microsphere-based single base chain extension assays. DNA fragments containing the polymorphic site to be typed were amplified either individually or by multiplexed PCR (step 1). The PCR products containing a SNP site were pooled and treated with SAP and exonuclease I (step 2). After heat inactivation of the enzymes, the PCR products were used in the SBCE reactions (step 3) as described in Methods. For every SNP, one capture oligonucleotide probe with a unique ZipCode sequence was designed and used to assay the two alleles in each of two separate wells with a different labeled ddNTP per well. Multiplexed SNP analysis could be achieved by the employment of different ZipCode sequences for different SNPs in the presence of pooled PCR products. After the completion of the SBCE reaction, ∼1200 of each type of microsphere [with an attached oligonucleotide encoding the complement to the ZipCode sequences and a common luciferase sequence (SeqLUC)] were added to the completed SBCE reactions. The hybridization reactions were carried out at 40°C in the presence of NaCl for >2 hr (step 4). The microspheres were then subjected to flow cytometric analysis (step 5). Minimums of 100 of each type of microsphere were read and the mean value of MESF was used for determining the genotypes. The fluorescence signal of the corresponding microsphere without SBCE reactions (microsphere alone) or SBCE reactions without AmpliTaq FS were subtracted from the MESF values.
Figure 2.
SNP analysis in multiplexed reactions. PCR products were amplified individually from genomic DNA with either homozygous genotypes (CC and TT) or heterozygous genotype (CT). PCR products were then pooled according to their known genotypes into three separate groups. For example, one pool contained the homozygous PCR products (CC) from the four SNPs and so on. The three pools (15 ng of each PCR products) were used as templates and assayed separately for either A or G (striped columns) incorporation with the antisense probe as described in Methods. The MESF values from each of the three genotypes for each SNP are grouped together and shown. About 10,000 microspheres were pretreated with BSA at 1 mg/ml for 45 min and then added to each reaction to capture the SBCE products. The fluorescent intensity of the microspheres is represented by the MESF values on the y-axis. A pair of numbers from the A and G reactions determine the genotypes of the samples analyzed. Genotypes of the DNA samples were labeled as CC, CT, and TT. The absence of the PCR products for SNP11 and SNP20 for the TT allele is indicated by (TT).
Optimization of the Microsphere-Based SBCE Reactions
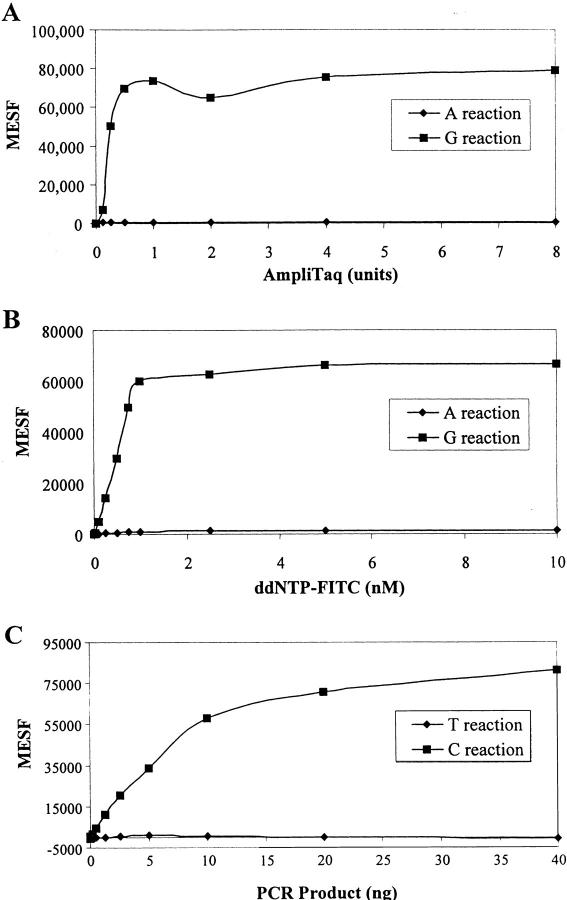
When it is necessary for large numbers of SNPs to be assayed in thousands of DNA samples, a reliable robust assay with minimal reagent costs will be essential. Therefore, several experiments were performed to optimize reaction conditions. Figure 3A shows a typical titration curve of AmpliTaq FS for SNP18 in a multiplex reaction of four SNPs (the same SNPs were used as described in the previous multiplex experiment). A homozygous mixture of PCR products (CC) of the four SNPs was used as template and was assayed for alleles A and G with the antisense capture oligonucleotide. As expected, the specific signal of the G reaction was very high while the A reaction signal remained low. Similar results were obtained for the other three SNPs. There is no significant increase of signal between 0.5 to 8 units of the DNA polymerase used (Fig. 3A). We believe that this may be due to competition of excess unlabeled capture probes over labeled capture probe for complementary ZipCode sites on the fluorescent microspheres.
Figure 3.
Optimization of SBCE reactions. The MESF values in the y-axis represent the mean fluorescence per microsphere. (A) Titration of the AmpliTaq FS enzyme for SNP18 in a multiplex reaction of four SNPs as used in Figure 2. A mixture of 15 ng PCR products containing homozygous CC genotypes was used as template for either G (█) or A (♦) reactions and the assays were performed with the anti-sense probe as described in Methods. A total of 10,000 microspheres were used for each reaction. The value obtained from the reaction in the absence of enzyme was subtracted from the data points. (B) Titration of ddNTPs for SNP18 was done using the same multiplex conditions as in (A). The experiments were performed with various amounts of fluorescent-labeled ddNTP. The ratio of labeled to unlabeled ddNTPs was kept constant at 1:3. The signals remained fairly consistent for the G reactions (█,) at and above 0.75 nm of ddGTP; A reactions (♦) remained near 0. The results for the other three SNPs are nearly identical to the results shown here. (C) Effect of PCR products on the enzymatic activities. A PCR product (250 bp) generated from a homozygous (CC) DNA sample for SNP18 was used as template for assaying the incorporation of either C (█) or T (♦) nucleotides with the sense capture oligonucleotide.
Figure 3B displays the signal strengths of SNP18 at various concentrations of ddNTP-FITC. The reactions were performed in the presence of three other SNPs (as in Fig. 3A) and the results for the four SNPs were nearly identical. PCR product amplified from a homozygous (CC) DNA sample was used as template and the antisense capture oligonucleotide was used as primer. Specific incorporation of ddGTP–FITC was found to generate strong signal while the signal for the A reaction was near the background level. Signals were found to remain constant as the concentration of ddNTP-FITC was reduced from 10 to 1 μm. A near linear increase of specific signal (G reaction) was observed when the ddNTP was at a much lower concentration (from 20 to 750 nm) in the SBCE reaction.
A key component of the microsphere-based SBCE system is the capture oligonucleotide, which is used both as the primer for the base incorporation and as the anchor for the resultant SBCE product to be hybridized to the appropriate microsphere. Various concentrations of the capture oligonucleotide were analyzed under standard conditions. No significant difference was observed between 10 and 100 nm. When the capture oligonucleotide concentration increased to 125 nm, the signals were found to be reduced significantly as excess nonextended oligonucleotide primers reduced the binding of extended primers to the microspheres.
The level of PCR amplification varies and is dependent on, among other factors, primers and template sequences. This variability is particularly true for multiplex PCRs and it is therefore hard to control and predict. For this reason, the sensitivity and tolerance of the microsphere-SBCE assay were tested with various amounts of PCR products under standard conditions (Fig. 3C). In this experiment, PCR product amplified from homozygous (CC) genomic DNA was used and assayed for either the specific incorporation of a C nucleotide or the nonspecific incorporation of a T nucleotide. While the nonspecific T incorporation remained near zero, the signal from the C reaction was found to increase with increasing quantity of PCR product (up to 40 ng; Fig. 3C). The specific signals were proportional to the amount of PCR products used, up to 2.5 ng. The correct genotypes were generated in the presence of as little as 0.5 ng of PCR product, where the MESF values for the C and T reactions were 4400 and 200, respectively. This suggests that our assay system is fairly sensitive and can tolerate up to an 80-fold variation of template material. These results have significant ramifications for multiplexed PCR and high-throughput genotyping efforts.
Validation of the Microsphere-Based SBCE Assays
It is well known that one allelic variant of the apolipoprotein, APOE4, is a significant susceptibility allele or risk factor for younger age onset of Alzheimer's disease (Saunders et al. 1993; Strittmatter et al. 1993). Over 100 SNPs have been developed around the APOE gene for association studies (Lai et al. 1998). These SNPs were identified by DNA sequencing of amplicons from the seven CEPH DNAs; therefore, nearly all of the genotypes for the SNPs are available (Lai et al. 1998). A total of 58 SNPs were selected randomly from this set, and SBCE assay probes were synthesized. A set of 58 unique cZipCode sequences, validated empirically for non-cross-reactivity, were coupled to 58 microspheres (of 64 possible) for capturing each of the SNPs (Table 1).
Table 1.
Compatible ZipCode DNA Sequences
| ZipCode | DNA sequence | ZipCode | DNA sequence | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | G | A | T | G | A | T | C | G | A | C | G | A | G | A | C | A | C | T | C | T | C | G | C | C | A | 35 | A | C | G | A | C | T | G | C | G | A | G | G | T | G | C | G | G | T | A | A | G | C | A | C | A |
| 2 | C | G | G | T | C | G | A | C | G | A | G | C | T | G | C | C | G | C | G | C | A | A | G | A | T | 36 | G | C | G | A | T | C | G | C | C | G | G | G | A | G | A | T | A | T | A | C | C | C | A | A | C |
| 3 | G | A | C | A | T | T | C | G | C | G | A | T | C | G | C | C | G | C | C | C | G | C | T | T | T | 37 | T | C | G | T | G | C | C | G | G | A | C | T | C | G | A | G | C | A | C | C | A | A | T | A | C |
| 4 | C | G | G | T | A | T | C | G | C | G | A | C | C | G | C | A | T | C | C | C | A | A | T | C | T | 38 | G | C | T | T | T | A | G | C | A | C | C | G | C | G | A | T | G | G | C | G | T | A | G | A | C |
| 5 | G | C | T | C | G | A | A | G | A | G | G | C | G | C | T | A | C | A | G | A | T | C | C | T | C | 39 | C | A | G | C | C | G | C | G | G | T | A | C | T | G | A | A | T | G | C | G | A | T | G | C | T |
| 6 | C | A | C | C | G | C | C | A | G | C | T | C | G | G | C | T | T | C | G | A | G | T | T | C | G | 40 | C | C | C | C | G | G | A | T | A | G | C | T | G | A | C | G | A | G | G | C | T | T | A | C | G |
| 7 | C | G | A | C | T | C | C | C | T | G | T | T | T | G | T | G | A | T | G | G | A | C | C | A | C | 41 | T | C | C | G | G | A | C | A | G | G | T | T | G | G | G | G | T | G | C | G | T | T | T | G | G |
| 8 | C | T | T | T | T | C | C | C | G | T | C | C | G | T | C | A | T | C | G | C | T | C | A | A | G | 42 | C | G | T | A | G | A | G | C | A | A | C | G | C | G | A | T | A | C | C | C | C | C | G | A | C |
| 9 | G | G | C | T | G | G | G | T | C | T | A | C | A | G | A | T | C | C | C | C | A | A | C | T | T | 44 | A | G | C | A | G | C | A | G | T | G | A | C | A | A | T | G | C | C | A | C | C | G | C | C | G |
| 10 | G | A | A | C | C | T | T | T | C | G | C | T | T | C | A | C | C | G | G | C | C | G | A | T | C | 46 | T | C | G | C | C | C | G | C | G | G | A | C | A | C | C | G | A | G | A | A | T | T | C | G | A |
| 12 | T | T | T | C | G | G | C | A | C | G | C | G | C | G | G | G | A | T | C | A | C | C | A | T | C | 48 | G | A | G | G | C | A | G | A | T | C | C | G | T | A | G | G | C | G | G | G | T | G | C | A | T |
| 14 | C | T | C | G | G | T | G | G | T | G | C | T | G | A | C | G | G | T | G | C | A | A | T | C | C | 49 | G | C | G | A | T | A | G | C | C | A | G | T | G | C | C | G | C | C | A | A | T | C | G | T | C |
| 15 | T | C | A | A | C | G | T | G | C | C | A | G | C | G | C | C | G | T | C | C | T | G | G | G | A | 50 | A | G | C | G | G | T | C | A | C | C | A | T | G | G | C | C | A | C | G | A | A | C | T | G | C |
| 16 | G | C | G | A | A | G | G | A | A | C | T | C | G | A | C | G | T | G | G | A | C | G | C | C | G | 51 | T | T | G | C | A | A | C | A | G | C | A | G | C | C | C | G | A | C | T | C | G | A | C | G | G |
| 17 | C | G | G | G | G | A | T | A | C | C | G | A | T | C | T | C | G | G | G | C | G | C | A | C | A | 52 | T | G | A | C | T | C | C | G | G | C | G | A | T | A | C | G | G | G | C | T | C | C | G | A | A |
| 18 | G | G | A | G | C | T | T | A | C | G | C | C | A | T | C | A | C | G | A | T | G | C | G | A | T | 53 | A | C | C | G | G | C | T | A | C | C | T | G | G | T | A | T | C | G | G | T | C | C | C | G | A |
| 19 | C | G | T | G | G | C | G | G | T | G | C | G | G | A | G | T | T | T | C | C | C | C | G | A | A | 54 | G | A | G | C | G | A | G | C | G | G | G | C | A | A | A | C | G | C | C | A | G | T | A | C | T |
| 20 | C | G | A | T | C | C | A | A | C | G | C | A | C | T | G | G | C | C | A | A | A | C | C | T | A | 55 | A | G | T | C | G | A | A | G | T | G | G | G | C | G | G | C | G | T | C | A | G | A | C | T | C |
| 21 | C | T | G | A | A | T | C | C | T | C | C | A | A | C | C | G | G | G | T | T | G | T | C | G | A | 56 | C | A | C | C | A | C | C | A | G | T | G | C | C | G | C | T | A | C | C | A | C | A | A | C | G |
| 22 | T | T | C | G | G | C | G | C | T | G | G | C | G | T | A | A | A | G | C | T | T | T | T | G | G | 57 | C | C | G | T | G | T | T | A | A | C | G | G | C | G | C | G | A | C | G | C | A | A | G | G | A |
| 23 | G | T | A | A | A | T | C | T | C | C | A | G | C | G | G | A | A | G | G | G | T | A | C | G | G | 58 | G | A | G | T | G | A | A | C | G | C | A | G | A | C | T | G | C | A | G | C | G | A | G | G | C |
| 24 | C | C | G | G | C | T | T | T | G | A | A | C | T | G | C | T | C | A | C | C | G | A | T | C | T | 59 | C | G | G | C | G | G | T | C | T | T | C | A | C | G | C | T | C | A | A | C | A | G | C | A | G |
| 27 | A | C | T | A | C | G | C | A | A | C | A | C | C | G | A | A | C | G | G | A | T | A | C | C | C | 60 | G | T | T | G | G | G | C | C | C | G | A | G | C | A | C | T | G | C | A | A | G | C | A | C | C |
| 28 | G | G | A | C | C | A | A | T | G | G | T | C | C | C | A | T | T | G | A | C | C | A | G | G | T | 61 | T | C | G | G | C | G | T | A | C | G | A | G | C | A | C | C | C | A | C | A | C | C | C | A | G |
| 29 | C | A | A | C | G | C | T | G | A | G | C | G | C | G | T | C | A | C | T | G | A | C | A | T | A | 62 | C | C | C | C | A | A | A | C | G | T | A | C | C | A | A | G | C | C | C | G | C | G | T | C | G |
| 31 | G | A | G | A | C | A | A | A | G | G | T | C | T | G | C | G | C | C | A | G | C | A | C | C | A | 63 | A | T | G | G | C | A | C | C | G | A | C | G | G | C | T | G | G | C | A | C | A | C | C | A | C |
| 32 | T | G | G | C | C | A | C | A | C | T | G | T | C | C | A | T | T | T | G | C | G | C | G | G | T | 64 | A | G | C | C | G | C | G | A | A | C | A | C | C | A | C | G | A | T | C | G | A | C | C | G | G |
| 33 | C | C | T | T | G | C | G | A | C | G | T | G | T | C | A | A | G | T | T | G | G | G | G | T | C | 65 | C | G | C | G | C | G | C | A | G | C | T | G | C | A | G | C | T | T | G | C | T | C | A | T | G |
| 34 | A | G | G | T | T | A | G | G | G | T | C | G | C | G | C | C | A | A | A | C | T | C | T | C | C | 66 | T | A | C | C | G | G | C | G | G | C | A | G | C | A | C | C | A | G | C | G | G | T | A | A | C |
A typical set of these experiments to analyze these 58 SNPs is described below. Each SNP was first amplified individually using each of seven CEPH DNAs as target in 406 PCR reactions (58 SNPs × 7 CEPH targets). Equal masses of PCR products were pooled in groups of 12 SNPs for each of the CEPH DNAs to form 35 target pools. Four microsphere-based SBCE reactions were performed on the pooled PCR products—one reaction for each nucleotide. Of the total 58 SNPs, 55 were converted successfully to this assay format on the first pass. The failure of two SNPs was traced to problems with oligonucleotides; one capture probe contained an incorrect sequence and one PCR primer set amplified the wrong amplicon. A third SNP failure, a homozygous GG, showed greater incorporation of G than the other three nucleotides, but signal intensity was only 2200 MESF and the signal-to-noise ratio was <2. This SNP was later rescued successfully by redesigning its capture probe to be complementary to the opposite strand. In these experiments MESF values <3000 were assumed to be nonspecific background. In general, we have not experienced problems with high nonspecific background, rather our failed SNPs stem from low positive signals, where poor signal-to-noise ratios make genotypic calls questionable.
Table 2 shows the signal intensity of each of the four alleles in the seven CEPH DNAs for one 12-SNP pool, generating 84 genotypes. For SNP503 with A and G alleles, the genotypes easily can be read as GG, AG, GG, AG, GG, AG, and GG for the seven CEPH DNA samples, based upon the intensity of the 4 bases (Table 2). Because of the dramatic difference between the signal and noise, all of the remaining 77 genotypes could be determined easily as well (Table 2). The 12 SNPs represent several different types of base substitutions (AG, AT, CG, CT, and GT). All of the five types of SNPs examined can be analyzed by assaying the four bases.
Table 2.
Calculated MESF Values for Multiplexed Analysis
| DNA sample | ddNTP terminator reaction | SNP analyzeda | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 503 | 504 | 505 | 506 | 507 | 509 | 510 | 511 | 512 | 513 | 514 | 515 | ||
| CEPH1 | A | −40 | 989 | 15,415 | 2,866 | 1,727 | 23,113 | 1,800 | 2,051 | 2,986 | 4,750 | 2,114 | 37,943 |
| C | 1,118 | 3,062 | 2,341 | 4,263 | 3,128 | 3,060 | 22,545 | 26,298 | 4,676 | 34,825 | 2,862 | 2,502 | |
| G | 30,260 | 17,380 | 10,218 | 40,707 | 44,653 | 854 | 1,713 | 2,077 | 18,400 | 3,360 | 31,014 | 2,052 | |
| T | 1,677 | 2,997 | 766 | 4,868 | 2,630 | 3,428 | 1,862 | 3,097 | 2,139 | 31,556 | 1,350 | 3,399 | |
| CEPH2 | A | 24,324 | 1,281 | 1,158 | 36,042 | 2,001 | 14,220 | 2,671 | 2,695 | 3,273 | 5,775 | 31,821 | 20,874 |
| C | 682 | 2,983 | 1,701 | 6,047 | 56,597 | 1,108 | 23,931 | 27,097 | 4,258 | 35,886 | 2,979 | 813 | |
| G | 13,750 | 8,003 | 20,345 | 19,321 | 19,224 | 616 | 1,348 | 2,082 | 2,544 | 3,418 | 15,880 | 1,400 | |
| T | 2,681 | 10,827 | 781 | 3,629 | 2,011 | 24,111 | 2,846 | 3,200 | 17,666 | 36,343 | 3,463 | 30,878 | |
| CEPH3 | A | −300 | 1,404 | 31,515 | 8,101 | 1,950 | 13,979 | 2,155 | 3,071 | 3,236 | 5,454 | 1,392 | 41,198 |
| C | 852 | 2,574 | 616 | 5,550 | 8,467 | 1,321 | 11,044 | 22,882 | 1,511 | 4,293 | 2,145 | 1,331 | |
| G | 27,595 | 15,679 | 1,128 | 41,843 | 43,402 | 288 | 886 | 1,563 | 10,563 | 3,606 | 31,904 | 1,612 | |
| T | 1,154 | 2,407 | 1,681 | 4,091 | 1,661 | 22,340 | 10,387 | 3,386 | 13,856 | 63,945 | 1,021 | 3,665 | |
| CEPH4 | A | 21,336 | 1,409 | 14,919 | 3,654 | 1,707 | 29,352 | 2,341 | 2,062 | 2,487 | 5,333 | 1,090 | 39,260 |
| C | 747 | 1,235 | 992 | 5,141 | 2,310 | 1,788 | 30,619 | 12,469 | 2,622 | 5,754 | 2,299 | 1,856 | |
| G | 14,173 | 8,235 | 10,605 | 49,175 | 49,640 | 1,174 | 2,042 | 2,679 | 10,182 | 4,703 | 31,362 | 1,152 | |
| T | 1,843 | 10,360 | 1,424 | 5,601 | 3,083 | 4,322 | 3,176 | 10,497 | 11,547 | 57,578 | 1,695 | 3,436 | |
| CEPH5 | A | 667 | 1,509 | 16,122 | 3,563 | 1,332 | 27,454 | 1,379 | 2,370 | 4,242 | 5,533 | 2,124 | 38,337 |
| C | 621 | 3,295 | −225 | 7,332 | 2,046 | 3,560 | 30,488 | 22,694 | 3,809 | 32,136 | 2,506 | 2,943 | |
| G | 30,852 | 17,229 | 10,238 | 48,006 | 48,465 | 1,235 | 2,052 | 2,901 | 3,591 | 4,860 | 32,230 | 2,980 | |
| T | 2,125 | 2,449 | 2,170 | 5,201 | 2,163 | 3,127 | 3,233 | 3,685 | 25,453 | 34,144 | 2,932 | 3,118 | |
| CEPH6 | A | 21,077 | 1,331 | 14,439 | 37,356 | 1,884 | 12,900 | 2,253 | 2,886 | 3,023 | 5,775 | 1,520 | 33,546 |
| C | 2,873 | 2,006 | 1,083 | 6,681 | 57,081 | 2,337 | 30,141 | 22,979 | 4,060 | 7,013 | 2,746 | 3,118 | |
| G | 13,735 | 7,809 | 10,383 | 25,831 | 22,917 | 900 | 1,996 | 2,685 | 4,185 | 4,661 | 30,584 | 2,295 | |
| T | 1,939 | 10,925 | 516 | 5,166 | 2,234 | 22,665 | 2,799 | 4,301 | 27,736 | 69,067 | 2,289 | 2,794 | |
| CEPH7 | A | 466 | 1,012 | 31,337 | 37,161 | 2,412 | 12,493 | 2,495 | 3,040 | 3,820 | 6,882 | 1,848 | 39,483 |
| C | 1,154 | 2,472 | 2,215 | 5,889 | 54,698 | 2,098 | 37,761 | 23,440 | 3,622 | 7,953 | 2,947 | 3,590 | |
| G | 33,017 | 18,848 | 1,344 | 25,874 | 22,774 | 1,346 | 2,650 | 3,494 | 4,091 | 5,444 | 32,169 | 1,363 | |
| T | 2,428 | 1,276 | 1,304 | 5,636 | 1,828 | 21,973 | 2,774 | 4,218 | 26,748 | 67,520 | 1,186 | 2,820 | |
Twelve SNPs (SNP503–SNP515) were amplified separately and pooled. The pooled samples were then analyzed in a multiplexed single base chain reaction as described in Methods. MESF values indicating the presence of a particular allele are indicated in boldface type. Sequences of PCR primers and capture oligonucleotides will be provided upon request.
A total of 181 genotypes determined by SBCE from 55 SNPs (21 SNPs assayed in 7 DNAs and 34 SNPs in one DNA) were compared to their known genotypes as determined by either DNA sequencing or TaqMan analysis. All of the 181 genotypes generated from our assays were proven to be correct.
Fifty-two SNP Multiplex Reactions
To test the limit of higher multiplexing capacity, the same SNPs analyzed in the 12-plex experiments from a single DNA sample were assayed again in a 52 multiplex format. PCR products from 52 SNPs were pooled and assayed for the 4 bases. In general, the signal was lower than those in the 12-plex experiments. However, all of the 52 genotypes determined from this experiment were found to be the same as in the 12 SNP multiplex reactions. Therefore, all of the 52 genotypes could be confirmed in a single multiplex reaction.
DISCUSSION
In this report we describe a microsphere-based technology platform that could be used in a high-throughput format. The success of the microsphere-based SBCE system depends on three components: (1) accuracy of the allele discrimination reaction by the DNA polymerase, which has been well-established (Syvanen 1999), (2) specific hybridization of the products of SBCE reactions to their address-microspheres, and (3) sensitivity of flow cytometer readout of biological reaction signals on individual fluorescent microspheres. Data presented here demonstrate that the microsphere-based SBCE system is both reliable and efficient.
Several interesting features have been integrated into this microsphere-based readout technology platform. For example, conducting the enzymatic reactions in solution, as opposed to the microsphere surface, allows us to obtain the benefit of liquid-phase kinetics. Furthermore, each fluorescent microsphere requires only one unique cZipCode, and there is a nearly unlimited variability of DNA sequence that could be purchased readily and linked covalently to the microsphere surface. Such DNA sequences have been employed successfully as unique identifiers (molecular bar codes) in the analysis of Saccharomyces cerevisiae deletion strains (Shoemaker et al. 1996). The 20-base luciferase sequence, common to each cZipCode oligonucleotide, allows monitoring of oligonucleotide-to-microsphere coupling efficiency and therein, quality assurance (see Methods). The cZipCode also permits a single standard set of cZipCoded microspheres to be used repeatedly for analyzing multiple sets of SNPs. The dual function capture oligonucleotide is designed to have the same melting temperature for all ZipCode sequences, thereby allowing the specifically incorporated fluorescein-ddNTP to be captured by its appropriate cZipCode-linked microspheres.
The microsphere-based SBCE system offers several distinct advantages over the many other methods reported in the literature. First, this technology is highly suitable for large-scale genetic analysis due to its multiplexing capability. Although data for this study were generated on a FACSCalibur with individual loading, up to 96 assays can now be analyzed on the much less expensive LX100 (Luminex Corp., Austin, TX) equipped with an XY-plate reader. With the set of 25 microspheres available for the LX100, one fluorescent label per well, and an approximate reading time of the XY-plate reader at one microtiter plate per hour, ∼10,000 genotypes could be generated in an 8-hr day per machine. A 12-fold increase of throughput to 120,000 genotypes could be achieved through the combination of (1) a set of 100 microspheres, (2) automation that could allow extended operation, and (3) decreasing the number of the microspheres to be read. Bioinformatics tools, such as programs to design SBCE primers to avoid nonspecific extension, will be critical to achieve such a throughput.
Another inherent advantage in our microsphere-based system is the reduced cost for PCR amplification. The multiplexed SBCE reactions described here fit well with multiplex PCRs for sample amplification. By allowing SNPs to be amplified by multiplexed PCR, a further significant savings in cost is achieved. Our current cost for each genotype assay varies between 0.20 and 0.40, depending on certain cost parameters which includes labor, reagent costs, the total number of assays run, the number of simultaneous assays performed (multiplex factor), and whether many SNPs are assayed on few DNA samples, or few SNPs are assayed on many DNA samples. The fluorescent microsphere-SBCE technology is also very flexible and allows the relatively high-throughput and inexpensive genotyping capabilities to be available to many research laboratories, including those laboratories that currently have limited genotype throughput. In addition, because the resulting output consists of two numeric values per sample in an Excel spreadsheet, the genotypes can be assigned automatically using a simple computer program. Furthermore, due to the significant difference in values between signal and noise, genotypes can be assigned in the absence of a positive control.
In this report we describe a readout technology employing flow cytometric analysis of microspheres and allele discrimination using single base chain extension reactions. The microsphere-based system is also adaptable to allele detection using OLA. We have successfully developed this microsphere-based OLA assay for SNP analysis (Iannone et al. 2000).
A system based on competitive hybridization with sequence-specific probes utilizing fluorescent microspheres for multiplexed SNP analysis was first reported in 1997 by Fulton et al. Like other hybridization-based SNP systems (e.g., TaqMan), the success of this assay depends on the sequence-content surrounding the polymorphic sites. This requires extensive experience in probe design as well as optimization in genotyping applications. Therefore, application of this assay for SNP genotyping has been very challenging. By separating the allele detection in a solution-based reaction from the microspheres, the SBCE reaction conditions are practically universal and almost no optimization is required (Chen et al. 1999). This robustness is crucial for any high-throughput genotyping effort. With the employment of robotics for automation and bioinformatics tools for sample tracking and data management, the microsphere-based single base chain extension system will provide a new technology platform for high-throughput genotyping.
METHODS
AmpliTaq, AmpliTaq Gold, and AmpliTaq FS (catalog no. 361390) DNA polymerase were purchased from PE Applied Biosystems (Foster City, CA). KlenTaq was obtained from Ab Peptides, Inc. (St Louis, MO). PicoGreen for double strand DNA quantification was purchased from Molecular Probes (Eugene, OR). Shrimp alkaline phosphatase (SAP) and Exonuclease I (Exo I) were obtained from Amersham Pharmacia. Fluorescence labeled dideoxynucleotide triphosphates (ddNTPs) were obtained from NEN Life Science Products, Inc. (Boston, MA). Unlabeled ddNTPs were from Amersham Pharmacia. Unmodified oligonucleotides were purchased from Biosource International (Camarillo, CA). CEPH DNAs (NA07435, NA07445, NA10848, NA10849, NA07038A, NA06987A, and NA10846) are ordered from Coriell Cell Repositories (Camden, NJ). Oligonucleotides with 5′ amino groups were ordered from Oligos Etc. (Wilsonville, OR) or from PE Applied Biosystems. 2-[N-morpholino]ethanesulfonic acid (MES) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) were purchased from Sigma (St. Louis, IL) and Pierce (Rockford, IL), respectively. DNA polymerase was cloned from Thermatoga neapolitana (A. Nelsen, G. Purdy, D. Taylor, J. Chen and M. Weiner, in prep.) and expressed in Escherichia coli. The Klenow fragment (TneK), lacking the 5′ to 3′ exonuclease was used for SBCE reactions under the same assay conditions as for AmpliTaq (see below). Details of the cloning and expression of Tne, TneK, and TneK FS and their performance in the SBCE assay will be submitted elsewhere. Carboxylated fluorescent polystyrene microspheres were purchased from the Luminex Corp. (Austin, TX).
Incorporation of ddNTPs by DNA Polymerases
Unmodified double-stranded PCR product was used as template in our system. Several thermostable DNA polymerases were evaluated under thermocycling conditions for efficacy of fluorescein (FITC) labeled ddNTP incorporation. One PCR product containing a T/C polymorphism (SNP18) was analyzed with both sense and antisense capture oligonucleotides for T and C or A and G incorporation, respectively. AmpliTaq FS generated the highest signal and a ratio between the positive signal and nonspecific incorporation (noise) of >100-fold. AmpliTaq, KlenTaq, and TneK produced much weaker signals and a significantly reduced signal to noise ratio. Therefore, AmpliTaq FS is an appropriate choice for incorporating fluorescein-labeled ddNTPs under the conditions used.
Coupling of Oligonucleotides to Microspheres
Oligonucleotides with a 5′ amino group were coupled to the carboxyl group on the surface of the microspheres for capturing the SBCE reaction products. In these oligonucleotides, a carbon spacer (C15-18) was synthesized adjacent to the 5′ amino group to reduce the potential interference of the oligonucleotide hybridization by the microspheres. Next to the carbon spacer was a common 20-base luciferase sequence (CAGGCCAAGTAACTTCTTCG, SeqLUC) that was used to monitor the coupling efficiency of the oligonucleotides to the microspheres. Finally, a 25-base complementary ZipCode sequence (named cZipCode, see Table 1) was selected arbitrarily from the Mycobacterium tuberculosis genome and validated experimentally (see below). Carboxylated microspheres (2.5 × 106) in 62 μl of 0.1 m MES buffer were mixed with 5 nmoles of oligonucleotides in 0.1 m MES (6.25 μl). Freshly made 30 mg/ml EDC (10 μl) was added to the microspheres/oligo mixture and incubated at room temperature for 20 min. Two additional rounds of 10 μl of EDC were added at intervals of 20 min. The reaction mixture was mixed occasionally and sonicated during incubation to assure microsphere separation and suspension. After a total incubation period of 60 min, the microspheres were washed twice with 1 ml of PBS plus 0.02% Tween 20, rinsed with 150 μl of TE [Tris(hydroxymethyl)aminomethane hydrochloride (10 mm)/1 mm ethylenediamine-tetra-acetic acid (pH 8.0)], and resuspended in 250 μl TE. The number of the oligonucleotides coupled to the microspheres was assessed by hybridizing a fluorescent-labeled sequence that is complementary to the SeqLuc sequence. Microspheres with a minimum MESF value of 100,000 were used in SBCE experiments. We have found that the coupled microspheres stored at 4°C with minimum exposure to light could be successfully used after 4 months.
Validation of ZipCode and cZipCode Sequences
A set of 25-mer oligonucleotides was randomly selected from the M. tuberculosis genome. Oligonucleotides with an annealing temperature of 61°C–66°C as determined with a software program (OligoCalculator, http://www.pitt.edu/∼rsup/OligoCalc.html) and limited secondary structure (OligoTech software from Oligos Etc. Inc. and Oligo Therapeutics Inc., Wilsonville, OR) were further validated experimentally as a standard set of ZipCode sequences. A total of 58 unique capture probes were ligated individually to a single FITC-labeled oligonucleotide in OLA reactions (Iannone et al. 2000). Each capture probe was composed of one of the 58 ZipCodes at the 5′-end and a common SNP-specific sequence at the 3′end. After ligation, the reaction products were hybridized individually to a mix containing 58 unique fluorescent microspheres, each having one of 58 cZipCodes covalently attached. This 58 × 58 multiplexed matrix was analyzed by flow cytometry on the FACScalibur. For any given reaction, only one type of microsphere should have displayed a positive signal. Signals observed on multiple types of microspheres indicated cross-hybridization between ZipCodes. ZipCodes displaying spurious signals due to cross-hybridization were targeted for replacement. A second round of reactions established a set of 58 compatible ZipCodes. The sequences of the chosen 58 ZipCodes are shown in Table 1.
PCR Amplification
PCR reactions were performed in a 96-well plate on a GeneAmp 3700 thermal cycler (PE Biosystems). A typical 30 μl reaction mixture contained 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 1.5 mm MgCl2, 0.1 mm dNTPs, 0.2 μm of each primer, AmpliTaq Gold DNA polymerase (1.5 units) and 20 ng genomic DNA. The reaction mixture was held at 95°C for 10 min to activate the DNA polymerase and the amplification was carried out for 9 cycles at 94°C for 10 sec, 61°C for 45 sec, and 72°C for 90 sec, 9 cycles at 94°C for 10 sec, 56°C for 45 sec, and 72°C for 90 sec, and another 25 cycles at 94°C for 10 sec, 61°C for 45 sec, and 72°C for 90 sec. After another 5 min extension at 72°C, the reaction mixture was held at 4°C.
Quantitation of PCR Products, Primer, and dNTP Degradation
PCR products were quantified using the PicoGreen binding assay according to the manufacturer's instructions (Molecular Probes, Eugene, OR). The fluorescence intensity was measured using a CytoFluor MultiWell Plate Reader Series 4000 (PE Biosystems) and the quantity was calculated against DNA standards of known quantities. To degrade the PCR primers and dNTPs, 1 unit of SAP and 2 units of E. coli exonuclease I were added directly to 10 μl of PCR reaction mixture. The reaction was incubated at 37°C for 30 min, then at 99°C for 15 min for enzyme inactivation. Some PCR products were cleaned with the Qiagen Qiaquick kit (Qiagen, Valencia, CA).
SBCE Reactions
To either single or pooled PCR products (10–20 ng each), a SBCE reaction mixture was added to a total volume of 10 μl. The mixture consisted of 80 mm Tris-HCl (pH 9.0), 2 mm MgCl2, 100 nm of capture oligonucleotide, 3 units of AmpliTaq FS (PE Biosystems), 10 μm of each allele specific FITC-labeled ddNTP, and 30 μm of other three unlabeled ddNTPs. The reaction mixture was incubated at 96°C for 2 min followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. Reactions were held at 4°C prior to the addition of microspheres.
Hybridization of SBCE Reaction Mixture to the Microsphere
After the SBCE reactions, each of the allele-specific extension products was captured by its corresponding microspheres containing the cZipCode complementary sequence. A pool of different microspheres was concentrated by centrifugation at 1100g for 5 min. Approximately 1200 of each fluorescent microsphere were added to the 10 μl of SBCE reaction mixture for a final volume of 15 μl. The concentrations of NaCl and EDTA were adjusted to 1 m and 20 mm respectively. The mixture was incubated at 40°C for ≥2 hr. Microspheres were washed by the addition of 200 μl of 2× SSC [1× SSC is 8.77 grams of NaCl plus 4.41 grams of sodium citrate per liter (pH 7.0)], 0.02% Tween 20 at room temperature. After centrifugation at 1100g for 6 min, the pelleted microspheres were resuspended in 250 μl of 2× SSC, 0.02% Tween 20 for flow cytometry analysis.
Flow Cytometric Analysis and MESF Conversions
Flow cytometry uses a combination of fluidics, optics, and electronics to detect and measure the fluorescence associated with particles. A FACSCalibur flow cytometer (Becton Dickinson; San Jose, CA) measures the green, orange, and red fluorescence emitted from each particle as the microspheres pass in single file before a laser (488 nm). The fluorescence associated with each particle is evaluated using Luminex Lab MAP hardware and software (Luminex Corp). Each microsphere set is identified by its unique orange and red fluorescence profile from fluorochromes embedded in the microsphere. The signal intensity of the green fluorescence is associated with the SBCE biological reaction on the surface of the microsphere. The following green fluorochromes (coupled to ddNTP) may be utilized in this system: FITC, BODIPY, and Alexa 488 (used as a strepavidin-coupled fluorochrome with biotinylated ddNTPs). Spectral overlap (green fluorescence spilling-over into the orange and/or red detection bandwidth) was subtracted using electronic compensation (provided as part of the Luminex Lab MAP software). A minimum of 100 microspheres was analyzed per data point.
MESF values were calculated from raw fluorescence values (MFI or mean fluorescence intensity) using Quantum Fluorescence Kit and QuickCal software (Sigma, St. Louis, MO). A calibration curve was generated using five standard control microsphere populations, each containing a known MESF value. Background fluorescence was determined by analyzing the fluorescence associated with the microspheres alone and/or from microspheres plus SBCE reactions without DNA polymerase. In all figures, the green background fluorescence contributed by the microspheres has been subtracted.
Acknowledgments
We thank Quan Nguyen, Arash Afshari, Eric Lai, and Michael Wagner for reagents and helpful discussion and Terri Fleming for the bioinformatics support for high-throughput operations. Thanks also, to the Glaxo Wellcome Sequencing Core Facility for their service. We especially want to thank Dr. James Niedel and the GW Research and Development Executive Committee who have provided the opportunity and encouragement for the development of the Genetics Directorate. We also thank the Exploratory Discovery Board and Dr. Allan Baxter for encouragement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jc19570@glaxowellcome.com; mw32319@glaxowellcome.com; FAX (919) 483-0315.
REFERENCES
- Chen XN, Zehnbauer B, Gnirke A, Kwok PY. Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc Natl Acad Sci. 1997;94:10756–10761. doi: 10.1073/pnas.94.20.10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XN, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- Fu DJ, Tang K, Braun A, Reuter D, Darnhofer-Demar B, Little DP, O'Donnell MJ, Cantor CR, Koster H. Sequencing exons 5 to 8 of the p53 gene by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:381–384. doi: 10.1038/nbt0498-381. [DOI] [PubMed] [Google Scholar]
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman, J. JR. Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- Gilles PN, Wu DJ, Foster CB, Dillon PJ, Chanock SJ. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nat Biotechnol. 1999;17:365–370. doi: 10.1038/7921. [DOI] [PubMed] [Google Scholar]
- Iannone MA, Taylor JD, Chen JW, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry. 2000;39:131–140. [PubMed] [Google Scholar]
- Kettman JR, Davis T, Chandler D, Oliver KG, Fulton RJ. Classification and properties of 64 multiplexed microsphere sets. Cytometry. 1998;33:234–243. [PubMed] [Google Scholar]
- Lai E, Riley J, Purvis I, Roses A. A 4-MB high-density single nucleotide polymorphism-based map around human APOE. Genomics. 1998;54:31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Marmaro J, Todd JA. Towards fully automated genome-wide polymorphism screening. Nat Genet. 1995;9:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- Marshall E. Drug firms to create public database of genetic mutations. Science. 1999;284:406–467. doi: 10.1126/science.284.5413.406. [DOI] [PubMed] [Google Scholar]
- McDade RL, Fulton RL. True multiplexed analysis by computer-enhanced flow cytometry. Med Dev Diag Indust. 1997;19(4):75–82. [Google Scholar]
- McHugh TM. Flow microsphere immunoassay for the quantitative and simultaneous detection of multiple soluble analytes. Methods Cell Biol. 1994;42:575–595. doi: 10.1016/s0091-679x(08)61096-1. [DOI] [PubMed] [Google Scholar]
- Nikiforov TT, Rendle RB, Goelet P, Rogers YH, Kotewicz ML, Anderson S, Trainor GL, Knapp MR. Genetic bit analysis: A solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 1994;22:4167–4175. doi: 10.1093/nar/22.20.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Walsh PS, Levenson CH, Erlich HA. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Nishimura DY, Stone EM. Novel approaches to linkage mapping. Curr Opin Genet Dev. 1995;5:335–341. doi: 10.1016/0959-437x(95)80048-4. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen AC. From gels to chips: “Minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum Mutat. 1999;13:1–10. doi: 10.1002/(SICI)1098-1004(1999)13:1<1::AID-HUMU1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Syvanen AC, Aalto-Setala K, Harju L, Kontula K, Soderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]