Summary
SIRT1 is a NAD+-dependent enzyme that affects metabolism by deacetylating key transcriptional regulators of energy expenditure. Here we tested whether deletion of PARP-2, an alternative NAD+ consuming enzyme, impacts on NAD+ bioavailability and SIRT1 activity. Our results indicate that PARP-2 deficiency increases SIRT1 activity in cultured myotubes. However, this increase was not due to changes in NAD+ levels, but to an increase in SIRT1 expression, as PARP-2 acts as a direct negative regulator of the SIRT1 promoter. PARP-2 deletion in mice increases SIRT1 levels, promotes energy expenditure, and increases mitochondrial content. Furthermore, PARP-2−/− mice were protected against diet-induced obesity. Despite being insulin sensitized, PARP-2−/− mice were glucose intolerant due to a defective pancreatic function. Hence, while inhibition of PARP activity promotes oxidative metabolism through SIRT1 activation, the use of PARP inhibitors for metabolic purposes will require further understanding of the specific functions of different PARP family members.
Keywords: SIRT1, poly(ADP-ribose) polymerase-2 (PARP-2), peroxisome proliferator activated receptor coactivator 1α (PGC-1α), NAD+
Introduction
SIRT1 has recently emerged as a promising target in the battle against a number of metabolic disorders, including obesity and type 2 diabetes (Banks et al., 2008; Pfluger et al., 2008; Yu and Auwerx, 2009). The ability of NAD+ levels to control SIRT1 activity gave raise to the hypothesis that artificial modulation of NAD+ levels could be used to regulate SIRT1 (Houtkooper et al., 2010; Sauve, 2009). Indeed, most physiological situations where NAD+ is increased, correlate with higher SIRT1 activity (Canto et al., 2009; Houtkooper et al., 2010; Sauve, 2009). One interesting possibility to artificially modulate NAD+ levels relies on the inhibition of alternative NAD+ consumers, such as the poly(ADP-ribose) polymerases (PARPs) (Houtkooper et al, 2010). Confirming this hypothesis, a number of different labs have shown that the activity of the PARP-1 enzyme, a major cellular NAD+-consumer, and SIRT1 are interrelated due to the competition for the same limiting intracellular NAD+ pool (Kolthur-Seetharam et al., 2006; Pillai et al., 2005; Rajamohan et al., 2009). It is, however, not known whether other PARP family members, have similar effects on SIRT1 activity and global metabolism.
The poly(ADP-ribose) polymerase-2 (PARP-2) is a 66.2 kDa nuclear protein with PARP activity, capable of binding to aberrant DNA forms (Ame et al., 1999). DNA binding of PARP-2 results in its activation and PARP-2 subsequently catalyzes the formation of poly(ADP-ribose) polymers (PAR) onto itself and to different acceptor proteins (Ame et al., 1999; Haenni et al., 2008; Yelamos et al., 2008). PARP-2 has a structurally similar catalytic domain (amino acids 202-593) as PARP-1 (Oliver et al., 2004), the founding member of the PARP family. However, while it is known that PARP-1 activity critically influences NAD+ bioavailability (Sims et al., 1981), the possible effects of a secondary PARP activity, like PARP-2, on intracellular NAD+ levels and global metabolism in cells or organs has not been fully determined yet.
Previous studies already provide evidence that PARP-2 is involved in metabolic homeostasis as it regulates the peroxisome proliferator-activated receptor γ (PPARγ), therefore influencing adipocyte differentiation (Bai et al., 2007). The potential relevance of PARP-2 for NAD+ homeostasis, which would impact on SIRT1 activity and global metabolism, prompted us hence to fully examine the metabolic phenotype of germline PARP-2−/− mice. We show here how the absence of PARP-2 activates SIRT1 and promotes mitochondrial biogenesis in muscle. However, our data also reveals that the absence of PARP-2 leads to pancreatic failure upon high-fat feeding, underscoring the possibility of developing drugs that selectively inhibit specific PARP proteins for metabolic indications.
Results
PARP-2 regulates oxidative metabolism by repressing SIRT1 transcription
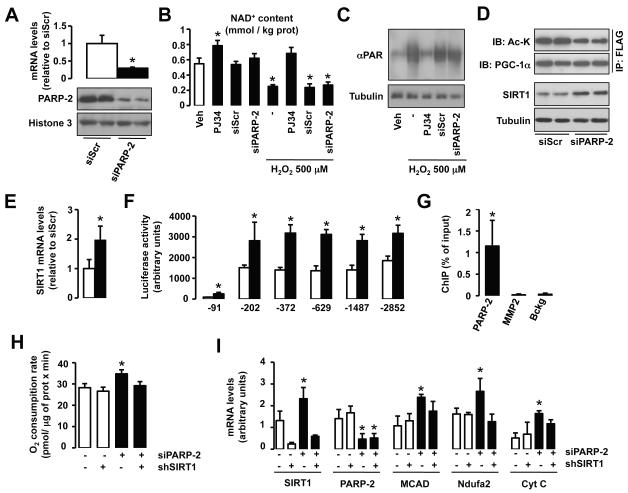
In order to examine the potential role of PARP-2 as a regulator of SIRT1 activity, we generated C2C12 myocytes stably transfected with either a scrambled or a PARP-2 shRNA. PARP-2 mRNA and protein content is reduced by 80 % in myotubes from cells carrying the PARP-2 shRNA (Fig.1A). We next evaluated whether this deficiency in PARP-2 activity affects NAD+ homeostasis. While inhibition of total PARP activity with the inhibitor PJ34 leads to increased intracellular NAD+ content, a reduction in PARP-2 by itself did not affect total (Fig.1B) or mitochondrial (Fig.S1A) NAD+ levels. Similarly, knocking down PARP-2 did not prevent H2O2-induced NAD+ depletion, while global inhibition of PARP activity with PJ34 did (Fig.1B). To further sustain our observations we analyzed the impact of the PARP-2 knock-down on global PARP activity by checking H2O2-induced protein PARylation. While PJ34 completely reversed H2O2-induced PARylation, the knock-down of PARP-2 could not prevent protein hyperPARylation (Fig.1C). These results confirm that PARP-2 is a secondary PARP activity in the cell, as already demonstrated previously (Ame et al., 1999; Shieh et al., 1998). Furthermore, it also suggests that PARP-2 depletion has little impact on NAD+ homeostasis.
Fig.1. PARP-2 regulates oxidative metabolism by acting as a transcriptional repressor of SIRT1.
(A) PARP-2 protein and mRNA levels were analyzed in C2C12 myotubes carrying a stably transfected scramble or PARP-2 shRNA. (B) NAD+ content was evaluated in C2C12 myotubes treated with PJ34 (24 hrs, 1 mM) or carrying a stable transfection of a scramble or a PARP-2 shRNA. H2O2 treatment was performed for 1 hr. (C) Total protein extracts from C2C12 mytotubes treated as in (B) were used to test total PARylation (D) Scramble or PARP-2 shRNA were stably transfected in C2C12 myotubes that were infected with FLAG-PGC-1α. After 48 hr, total protein extracts were obtained and used for FLAG immunoprecipitation and to test the markers indicated. (E) SIRT1 mRNA levels were analyzed in C2C12 myotubes carrying a stable transfection with either scramble or a PARP-2 siRNA. (F) The activity of nested deletions of the SIRT1 promoter was measured after PARP-2 depletion in C2C12 cells. (G) The presence of PARP-2 on the SIRT1 (−1 - −91) and K19 promoter was assessed in C2C12 cells by ChIP assays. (H-I) O2 consumption (H) and mRNA levels of the markers indicated (I) were measured in C2C12 myotubes carrying a stable transfection with either a scramble (−) or a PARP-2 (+) shRNA and infected with adenovirus encoding for either a scramble (−) or a SIRT1 (+) shRNA. Unless otherwise indicated, white bars represent scramble shRNA transfected myotubes and black bars represent PARP-2 shRNA transfected myotubes. All results are expressed as mean ± SD. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
Given the absence of an impact on NAD+ homeostasis, it was surprising to observe that myotubes in which PARP-2 had been knocked down, displayed higher SIRT1 activity, as demonstrated by reduced PGC-1α acetylation (Fig.1D, top panels). We could not find any direct interaction between PARP-2 and SIRT1 (Fig.S1B), indicating that changes in SIRT1 activity are not likely to happen through direct post-translational modification by PARP-2. Rather, the increase in SIRT1 activity was linked to increased SIRT1 content (Fig.1D, bottom panels). The increase in SIRT1 protein was concomitant to an increase in SIRT1 mRNA levels (Fig.1E). To explore why SIRT1 mRNA levels were increased by transcriptional induction, we used a reporter construct in which serial deletions of the mouse SIRT1 promoter region controlled luciferase expression (Fig.1F). These studies demonstrated that knocking down PARP-2 promoted a ~2-fold increase in SIRT1 transcription through the very proximal promoter region (−91 bp), an effect that was maintained for the whole upstream regulatory region that was analyzed (Fig.1F). In chromatin immunoprecipitation (ChIP) assays, PARP-2 was shown to bind directly to the proximal SIRT1 promoter (region comprised between the transcription start site and −91 bp) in C2C12 myotubes (Fig.1G). The direct binding of PARP-2 on the SIRT1 promoter was also observed in a non-murine cell line, like 293HEK cells (Fig.S1C), as this proximal −91 bp region is extremely conserved along evolution (Fig.S1D). All these results suggest that PARP-2 acts as a direct negative regulator of the SIRT1 promoter. Consequently, a reduction of PARP-2 levels induces SIRT1 transcription, leading to higher SIRT1 protein levels and activity. An expected consequence of this increase in SIRT1 activity is that a reduction in PARP-2 content should lead to higher mitochondrial gene expression, by the activation of PGC-1α through deacetylation, and to increased O2 consumption. This hypothesis turned out to be correct, as cellular O2 consumption was increased in PARP-2 knock-down cells (Fig.1H), concomitant to the increase in expression of genes related to lipid and mitochondrial metabolism, such as Medium Chain Acyl coenzyme A Dehydrogenase (MCAD), NADH Dehydrogenase [Ubiquinone] 1 alpha subcomplex subunit 2 (Ndufa2) and Cytochrome C (Cyt) (Fig.1I). Furthermore, using adenoviruses encoding for a shRNA for SIRT1, we demonstrated that the increase in SIRT1 activity contributed in a major fashion to the oxidative phenotype of PARP-2 deficient myotubes (Fig.1H-I).
General physiological characterization of the PARP-2−/− mice
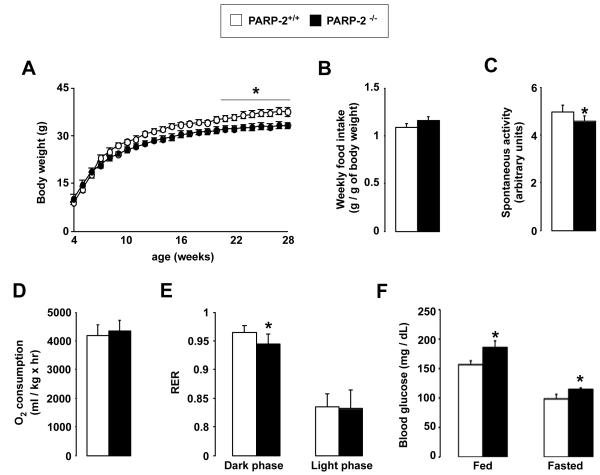
All the experiments above illustrate that reducing PARP-2 activity might be useful to increase SIRT1 activity and, consequently, potentiate oxidative metabolism. In order to gain further insight in this mechanism, we next examined the metabolic profile of the PARP-2−/− mice. PARP-2−/− mice were smaller and leaner then their PARP-2+/+ littermates (Fig.2A). The fact that there was no difference in food intake between the PARP-2−/− and PARP-2+/+ mice (Fig.2B) and that spontaneous locomotor activity was lower in the PARP-2−/− mice (Fig.2C), suggested that the difference in weight gain was due to altered energy expenditure (EE). Indirect calorimetry, however, only indicated a slight tendency towards a higher O2 consumption in chow fed PARP-2−/− mice compared to wild type littermates under basal conditions (Fig.2D). Interestingly, RQ values indicate that during the dark phase PARP-2−/− mice use lipid substrates as energy source at proportionally higher rates than the PARP-2+/+ littermates (Fig.2E). Strikingly, PARP-2−/− mice were mildly hyperglycemic in both fed and fasted states (Fig.2F), linked to a tendency towards lower blood insulin levels in both fed (2.52 ± 0.24 μg/L for PARP-2+/+ vs. 1.77 ± 0.13 μg/L for PARP-2−/− mice) and fasted (0.77 ± 0.07 μg/L for PARP-2+/+ vs. 0.71 ± 0.11 μg/L for PARP-2−/− mice) states. Overall, these results illustrate that PARP-2 deletion promotes an increase in the use of fat as main energy source, associated with a leaner phenotype.
Fig. 2. General physiologic characteristics of PARP-2−/− mice.
(A) PARP-2+/+ and −/− male mice (n=15/13) were weighed weekly and (B) food consumption was measured. (C-E) PARP-2+/+ and −/− male mice on a chow diet (n=6/6, age of 3 months) were subjected to indirect calorimetry, where (C) locomotor activity, (D) O2 consumption and (E) RER were determined. (F) Fed and fasted blood glucose levels. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
Higher SIRT1 activity, mitochondrial content and oxidative profile in PARP-2−/− tissues
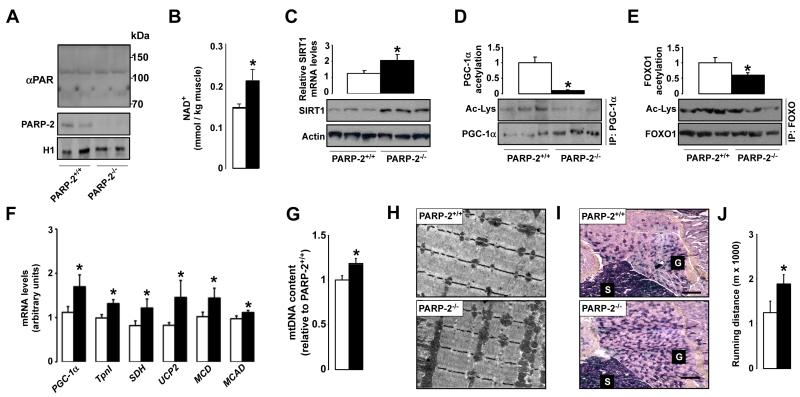
At the molecular level, PARP-2 deletion was not linked to higher DNA damage in either young or old mice (Fig.S2A). In line with these in vitro data, we could not detect a significant change in protein PARylation in PARP-2−/− mice, as determined by western blotting (Fig.3A). In contrast to the data from C2C12 myotubes, PARP-2−/− muscles contained more NAD+ (Fig.3B). The data from cultured myotubes suggests that the increase in NAD+ levels observed in muscle tissue might be secondary to the leaner phenotype rather than a direct consequence of the reduction in PARP-2 function per se. In line with the role of PARP-2 as a negative regulator of the SIRT1 promoter, SIRT1 mRNA and protein levels were increased in muscles from PARP-2−/− mice (Fig.3C). The combination of higher NAD+ and higher SIRT1 protein provides an excellent scenario to increase SIRT1 activity. Confirming this hypothesis, the acetylation levels of two different SIRT1 substrates, the peroxisome proliferator-activated receptor (PPAR)γ coactivator-1α (PGC-1α) (Fig.3D) and the forkhead box O1 (FOXO1) transcription factor (Fig.3E), were markedly decreased in muscles from PARP-2−/− mice. Importantly, the acetylation status of SIRT2 and SIRT3 targets, such as tubulin and Ndufa9, respectively, was not affected by PARP-2 deletion, indicating that PARP-2−/− deletion is not affecting the activity of the closest SIRT1 homologs (Fig.S2B). PGC-1α and FOXO1 are transcriptional activators strongly linked to the regulation of mitochondrial biogenesis and oxidative metabolism. Consequent to their activation through deacetylation, the expression of transcriptional regulators of oxidative metabolism (PGC-1α), of biomarkers of oxidative muscle fibers (troponin I (tpnI)), and of mitochondrial proteins (succinate dehydrogenase (SDH), uncoupling protein 2 (UCP2)) and lipid oxidation enzymes (malonyl-CoA decarboxylase (MCD), MCAD) were increased in gastrocnemius muscle of the PARP-2−/− mice (Fig.3F). The increase in mitochondrial content was further evidenced by the higher mitochondrial DNA content (Fig.3G) and by the more prominent mitochondria observed upon transmission electron microscopy analysis of the gastrocnemius muscle (Fig.3H). The increased mitochondrial biogenesis clearly promoted a more oxidative phenotype of the PARP-2−/− muscles, as reflected by the prominent increase in SDH positive oxidative muscle fibers (Fig.3I). As a physiological consequence of this increased oxidative muscle profile, PARP-2−/− mice performed much better than their PARP-2+/+ littermates on a treadmill endurance test (Fig.3J). As a whole, these results indicate that PARP-2 deletion promotes mitochondrial biogenesis in muscle, increasing the oxidative and endurance profile of the fibers.
Fig.3. PARP-2−/− muscles have higher SIRT1 activity, mitochondrial content and oxidative profile.
(A) PARylation and PARP-2 levels in gastrocnemius muscle were determined by western blot. PARP-2 levels were determined in nuclear extracts, and histone 1 (H1) was used as loading control. (B) NAD+ levels in gastrocnemius muscle of 4-months old PARP-2+/+ and −/− male mice (n=4 and 8, respectively) were determined by HPLC/MS (C) SIRT1 mRNA and protein levels were determined in total muscle mRNA or protein extracts. (D) PGC-1α and (E) FOXO1 acetylation lysine levels were examined after immunoprecipitation. Quantifications are shown on top of the respective images. (F) Gene expression of the indicated genes in the gastrocnemius muscle of PARP-2+/+ and −/− mice was evaluated by RT-qPCR. (G) Quantification of mitochondrial DNA by qPCR (H) Transmission electron micrographs and (I) SDH staining of representative gastrocnemius muscle sections show increased mitochondrial content (PARP-2+/+ and −/− male mice n=15 and 13, respectively; age of 7 months). Scale bar in (I) = 100 μm (J) Endurance treadmill test was performed as described. White bars represent PARP-2+/+ mice, while black bars represent PARP-2−/− mice. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
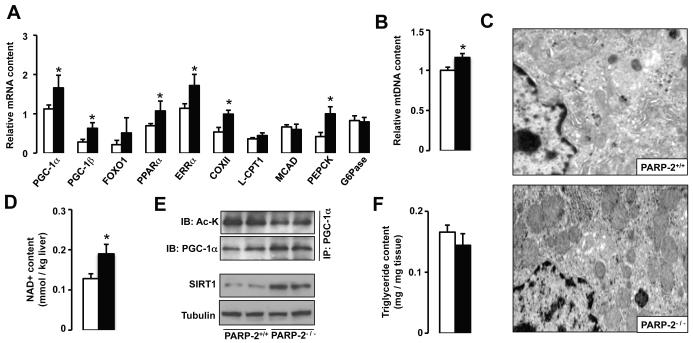
We also explored whether PARP-2 deletion could also influence mitochondrial biogenesis in other highly metabolic tissues, such as in brown adipose tissue (BAT) and liver. In BAT, despite higher SIRT1 content (Fig.S3A), we were unable to detect changes in the expression of the main metabolic genes (Fig.S3B). Supporting the minor impact of PARP-2 deletion on BAT function, body temperature dropped similarly in PARP-2+/+ and PARP-2−/− mice upon cold exposure (Fig.S3C). This suggested that BAT is unlikely to contribute significantly to the differences in EE observed in the PARP-2−/− mice. In contrast, PARP-2 deletion had strong effects on the expression of diverse regulators of mitochondrial metabolism in the liver, including PGC-1α, PGC-1β, FOXO1, PPARα, estrogen-related receptor α (ERRα) and Cytochrome C oxidase subunit II (COXII) (Fig.4A). Consistently, PARP-2−/− livers displayed a higher mitochondrial content, as evidenced by the increase in mitochondrial DNA levels (Fig.4B) and by the appearance of more mitochondria upon electron microscopy (Fig.4C). As in muscle, liver NAD+ content was higher in PARP-2−/− mice (Fig.4D), which, together with the higher amounts of SIRT1 protein, translated into increased SIRT1 activation (Fig.4E). In line with what was observed in muscle, no changes in the activity of SIRT2 and SIRT3, the closest SIRT1 homologs, was detected (Fig.S4A). The observation that PARP-2−/− livers had a tendency towards a reduced triglyceride content both upon oil red O staining (Fig.S4B) and direct biochemical measurement (Fig.4F) is consistent with the induction of oxidative metabolism. Despite the increase in phosphoenolpyruvate carboxykinase (PEPCK) expression in PARP-2−/− mice and the increased capacity of liver for oxidative metabolism, PARP-2−/− mice responded similar to PARP-2+/+ littermates upon a pyruvate-tolerance test (Fig.S4C), probably due to the similar expression of another rate-limiting enzyme, the glucose-6-phosphatase (G6Pase) (Fig.4A).
Fig.4. PARP-2−/− mice display higher mitochondrial content in liver.
(A) mRNA expression analysis in livers from PARP-2+/+ and −/− male (n=16/13, respectively; 6 months of age) mice fed a chow diet. (B) Relative liver mitochondrial DNA (mtDNA) content was estimated by RT-qPCR. (C) Transmission electron microscopic images of liver sections demonstrate higher mitochondrial number in PARP-2−/− mice. (D) Total intrahepatic NAD+ content was measured by HPLC/MS. (E) Total liver protein extracts were used to evaluate SIRT1 protein levels and immunoprecipitate PGC-1α to examine PGC-1α acetylation levels. (F) Liver triglyceride content was estimated after methanol/chloroform lipid extraction as described. White bars represent PARP-2+/+ mice, while black bars represent PARP-2−/− mice. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
PARP-2−/− mice are protected against diet-induced obesity and insulin resistance
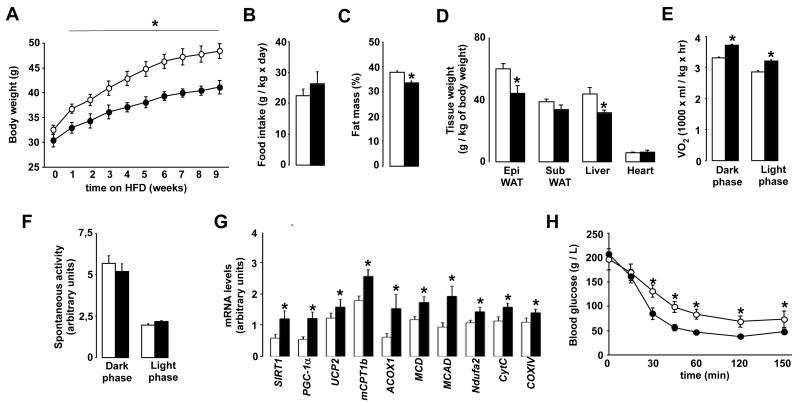
The increased mitochondrial biogenesis and oxidative phenotype observed in the skeletal muscle and liver of PARP-2−/− mice incited us to test how these mice would respond to high-fat diet (HFD) feeding. PARP-2−/− mice were protected against weight gain when fed a HFD (Fig.5A), despite a similar food intake (Fig.5B). This leaner phenotype was associated with a reduced body fat mass, as evidenced by Echo-MRI analysis (Fig.5C). This reduction in fat content was clearly more pronounced (~20% decrease) in the epidydimal fat depots, which is equivalent to visceral fat in man, than in the subcutaneous fat pads (Fig.5D). The weight of the PARP-2−/− livers was also markedly reduced (Fig.5D), consequent to a lower triglyceride accumulation (Fig.S5A-B). Accentuating what was observed in chow-fed mice, PARP-2−/− mice on high fat diet displayed now significantly higher O2 consumption rates (Fig.5E). The increase in VO2 was not due to increased activity (Fig.5F), indicating that high-fat fed PARP-2−/− mice have higher basal EE. As expected, the expression of the transcriptional regulators governing EE (SIRT1, PGC-1α), was increased in gastrocnemius from PARP-2−/− mice when compared to their PARP-2+/+ littermates after the HFD (Fig.5G). The expression of several genes involved in fatty acid uptake and oxidation (muscle carnitine palmitoyltansferase 1b (mCPT1b), peroxisomal acyl-coenzyme A oxidase 1 (ACOX1), MCD, MCAD), mitochondrial electron transport and oxidative phosphorylation (Ndufa2, Cyt C, COXIV) followed a similar pattern as these transcriptional regulators and were maintained at a higher level in the PARP-2−/− muscle (Fig.5G). Consequent to the much leaner and oxidative phenotype, PARP-2−/− mice remained more insulin-sensitive than their wild-type littermates after high-fat feeding (Fig.5H), and their endurance performance was markedly better (data not shown). These results clearly indicate that PARP-2−/− mice are protected from body weight gain and insulin resistance upon high-fat feeding, linked to a better muscle oxidative phenotype.
Fig.5. PARP-2−/− mice are protected against diet-induced body weight gain and insulin resistance.
(A) 6 month old PARP-2+/+ and −/− male mice (n= 7 and 9, respectively) fed on high fat diet were weighed weekly (B) Food intake was monitored during high-fat feeding. (C) Body fat mass composition was evaluated through EchoMRI. (D) The weight of the tissues indicated was determined upon autopsy at the end of the high-fat feeding period. (E) VO2 and (F) spontaneous activity was determined by indirect calorimetry. Quantification of the mean values during light and dark phases are shown. (G) mRNA expression levels in gastrocnemius muscles from PARP-2+/+ and −/− mice after 12 weeks of high-fat diet was determined by qRT-PCR (H) Glucose excursion after an intraperitoneal insulin tolerance test. White bars and circles represent PARP-2+/+ mice, while black bars and circles represent PARP-2−/− mice. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
Increased SIRT1/FOXO1 activity renders PARP-2−/− mice glucose intolerant after high fat feeding due to pancreatic dysfunction
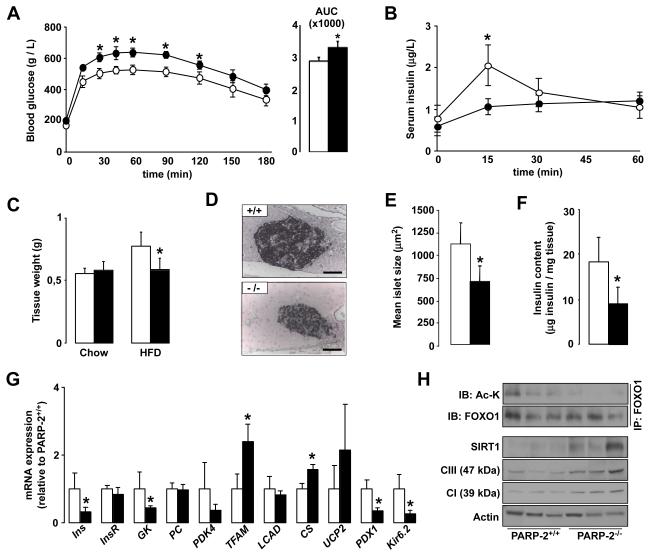
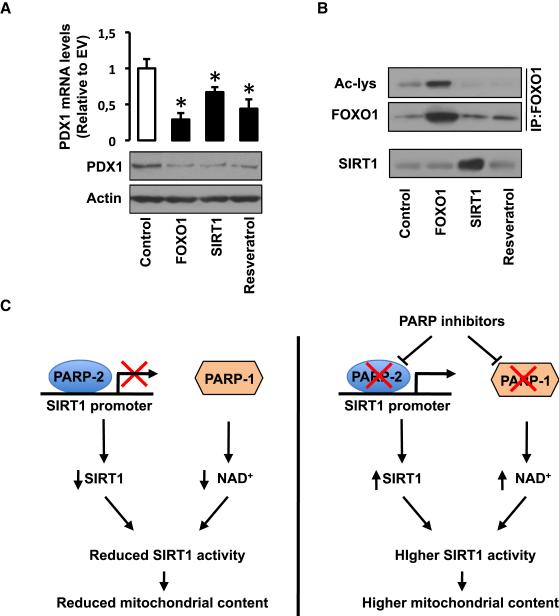
To our surprise, despite their lower body weight and higher insulin sensitivity, PARP-2−/− mice were more glucose intolerant compared to their PARP-2+/+ littermates after high-fat feeding (Fig.6A), and still displayed fasting hyperglycemia (172.44 ± 20,11 mg/dL for PARP-2+/+ vs. 203.34 ± 10.26 mg/dL for PARP-2−/−). The fact that PARP-2−/− mice are also more insulin sensitive (Fig.5H) suggested that this glucose intolerance could be related to defects in the insulin-release upon a glucose load. Confirming this hypothesis, the insulin peak after an intraperitoneal glucose injection in PARP-2+/+ mice was blunted in PARP-2−/−, mice (Fig.6B). Furthermore, fasting blood insulin levels were lower in PARP-2−/− mice (0,87 ± 0,24 μg/L for PARP-2+/+ vs. 0,58 ± 0,16 μg/L for PARP-2−/− mice). These observations led us to examine the pancreas from PARP-2−/− mice. High-fat diet increased the pancreatic mass in wild-type mice, but not in PARP-2 deficient mice (Fig.6C). Histological analysis of the pancreas of PARP-2+/+ and PARP-2−/− mice revealed that islet size was smaller in PARP-2−/− mice after high-fat feeding (Fig.6D and 6E). This reduction in islet size translated into a robust reduction in pancreatic insulin content (Fig.6F). When pancreatic gene expression was analyzed in pancreas from PARP-2+/+ and PARP-2−/− mice, it became evident that, in addition to an increase in some mitochondrial-related genes (mitochondrial transcription factor A (TFAm), citrate synthase (CS)), the pancreas of PARP-2−/− mice had severe reductions in the expression of a number of key genes for pancreatic function (such as glucokinase (GK) and Kir6.2) and proper β-cell growth (pancreatic and duodenal homeobox 1 (pdx1)) (Fig.6G). Given the reduced insulin content and pdx1 expression it was also not surprising that expression of the insulin gene (Ins) was decreased in the PARP-2−/− pancreas (Fig.6G). As in other tissues, PARP-2 deletion led to higher SIRT1 protein levels in pancreas (Fig.6H), which translated not only into higher mitochondrial protein content, as manifested by complex I (39 kDa sbunit) and complex III (47 kDa subunit) levels, (Fig.6H) but also in the constitutive deacetylation of FOXO1 (Fig.6H). NAD+ levels were similar in pancreas form PARP-2+/+ and −/− mice (Fig.S6A). The deacetylation and activation of FOXO1 could underpin the pancreatic phenotype of the PARP-2−/− mice, as FOXO1 activity compromises insulin content and pancreatic growth by acting as a negative regulator of pdx1 (Kitamura et al., 2002). To further consolidate the hypothesis that higher SIRT1 and/or FOXO1 function is detrimental for pancreatic function by the downregulation of pdx1, we tested whether activation of FOXO1 or SIRT1 could decrease pdx1 expression in the MIN6 mouse pancreatic β-cell line. Overexpression of either FOXO1 or SIRT1 was enough to decrease pdx1 mRNA and protein levels (Fig.7A). Similarly, resveratrol treatment, which indirectly activates endogenous SIRT1, leading to FOXO1 deacetylation (Fig.7B), also decreased pdx1 content (Fig.7A). Altogether, these results illustrate that PARP-2−/− mice have impaired pancreatic hyperplasia upon HFD, due to the lower expression of key genes involved in pancreatic β-cell proliferation and function, such as pdx1, as a consequence of higher SIRT1 and/or FOXO1 activity.
Fig.6. Pancreatic abnormalities render PARP-2−/− mice glucose intolerant after high-fat feeding.
(A) Plasma glucose levels during an intraperitoneal glucose tolerance test (IPGTT) in 9-month old PARP-2+/+ and −/− male mice (n=7 and 9, respectively) fed a high fat diet for 12 weeks. The area under the curve of the glucose curves is shown at the right. (B) Insulin levels during the first hour of the IPGTT in (A). (C) Comparison of total pancreas weight between PARP-2+/+ and PARP-2−/− mice on chow and high-fat diet. (D) Pancreas from PARP-2+/+ and PARP-2−/− mice after high-fat diet were stained for insulin (scale bar = 50 μm) and (E) Mean islet size was quantified. (F) Total insulin content in pancreas was measured as described. (G) Gene expression in the pancreas of PARP-2+/+ and PARP-2−/− mice was measured by RT-qPCR. (H) Pancreatic total protein extracts were used to test the abundance of SIRT1, and subunits from the respiratory complexes I and III. FOXO1 was also immunoprecipitated to determine relative FOXO1 acetylation levels. Through the figure, white bars and circles represent PARP-2+/+ mice, while black bars and circles represent PARP-2−/− mice. * indicates statistical difference vs. PARP-2+/+ mice at p<0.05
Figure 7. Increased SIRT1/FOXO1 function reduces pdx1 expression.
(A-B) MIN6 cells were transfected with either empty vector (Control), human FLAG-FOXO1 or human FLAG-SIRT1. Additionally, one empty vector group was treated with resveratrol (50 mM, 5 hrs per day for 2 days). Then, total protein and RNA extracts were obtained to test (A) pdx1 mRNA and protein levels, (B) total FOXO1 and acetylated FOXO1 levels, as well as SIRT1 levels. (C) Scheme illustrating how the activation of PARP enzymes can downregulate SIRT1 function through different means. On one hand, PARP-1 activation limits SIRT1 activity by decreasing NAD+ bioavailability and, on the other, we now report how PARP-2 acts as a negative regulator of the SIRT1 promoter. The PARP-1 and PARP-2 knock-out mouse models indicate that PARP inhibition can be a useful to promote SIRT1 activity and enhance mitochondrial content. This strategy might be exploited therapeutically in the context of metabolic disease, commonly linked to impaired mitochondrial content or function.
Discussion
In our present study we have shown that PARP-2 is a negative regulator of the SIRT1 promoter and that the reduction or ablation of PARP-2 expression induces SIRT1 expression, protein content and activity. As expected from the activation of SIRT1 (Canto et al., 2009; Lagouge et al., 2006), PARP-2−/− mice displayed higher whole body energy expenditure and were protected against high-fat diet induced obesity. Previous studies on PARP-1, the predominant PARP activity in most tissues, have demonstrated that the modulation of intracellular NAD+ levels by PARP-1 critically influences SIRT1 activity (Kolthur-Seetharam et al., 2006; Pillai et al., 2005; Rajamohan et al., 2009; Bai et al., 2011). Our data demonstrates for the first time that deletion of an alternative PARP activity, PARP-2, also impacts on SIRT1 activity. Unexpectedly, our results clearly indicate that, unlike what happens with PARP-1 (Bai et al., 2011), the impact of PARP-2 on SIRT1 activity is not necessarily based on changes in NAD+ content. This is in line with previous evidence indicating that PARP-2 is not a major PARP enzyme in the cell, and, therefore, is not likely to significantly influence NAD+ homeostasis (Ame et al., 1999; Shieh et al., 1998). Rather, PARP-2 impacts on SIRT1 by directly interacting with the proximal SIRT1 promoter, where it acts as a potent transcriptional repressor. Accordingly, in all models and tissues tested, PARP-2 deletion or depletion induces SIRT1 mRNA expression, protein content and activity.
PARP-2 can act as a transcriptional regulator (Yelamos et al., 2008). We have previously shown that PARP-2 acts as a positive regulator of PPARγ, leading to triglyceride accumulation (Bai et al., 2007). In line, and supported by the present data, PARP-2 deficiency led to decreased lipid accumulation. Our current work furthermore expands the potential mechanisms by which PARP-2 activity might impact on lipid accumulation. First, the activation of SIRT1 in PARP-2−/− mice would provide a second way to reduce PPARγ activity, as SIRT1 docks the transcriptional corepressors NCoR and SMRT to PPARγ regulated promoters (Picard et al., 2004). Second, and most importantly, the increase in energy expenditure and fat burning observed in PARP-2−/− mice would also prevent fat accumulation. Reinforcing this second hypothesis, the reduced size of adipose depots is not associated with lipid accumulation in other tissues, such as liver (Fig.S5A-B), indicating that enhanced fat burning rather than fat redistribution accounts for the decreased fat mass.
Our results illustrate how pharmacological approaches to promote oxidative metabolism by inhibiting PARP activity could benefit from targeting specific PARP enzyme forms. Certainly, PARP-2 deletion recapitulates all the previous beneficial effects observed with different strategies aimed to promote SIRT1 activation, i.e. enhanced oxidative metabolism, endurance phenotype and protection against body weight (Feige et al., 2008; Lagouge et al., 2006). These positive outcomes seen in the absence of PARP-2 are, however, overshadowed by the deleterious effects that the absence of PARP-2 has on pancreatic function. Our results show that, upon high-fat feeding, pancreatic proliferation and insulin production are blunted in PARP-2−/− mice. This could be a consequence of the impaired expression of pdx1, a crucial regulator of pancreatic growth and insulin expression (Kaneto et al., 2008), as well as of other genes that fine-tune pancreatic function, such as Kir6.2 and glucokinase (MacDonald et al., 2005). While many reasons could account for such defect, a likely possibility is that the increased SIRT1 activity in PARP-2−/− pancreas activates FOXO1 (see Fig 6H) (Brunet et al., 2004; Frescas et al., 2005), which is known to downregulate pdx1 expression (Kawamori et al., 2006). Interestingly, decreased FOXO1 activity, through its nuclear exclusion, is necessary for the compensatory pancreatic proliferation observed in situations of insulin resistance, as induced by high fat diet (Buteau and Accili, 2007). FOXO1 activation also is detrimental for the pancreatic proliferative effects of hormones such as glucagon-like peptide 1 (GLP-1) (Buteau and Accili, 2007). Hence, the constitutive activation of FOXO1 provides a plausible mechanism to explain the pancreatic phenotype of the PARP-2−/− mice upon HFD. Given that overexpression or activation of SIRT1 or overexpression of FOXO1 in the MIN6 β-cell line recapitulates the downregulation of pdx1 in the PARP-2−/− mice, it is surprising that this phenotype was not observed in other models of SIRT1 activation in the pancreas (Banks et al., 2008; Moynihan et al., 2005). Unfortunately, the fact that FOXO1 deacetylation, as a read-out of effective pancreatic SIRT1 activation, was not tested in those studies (Banks et al., 2008; Moynihan et al., 2005) makes it difficult to compare it with our work. Another possibility would be that, besides SIRT1-mediated FOXO deacetylation, other PARP-2 specific functions might contribute to the pancreatic alterations in the PARP-2−/− mice. For example, PARPs can modulate the enzymatic activities of a number of proteins through direct interaction or by poly(ADP-ribosyl)ation (Yelamos et al., 2008). In this sense, PARP-1 is known to directly interact, poly(ADP-ribosyl)ate and inhibit FOXO (Sakamaki et al., 2009), but we could not demonstrate an interaction between PARP-2 and FOXO in the pancreas (Fig.S6B). Notably, we also could not find evidence for a direct interaction between PARP-2 and SIRT1 (Fig.S1B) or poly(ADP-ribosyl)ation of SIRT1 (Bai et al, 2011). Furthermore, PARP-2 deletion affects blood adipokine levels (Bai et al, 2007), which might also impact pancreatic function. Unraveling the additional mechanisms by which PARP-2 influences pancreatic function warrants future investigation. Importantly, it must be noted that this pancreatic phenotype is specific for the PARP-2−/− mice, as PARP-1−/− mice, while sharing many common characteristics with the PARP-2 mutants, do not develop glucose intolerance after high-fat feeding (Bai et al, 2011) and islet size and insulin content are similar between wild-type and PARP-1−/− littermates upon high fat feeding (Fig.S6C-D), indicating that the pancreatic phenotype is specific to PARP-2 deletion.
In conclusion, our results complement the picture by which PARP activation is detrimental for mitochondrial function by decreasing SIRT1 activity (Fig.7C). On the one hand, we recently reported that the activation of PARP-1 depletes intracellular NAD+, which limits SIRT1 activity (Bai et al, 2011). On the other hand, we describe here how PARP-2 acts as a negative regulator of SIRT1 expression. In combination, these results prompt the speculation that PARP inhibitors could be used not only for cancer, but also in the context of metabolic disease to enhance mitochondrial content.
Experimental procedures
Materials
All chemicals were from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise.
Animal studies
All animal experiments were carried out according to local national and EU ethical guidelines. Homozygous PARP-2−/− and littermate PARP-2+/+ mice (Menissier-de Murcia et al., 2003) and PARP-1−/− and PARP-1+/+ mice (Menissier-de Murcia et al., 1997), on a mixed C57BL/6J / SV129 background (87.5%/12.5%) background, from heterozygous crossings were used. Mice were housed separately, had ad libitum access to water and food, and were kept under a 12/12 h dark-light cycle. Mice were selected for the study at the age of 4 weeks and were kept on chow diet (10 kcal% of fat, Safe, Augy, France). For a part of the animals, food was changed by a high fat diet (HFD, 60 kcal% of fat, Research Diets, New Brunswick, NJ, USA) at the age of 16 weeks. Each week on the same day mice were weighed and the food-consumption was measured. O2 consumption, CO2 production and actimetry were measured by CLAMS of Colombus Instruments, total body fat was measured by echoMRI as described (Lagouge et al., 2006; Watanabe et al., 2006). Endurance test was performed as described in (Canto et al., 2009). Intraperitoneal glucose and insulin tolerance tests (IPGTT and IPITT, respectively) were performed as previously described (Feige et al, 2008). The animals were killed after 6 h of fasting by CO2 asphyxiation and tissues were collected. Total body fat content was then examined at autopsy, by weighing the subcutaneous, gonadal, mesenterial, retroperitoneal and BAT associated fat depots. Liver triglyceride was determined after Folch extraction using a commercial triglyceride kit (Roche). Pancreas and plasma insulin content was determined from acidic extracts using a commercial ELISA kit (Mercodia) (Champy et al., 2004).
Histology and microscopy
Haematoxylin-eosin (HE), Oil Red-O and succinate-dehydrogenase (SDH) stainings were performed on 7 μm tissue sections as described (Lagouge et al., 2006; Picard et al., 2002). Immunohistochemistry was performed on 7 μm frozen tissue sections using anti-insulin antibody (Dako) Aspecific binding of the secondary antibody was controlled on sections where the primary antibody was omitted. Islet size was determined on insulin-stained sections. Tunel assay was performed using a commercial kit (Millipore). Transmission electron microscopy (TEM) was performed on glutaraldehyde-fixed, osmium tetroxyde stained ultrafine sections (Watanabe et al., 2006).
Cell culture
HEK293T cells were cultured as described in (Bai et al., 2007). The MIN6 mouse pancreatic β-cell line was grown in DMEM 4.5 g/L glucose with 15% inactivated FBS, 1% penicillin/streptomycin, 2 mM glutamine and 50 μM 2-mercaptoethanol. C2C12 cells were cultured and differentiated as described in (Canto et al., 2009). The MIN6 mouse pancreatic b-cell line, was MLentiviral short-hairpin (shRNA) and control scramble constructs against PARP-2 were from Sigma-Aldrich (MISSION Custom Vector) incorporating the interfering sequence described in (Bai et al., 2007). C2C12 cells were transduced with 20 MOI of virus and transduced clones were selected with 2.5 μg/ml puromycin. Puromycin selection was maintained during subsequent routine cell culture.
Constructs, reporter assays and chromatin immunoprecipitation
SIRT1 promoter constructs were described in (Nemoto et al., 2004), pSuper-siPARP2, pSuper-scrPARP2 and pBabe-PARP2 were described in (Bai et al., 2007). Adenovirus for SIRT1 knockdown is reported in (Rodgers et al., 2005). Reporter assays and chromatin immunoprecipitations (ChIP) were performed as in (Bai et al., 2007). Primers for ChIP are summarized in Supplementary Table S3.
mRNA preparation, reverse transcription and qPCR
Messenger RNA preparation, cDNA synthesis and RT-qPCR was performed as described in (Bai et al., 2007). Expression was normalized to the expression of cyclophyllin. Mitochondrial DNA was determined in qPCR reactions as described in (Lagouge et al., 2006). Primers are summarized in Supplementary Table S1 and S2.
NAD+ determination
NAD+ was determined by either a general alcohol-dehydrogenase – coupled colorimetric reaction (Canto et al., 2009) or, where indicated, by mass spectrometry, as decribed (Sauve et al, 2005)
Protein extraction and Western blotting
Protein extracts were prepared as described in (Canto et al., 2009), nuclear extraction was prepared as described (Edelman et al., 1965). Extracts were subjected to Western blotting as (Bai et al., 2007; Canto et al., 2009). Blots were probed with the following primary antibodies: SIRT1 (Millipore), actin (Sigma), PARP-2 (rabbit polyclonal antibody raised against full length mouse PARP-2) and H1 (kindly provided by S. Muller, IBMC, Strasbourg).
Determination of PGC-1α acetylation status
PGC-1α and FOXO1 acetylation was analyzed by immunoprecipitation as described in (Canto et al., 2009).
Statistical analysis
For the statistical analysis of the animal studies (body-weight gain, food-consumption) the data were checked for normal distribution. To determine significance Student‘s t-test was applied and the p<0,05 was considered as significant. Error bars represent SEM unless stated otherwise.
Supplementary Material
Acknowledgements
This work was supported by a Bolyai fellowship to PB, an EMBO fellowship to CC, Ambassade de France en Hongrie and Minsitére des Affaires Étrangeres, Hungarian Science and Technology Foundation (FR-26/2009), OTKA NNF78498, CNK80709, IN80481, Mecenatura (DE OEC Mec-1/2008), NIH (DK59820), the EU Ideas program (sirtuins; ERC-2008-AdG-23118), the EPFL, the Swiss National Science Foundation, the Association pour la Recherche Contre le Cancer, the Ligue Contre le Cancer, the Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche (ANR). The authors acknowledge the help of Zsolt Karányi in the statistical analysis and of Dr. Nadia Messadeq in the analysis of EM samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J.Biol.Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid × receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J.Biol.Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metabolism. 2011:XXX. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Buteau J, Accili D. Regulation of pancreatic beta-cell function by the forkhead protein FoxO1. Diabetes Obes Metab. 2007;9(Suppl 2):140–146. doi: 10.1111/j.1463-1326.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champy MF, Selloum M, Piard L, Zeitler V, Caradec C, Chambon P, Auwerx J. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm.Genome. 2004;15:768–783. doi: 10.1007/s00335-004-2393-1. [DOI] [PubMed] [Google Scholar]
- Edelman JC, Edelman PM, Kniggee KM, Schwartz IL. Isolation of skeletal muscle nuclei. J Cell Biol. 1965;27:365–377. doi: 10.1083/jcb.27.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int.J.Biochem.Cell Biol. 2008;40(10):2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneto H, Matsuoka TA, Miyatsuka T, Kawamori D, Katakami N, Yamasaki Y, Matsuhisa M. PDX-1 functions as a master factor in the pancreas. Front Biosci. 2008;13:6406–6420. doi: 10.2741/3162. [DOI] [PubMed] [Google Scholar]
- Kawamori D, Kaneto H, Nakatani Y, Matsuoka TA, Matsuhisa M, Hori M, Yamasaki Y. The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J Biol Chem. 2006;281:1091–1098. doi: 10.1074/jbc.M508510200. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Nakae J, Kitamura Y, Kido Y, Biggs WH, 3rd, Wright CV, White MF, Arden KC, Accili D. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- MacDonald PE, Joseph JW, Rorsman P. Glucose-sensing mechanisms in pancreatic beta-cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:2211–2225. doi: 10.1098/rstb.2005.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc.Natl.Acad.Sci.U.S.A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, hado De OR, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J.Biol.Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Konstatin B, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly (ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29(15):4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Sakamaki J, Daitoku H, Yoshimochi K, Miwa M, Fukamizu A. Regulation of FOXO1-mediated transcription and cell proliferation by PARP-1. Biochem Biophys Res Commun. 2009;382:497–502. doi: 10.1016/j.bbrc.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Sauve AA. Pharmaceutical strategies for activating sirtuins. Curr Pharm Des. 2009;15:45–56. doi: 10.2174/138161209787185797. [DOI] [PubMed] [Google Scholar]
- Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- Sims JL, Berger SJ, Berger NA. Effects of nicotinamide on NAD and poly(ADP-ribose) metabolism in DNA-damaged human lymphocytes. J Supramol Struct Cell Biochem. 1981;16:281–288. doi: 10.1002/jsscb.1981.380160308. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol.Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Yu J, Auwerx J. Protein deacetylation by SIRT1: An emerging key post-translational modification in metabolic regulation. Pharmacol Res. 2009;62(1):35–41. doi: 10.1016/j.phrs.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.