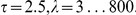Abstract
Very fast oscillations (VFO) in neocortex are widely observed before epileptic seizures, and there is growing evidence that they are caused by networks of pyramidal neurons connected by gap junctions between their axons. We are motivated by the spatio-temporal waves of activity recorded using electrocorticography (ECoG), and study the speed of activity propagation through a network of neurons axonally coupled by gap junctions. We simulate wave propagation by excitable cellular automata (CA) on random (Erdös-Rényi) networks of special type, with spatially constrained connections. From the cellular automaton model, we derive a mean field theory to predict wave propagation. The governing equation resolved by the Fisher-Kolmogorov PDE fails to describe wave speed. A new (hyperbolic) PDE is suggested, which provides adequate wave speed 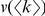 that saturates with network degree
that saturates with network degree 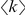 , in agreement with intuitive expectations and CA simulations. We further show that the maximum length of connection is a much better predictor of the wave speed than the mean length. When tested in networks with various degree distributions, wave speeds are found to strongly depend on the ratio of network moments
, in agreement with intuitive expectations and CA simulations. We further show that the maximum length of connection is a much better predictor of the wave speed than the mean length. When tested in networks with various degree distributions, wave speeds are found to strongly depend on the ratio of network moments 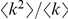 rather than on mean degree
rather than on mean degree 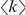 , which is explained by general network theory. The wave speeds are strikingly similar in a diverse set of networks, including regular, Poisson, exponential and power law distributions, supporting our theory for various network topologies. Our results suggest practical predictions for networks of electrically coupled neurons, and our mean field method can be readily applied for a wide class of similar problems, such as spread of epidemics through spatial networks.
, which is explained by general network theory. The wave speeds are strikingly similar in a diverse set of networks, including regular, Poisson, exponential and power law distributions, supporting our theory for various network topologies. Our results suggest practical predictions for networks of electrically coupled neurons, and our mean field method can be readily applied for a wide class of similar problems, such as spread of epidemics through spatial networks.
Introduction
Different types of networks are found across many scales, from metabolic networks in a single cell, to neural networks in brain, up to social and technological global networks. The theory of networks receives increasing attention since the pioneering works that formulated random graphs [1], and the recently discovered ubiquity of small-world networks [2] and scale-free networks [3]. Reviews on general theory of networks can be found in [4]–[6]. A comprehensive up-to-date review of spatial networks is given in [7].
Since its first formulation [1], the Erdös-Rényi (ER) graph became a cornerstone of network theory. An ER graph 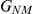 consists of
consists of 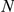 nodes and
nodes and 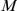 links (edges), and each link connects two nodes which are selected randomly. In a sufficiently large network, the number of links emanating from a node (degree) is a random variable with Poisson distribution
links (edges), and each link connects two nodes which are selected randomly. In a sufficiently large network, the number of links emanating from a node (degree) is a random variable with Poisson distribution 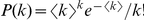 , where
, where 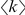 is the network mean degree,
is the network mean degree, 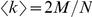 . Despite its advantages, the ER graph is not suitable for studying spatial phenomena because it is spatially homogeneous. However, in most real-world networks the connections are spatial and variable in length. Also, the maximum length of connection is usually limited by the available resources or other natural restrictions. To address this problem, spatial generalizations of the ER graph were suggested. For example, two nodes can be connected only if the distance between them is below threshold
. Despite its advantages, the ER graph is not suitable for studying spatial phenomena because it is spatially homogeneous. However, in most real-world networks the connections are spatial and variable in length. Also, the maximum length of connection is usually limited by the available resources or other natural restrictions. To address this problem, spatial generalizations of the ER graph were suggested. For example, two nodes can be connected only if the distance between them is below threshold 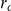 [8]. This model was used to simulate spatio-temporal activity in networks of electrically coupled neurons [9]. Another example is the Waxman model, in which the probability that two nodes are connected is a decreasing function of distance between the nodes [10]. The latter model was used to simulate the Internet [11].
[8]. This model was used to simulate spatio-temporal activity in networks of electrically coupled neurons [9]. Another example is the Waxman model, in which the probability that two nodes are connected is a decreasing function of distance between the nodes [10]. The latter model was used to simulate the Internet [11].
In many networks the nodes are excitable, meaning that active state can arise and propagate from one node to another if they are connected. In this way, action potentials propagate through neural networks, computer viruses spread in the Internet, and diseases are transmitted through transport networks. If the nodes are excitable, dynamical states propagate through a network both temporally and spatially, leading to waves and more complex patterns.
A case study in our work is the emergence of spatiotemporal patterns with very fast oscillations (VFO, 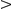 80 Hz) measured by electrocorticography [9], recorded in neocortex of patients prior to epileptic seizures (Figure 1A). There is growing experimental and theoretical evidence that VFO are caused by electrically coupled pyramidal neurons which are connected by gap junctions, thus providing direct excitation from one to another, which does not require synaptic transmission [9], [12], [13].
80 Hz) measured by electrocorticography [9], recorded in neocortex of patients prior to epileptic seizures (Figure 1A). There is growing experimental and theoretical evidence that VFO are caused by electrically coupled pyramidal neurons which are connected by gap junctions, thus providing direct excitation from one to another, which does not require synaptic transmission [9], [12], [13].
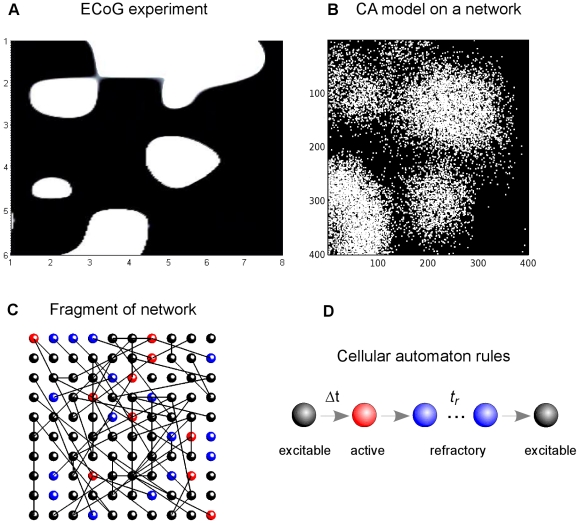
Figure 1. Neural network activity in experiments and in the cellular automaton model.
A. A snapshot of electrocorticographic (ECoG) data of brain activity, measured by 8×6 subdural array of electrodes. Data is interpolated between nodes, white areas correspond to high activity. B. A snapshot of activity from a cellular automaton model in an 400×400 network. The network is subject to noisy input from spontaneously activating cells (rate 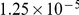 ). Active cells are white, refractory and excitable are black (simplified color code). C. Snapshot of activity in a 10×10 sub-network with detailed color code: red for active, blue for refractory, black for excitable nodes. Lines show links between nodes. D. Rules of the CA model: excitable node (black) may become active (red), if activated by a neighbor. After being activated, the node becomes refractory (blue) for a period of time
). Active cells are white, refractory and excitable are black (simplified color code). C. Snapshot of activity in a 10×10 sub-network with detailed color code: red for active, blue for refractory, black for excitable nodes. Lines show links between nodes. D. Rules of the CA model: excitable node (black) may become active (red), if activated by a neighbor. After being activated, the node becomes refractory (blue) for a period of time 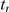 , after which it becomes excitable again. Data in A is a courtesy of Miles Whittington, recorded in Patient B of [29].
, after which it becomes excitable again. Data in A is a courtesy of Miles Whittington, recorded in Patient B of [29].
Nodes in such a network are dynamical excitable units. Although their intrinsic behavior can be complex and require detailed multi-compartment model of each neuron [14]–[16], understanding of the neural network spatiotemporal oscillations requires highly reduced models, such as cellular automata which capture three main states (resting, firing, refractory) with a minimal set of parameters [13]. The system can be described by a network version of Greenberg-Hastings cellular automaton (GHCA), a discrete model of an excitable medium [17]. Variations of GHCA have been used in many studies, including collective oscillations of pyramidal cells in the hippocampus [8], [13], [18], sensory networks [19], [20], and the evolution of HIV infection [21].
In the network version of GHCA that we use, cellular interactions can be distant rather than next-neighbor [8]. The cells are connected into Erdös-Rényi random graph with spatially constrained connections (hereafter SCC), so the distance between connected nodes is not greater than connectivity radius 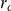 . Under random spontaneous activation of some of the resting cells, large networks demonstrate oscillatory dynamics with complex spatio-temporal activity driven by many interacting waves. Traub and coauthors [9] have recently demonstrated these patterns to be strikingly similar to those observed in ECoG recordings (Figure 1B). In the model, the complex patterns of activity arise when multiple waves are born from spontaneously activated cells and they grow and coalesce in time and space.
. Under random spontaneous activation of some of the resting cells, large networks demonstrate oscillatory dynamics with complex spatio-temporal activity driven by many interacting waves. Traub and coauthors [9] have recently demonstrated these patterns to be strikingly similar to those observed in ECoG recordings (Figure 1B). In the model, the complex patterns of activity arise when multiple waves are born from spontaneously activated cells and they grow and coalesce in time and space.
A single active node may generate an expanding circular wave of excitation, if the mean network connectivity is high enough. The wave maintains its shape and travels with constant speed, which is an important characteristic of system's excitability and depends on network topology. Knowledge of wave speed in excitable networks allows prediction of how fast the active state (neuronal activity, viral infection) propagates and invades the rest of the network. Although simulations can be done in each particular case, it is important to have a basic mean field theory (MFT) to understand and predict the network dynamics without simulations.
In this paper we derive an MFT to predict wave speed in a random (Erdös-Rényi type) network with spatially constrained connections (SCC), for given connectivity radius 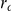 and mean degree
and mean degree 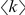 . The results are generalized to Erdös-Rényi type networks with various radii distributions, and further to non-Erdös-Rényi networks with various degree distributions, suggesting universality of our mean field theory.
. The results are generalized to Erdös-Rényi type networks with various radii distributions, and further to non-Erdös-Rényi networks with various degree distributions, suggesting universality of our mean field theory.
Results
The system
In the following section we describe the system and its dynamic properties, along with simulations that demonstrate its typical behavior.
2D network
We study a 2D network of excitable cells (nodes), which are are set on a uniform 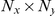 grid, with unit space between adjacent nodes (Figure 1C). Connections (links) are bidirectional, i.e. activity can be transmitted in both directions. Links can be long but limited by
grid, with unit space between adjacent nodes (Figure 1C). Connections (links) are bidirectional, i.e. activity can be transmitted in both directions. Links can be long but limited by 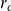 : within a circle of radius
: within a circle of radius 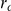 (network with round ‘footprint’) or within a square with side
(network with round ‘footprint’) or within a square with side 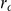 (network with square ‘footprint’) [8]. Generally, the connectivity radius
(network with square ‘footprint’) [8]. Generally, the connectivity radius 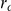 is much larger than 1, but much smaller than array dimensions
is much larger than 1, but much smaller than array dimensions 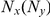 . The number of links per node (degree) follows the Poisson distribution
. The number of links per node (degree) follows the Poisson distribution 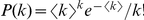 , whereas the distribution of link lengths is uniform in (
, whereas the distribution of link lengths is uniform in ( ), unless stated otherwise.
), unless stated otherwise.
Quasi-1D network
To treat the system analytically, we reduce it to a quasi one-dimensional network: the length of links is limited only along X but may be unlimited in the Y coordinate. In other words, it is a 2D network which has an interval footprint (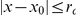 ). A quasi-1D network provides the same wave speed along X as the 2D network with square footprint (with same
). A quasi-1D network provides the same wave speed along X as the 2D network with square footprint (with same 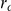 ), which is also consistent with 2D network with round footprint (simulations not shown).
), which is also consistent with 2D network with round footprint (simulations not shown).
Cellular automaton model
A node in excitable state 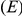 becomes firing
becomes firing 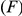 if one or more of its connected neighbors in the network are firing. After one time step
if one or more of its connected neighbors in the network are firing. After one time step 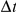 the firing node becomes refractory
the firing node becomes refractory  for a relatively long period
for a relatively long period 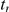 , after which it becomes excitable again. Thus each node rests in
, after which it becomes excitable again. Thus each node rests in 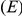 or undergoes a sequence of states
or undergoes a sequence of states 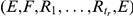 if activated by a neighbor (Figure 1D). Initially all nodes are in excitable state, except a small number of firing nodes that initiate the wave. Node states are updated simultaneously at each time step.
if activated by a neighbor (Figure 1D). Initially all nodes are in excitable state, except a small number of firing nodes that initiate the wave. Node states are updated simultaneously at each time step.
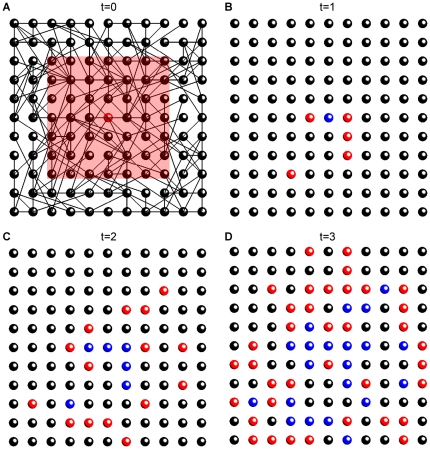
Initiation of wave in a small 2D network is shown in Figure 2 (first four time steps). Although directions of links are random, activity propagates outwards from the initial point because it is followed by refractory state, prohibiting backward propagation. Propagation of waves in large 2D networks is shown in Figure 3A–C. Waves in networks with round and square footprint do not differ qualitatively (not shown), because neighbors of each node are chosen as random lattice points from round or square neighborhood (respectively), which differ only in relatively small corner areas.
Figure 2. Initiation of wave in a CA model on a random network.
The first 4 time steps of wave initiation are shown for an 11×11 network. A. t = 0; B. t = 1; C. t = 2; D. t = 3. Colorcode: red for active, blue for refractory, black for excitable cells. Lines show links between cells, red square shows the connectivity footprint of the central cell (shown only in A). Parameters: 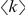 = 4,
= 4, 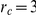 (small for demonstration purposes).
(small for demonstration purposes).
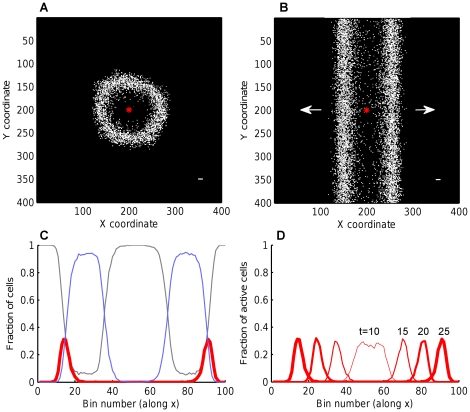
Figure 3. Traveling waves of activity in random networks.
Traveling waves emerging in the CA model on random networks with A. square, B. quasi-1D connectivity footprint. The cell which initiates the wave is shown by a red asterisk. Active cells are white, refractory and excitable cells are black. Directions of wave propagation are shown by arrows. C. A snapshot of wave (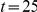 ) with spatial profiles of all three states: grey for excitable, bold red for active, light blue for refractory cell density. In the center, the wake of excitable cells (grey) grows by recovering from the refractory state (blue). D. Profiles of the active state at four time steps, showing two traveling waves emerged from a single active cell. Once formed, the speed and width of a wave remain constant. Profiles were calculated by averaging active cell counts over 100 bins along X. Parameters
) with spatial profiles of all three states: grey for excitable, bold red for active, light blue for refractory cell density. In the center, the wake of excitable cells (grey) grows by recovering from the refractory state (blue). D. Profiles of the active state at four time steps, showing two traveling waves emerged from a single active cell. Once formed, the speed and width of a wave remain constant. Profiles were calculated by averaging active cell counts over 100 bins along X. Parameters 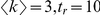 ,
, 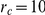 (
(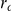 is shown in the bottom right corners in A,B).
is shown in the bottom right corners in A,B).
In the rest of the paper, we work with quasi-1D networks, because they allow transparent mean field analysis, and yet behave almost identically to 2D networks, with the only difference that wave fronts are linear rather than circular (Figure 3B vs. A). Next, we start from simpler case where all links have maximum length 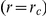 , and then analyse the case where links are distributed randomly
, and then analyse the case where links are distributed randomly 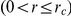 .
.
Mean field theory
Links of maximum length
In order to treat the system analytically, we assume that the network is regular (all nodes have same degree 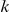 ), and all links have same length (
), and all links have same length (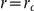 ). Later we will show that these crude assumptions provide a good approximation for more general cases.
). Later we will show that these crude assumptions provide a good approximation for more general cases.
We assume that each excitable cell can become firing by one (and only one) of its neighbors that is firing, which is true for wave front where firing cells are rare. Each firing cell is surrounded by excitable cells, and may produce at maximum 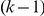 firing neighbors. One of
firing neighbors. One of 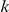 links is missing because it points to a cell from which firing has come. In other words, if a cell is excitable (probability
links is missing because it points to a cell from which firing has come. In other words, if a cell is excitable (probability 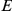 ), it can be activated by one of its neighbors (probability
), it can be activated by one of its neighbors (probability 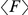 , spatial average of
, spatial average of 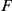 ). Otherwise, if a cell is already firing (
). Otherwise, if a cell is already firing (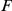 ) and there are excitable neighbors (prob.
) and there are excitable neighbors (prob. 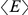 ), it produces
), it produces 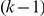 firing cells. These relations can be written as
firing cells. These relations can be written as
| (1) |
where 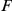 and
and 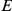 are the probabilities of a cell to be in firing or excitable state, respectively,
are the probabilities of a cell to be in firing or excitable state, respectively, 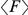 and
and 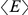 are spatially averaged
are spatially averaged 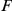 and
and 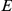 in the neighborhood of
in the neighborhood of  . This equation holds only for wave front, where firing cells are scarce, excitable cells are abundant, and there are no refractory cells yet.
. This equation holds only for wave front, where firing cells are scarce, excitable cells are abundant, and there are no refractory cells yet.
With the notion that most cells are excitable 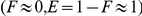 , the E-terms become unitary (linearized equation). In the quasi-1D system,
, the E-terms become unitary (linearized equation). In the quasi-1D system, 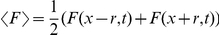 , and the equation simplifies to
, and the equation simplifies to
| (2) |
Taylor's expansion in  at the right hand side gives
at the right hand side gives
| (3) |
Taylor's expansion in  at the left hand side gives
at the left hand side gives
| (4) |
Parabolic (Fisher-Kolmogorov) equation
Taking into account only the first time derivative, we arrive at a linearized version of Fisher-Kolmogorov equation
| (5) |
where 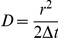 is a diffusion coefficient [22],
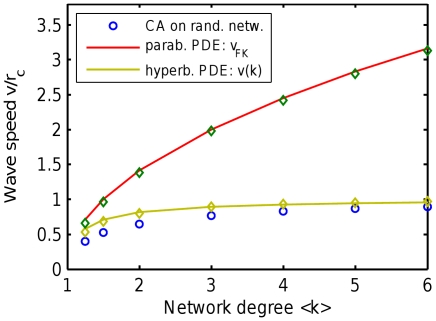
is a diffusion coefficient [22], 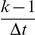 is the growth rate, and the second-order extinction term is omitted for wave speed analysis. The well-known [23] formula for Fisher-Kolmogorov wave speed
is the growth rate, and the second-order extinction term is omitted for wave speed analysis. The well-known [23] formula for Fisher-Kolmogorov wave speed 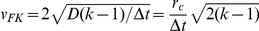 gives infinite growth of speed with mean degree
gives infinite growth of speed with mean degree 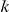 (Figure 4, upper line). Taking high-order terms into account in the right hand side of the PDE does not alter the principal behavior of wave speed (simulations not shown), demonstrating that parabolic PDEs are not suitable for wave speed prediction.
(Figure 4, upper line). Taking high-order terms into account in the right hand side of the PDE does not alter the principal behavior of wave speed (simulations not shown), demonstrating that parabolic PDEs are not suitable for wave speed prediction.
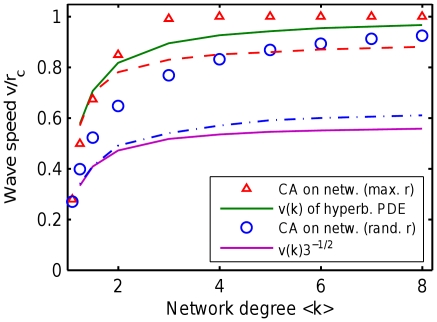
Figure 4. Wave speed predicted by the parabolic and hyperbolic PDEs compared to simulations of CA on random networks.
The parabolic (Fisher-Kolmogorov) PDE gives wave speed 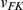 that indefinitely grows with network degree (red line and diamonds). In contrast, the suggested hyperbolic PDE (given in text) provides a reaso`nable wave speed
that indefinitely grows with network degree (red line and diamonds). In contrast, the suggested hyperbolic PDE (given in text) provides a reaso`nable wave speed 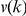 (given in text, shown by green line and diamonds). The
(given in text, shown by green line and diamonds). The 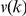 grows moderately and saturates to the maximum possible speed
grows moderately and saturates to the maximum possible speed 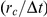 , in agreement with CA simulations (blue circles) and intuitive expectations. The solid lines show analytic formulae, the diamonds show simulations of corresponding full PDE systems.
, in agreement with CA simulations (blue circles) and intuitive expectations. The solid lines show analytic formulae, the diamonds show simulations of corresponding full PDE systems.
Hyperbolic equation
Keeping both first and second time derivatives in the left part of Eqn. 4, we obtain a hyperbolic equation for firing node density
| (6) |
Wave speed can be found by marginal stability analysis [24]. Substituting variable 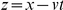 , we obtain the equation
, we obtain the equation 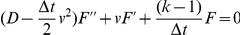 . Solution in the form
. Solution in the form 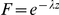 yields a characteristic equation
yields a characteristic equation 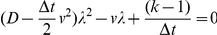 . The roots of the characteristic equation must be real, which gives the minimum wave speed
. The roots of the characteristic equation must be real, which gives the minimum wave speed
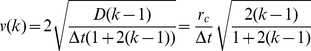 |
(7) |
We put the time step 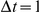 as in the CA model, and
as in the CA model, and 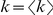 . This wave speed demonstrates qualitative agreement with the CA model on an ER SCC network (Figure 4, lower line). Most importantly,
. This wave speed demonstrates qualitative agreement with the CA model on an ER SCC network (Figure 4, lower line). Most importantly, 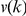 gradually saturates to the maximum possible speed
gradually saturates to the maximum possible speed 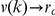 for high
for high 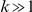 , in agreement with CA simulations and intuitive expectations.
, in agreement with CA simulations and intuitive expectations.
The wave speed 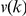 is shown in more detail in Figure 5 (upper solid line). One can see that
is shown in more detail in Figure 5 (upper solid line). One can see that 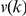 falls near to CA simulations on networks where all links have maximum length
falls near to CA simulations on networks where all links have maximum length 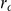 (Figure 5, triangles), as expected. Surprisingly,
(Figure 5, triangles), as expected. Surprisingly, 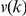 also approximates well the CA simulations on networks where links have random length
also approximates well the CA simulations on networks where links have random length 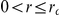 (Figure 5, circles), which is our primary model. This phenomenon is explained in the next section.
(Figure 5, circles), which is our primary model. This phenomenon is explained in the next section.
Figure 5. Wave speed derived from the hyperbolic PDE compared to CA simulations.
The wave speed 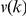 (red line, high) is derived assuming all links have maximum length. CA simulations are shown in two variants, with maximum-length links (red triangles) and generic random-length links (blue circles). The naive speed scaling
(red line, high) is derived assuming all links have maximum length. CA simulations are shown in two variants, with maximum-length links (red triangles) and generic random-length links (blue circles). The naive speed scaling 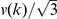 (blue line, low) is derived assuming that link lengths are uniformly distributed. This discrepancy is explained in Results, showing that maximum-length link is a better predictor of wave speed. The dashed lines show the high-order analysis, proving that the hyperbolic PDEs capture the wave behavior sufficiently well, and derivatives of order above 2 are not necessary.
(blue line, low) is derived assuming that link lengths are uniformly distributed. This discrepancy is explained in Results, showing that maximum-length link is a better predictor of wave speed. The dashed lines show the high-order analysis, proving that the hyperbolic PDEs capture the wave behavior sufficiently well, and derivatives of order above 2 are not necessary.
MFT for links with random length
The case where links have random length 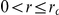 is derived similarly to the maximum-length case shown above. Recall that the spatial scale between nodes is unity,
is derived similarly to the maximum-length case shown above. Recall that the spatial scale between nodes is unity, 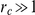 and
and 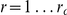 . Let all lengths be equally probable,
. Let all lengths be equally probable, 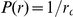 . All nodes have degree
. All nodes have degree 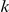 , so the fraction of nodes with at least one link of length
, so the fraction of nodes with at least one link of length  is
is 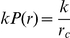 . The evolution of firing nodes density in time is given by
. The evolution of firing nodes density in time is given by
| (8) |
Integration over 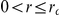 and omitting
and omitting 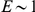 gives
gives
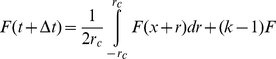 |
(9) |
Taylor's expansion up to second derivative of the function under the integral gives the equation
| (10) |
The latter equation is equivalent to Eqn. 3, with the only difference being a reduced radius (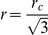 ). Therefore, the wave speed for the random-length links would be simply scaled by
). Therefore, the wave speed for the random-length links would be simply scaled by 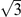 relatively to
relatively to 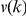 :
: 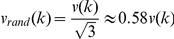 .
.
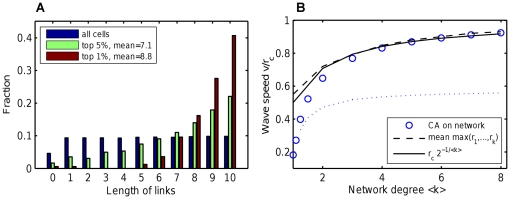
However, this naive scaling based on uniform radii distribution underestimates the wave speed of CA roughly by a factor of two (Figure 5, lower solid line, compare to circles). This happens because cells in the wave front have actually non-uniform distribution of links from which they have received their activation (Figure 6A). The distribution is strongly biased towards the longest links. A wave front generates a new wave front at the next time step by sending activity through the longest links out of available 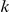 . To support this notion, we generated sets of
. To support this notion, we generated sets of 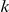 i.i.d. discrete random variables
i.i.d. discrete random variables  uniformly distributed in
uniformly distributed in  and computed their mean maxima
and computed their mean maxima 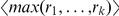 as a plausible estimate of wave speed, that is a contribution of activity propagation from each single node to a global propagation of wave per unit time. As one can see in Figure 6B (broken line), the mean maxima of
as a plausible estimate of wave speed, that is a contribution of activity propagation from each single node to a global propagation of wave per unit time. As one can see in Figure 6B (broken line), the mean maxima of 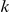 random radii gives a good measure of CA wave speed (circles), especially for high
random radii gives a good measure of CA wave speed (circles), especially for high 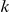 . These numerical calculations are supported by analytic formula for the expected mean of maxima (black solid line),
. These numerical calculations are supported by analytic formula for the expected mean of maxima (black solid line), 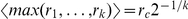 , derived for a uniform continuous distribution
, derived for a uniform continuous distribution 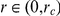 . This formula gives a very good prediction of CA wave speed at high
. This formula gives a very good prediction of CA wave speed at high 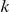 , demonstrating that the wave propagation is indeed mainly determined by the mean maximum of
, demonstrating that the wave propagation is indeed mainly determined by the mean maximum of 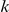 radii, which converges in the limit (
radii, which converges in the limit (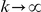 ) to the maximum possible radius
) to the maximum possible radius 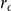 . In other words, it is not the average, but rather the maximum link length that determines the wave speed in a random network with random radii distribution.
. In other words, it is not the average, but rather the maximum link length that determines the wave speed in a random network with random radii distribution.
Figure 6. Role of maximum link lengths in wave propagation.
A. The distribution of link lengths between the cells at the wave front, and the cells which triggered their firing. The front cells (top 1 or 5 %) were selected by their positions in a wave. The mean distances are given in the legend, parameters 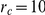 ,
, 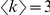 . B. Estimate of the wave speed by numerical estimate of mean maxima of
. B. Estimate of the wave speed by numerical estimate of mean maxima of 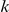 i.i.d radii taken from uniform distribution
i.i.d radii taken from uniform distribution 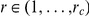 (broken line). The formula
(broken line). The formula 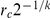 is the expected value of the mean maxima (solid line). The CA simulations of wave speed are shown by circles. As seen, the mean maxima give a good wave speed estimate, in contrast to naive scaling
is the expected value of the mean maxima (solid line). The CA simulations of wave speed are shown by circles. As seen, the mean maxima give a good wave speed estimate, in contrast to naive scaling 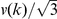 derived earlier from the PDE (dotted line).
derived earlier from the PDE (dotted line).
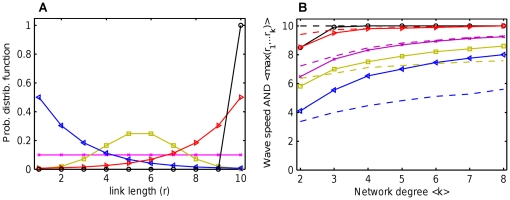
Role of link length distribution
To study the effects of other possible radii distributions, we simulated networks with five distributions (Figure 7A): the uniform radii distribution, the fixed-value distribution (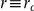 ), a bell-shaped and two exponential distributions (increasing and decreasing, respectively). The speeds of wave propagation in resulting random networks are qualitatively similar and always significantly higher than the average value of the corresponding distribution (Figure 7B). Broken lines in Figure 7B are mean maxima of
), a bell-shaped and two exponential distributions (increasing and decreasing, respectively). The speeds of wave propagation in resulting random networks are qualitatively similar and always significantly higher than the average value of the corresponding distribution (Figure 7B). Broken lines in Figure 7B are mean maxima of 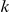 radii generated from each distribution, which served as a good estimate of wave speed in four out of five distributions. Note that the distribution for which our maximum link hypothesis works the least is the one with an exponentially small probability of reaching the maximum link length
radii generated from each distribution, which served as a good estimate of wave speed in four out of five distributions. Note that the distribution for which our maximum link hypothesis works the least is the one with an exponentially small probability of reaching the maximum link length 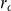 (see the purple line in Figure 7A).
(see the purple line in Figure 7A).
Figure 7. Effect of link lengths (radii) distribution on wave speed.
A. The radii distributions between nodes in the random networks: black o - fixed value, cyan x - uniform, red (blue) triangles - exponentially increasing (decreasing), green squares - bell-shaped distribution (see Methods for detailed formulae). B. Wave speeds in the networks with corresponding radii distributions (markers are consistent with panel A). Broken lines are computed mean maxima out of 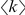 radii samples from each distribution, used as a plausible estimates of the true wave speeds (solid lines). Networks are Erdös-Rényi SCC, so the degree distribution (links per node) is Poissonian.
radii samples from each distribution, used as a plausible estimates of the true wave speeds (solid lines). Networks are Erdös-Rényi SCC, so the degree distribution (links per node) is Poissonian.
Although we can not provide an analytic estimate of the effective radius in each distribution, these simulations support the notion that wave speed is predominantly determined by the maximum rather than average radius, with maximum taken out of 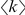 radius realizations.
radius realizations.
High-order analysis
The use of Taylor's approximation in the PDE derivation could potentially bring unwanted errors because 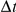 and
and  are in fact not small. We use a high-order analysis to estimate wave speed in exact terms, by looking for a traveling wave solution in the form
are in fact not small. We use a high-order analysis to estimate wave speed in exact terms, by looking for a traveling wave solution in the form 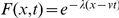 without using Taylor's expansion. See Methods and Algorithms for details. Figure 5 shows that wave speed
without using Taylor's expansion. See Methods and Algorithms for details. Figure 5 shows that wave speed 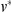 (upper dashed line) obtained by this method is close to
(upper dashed line) obtained by this method is close to 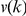 obtained from a hyperbolic PDE (upper solid line). This confirms that considering derivatives of order above 2 (in time and space) will not affect the wave speed qualitatively. Therefore, the hyperbolic PDE is both necessary and sufficient for qualitative wave speed prediction.
obtained from a hyperbolic PDE (upper solid line). This confirms that considering derivatives of order above 2 (in time and space) will not affect the wave speed qualitatively. Therefore, the hyperbolic PDE is both necessary and sufficient for qualitative wave speed prediction.
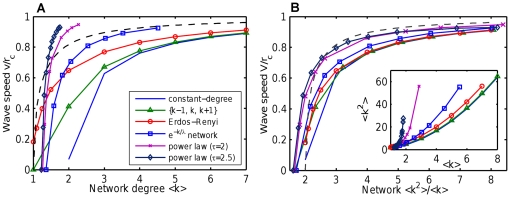
Role of degree distribution
The variation of node degrees depends on degree distribution of a network and strongly affects the wave speed. To ultimately simplify the network and add the variance in stages, we simulated the CA model on spatial networks the following degree distributions:
regular network (each node has same degree
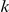 ),
),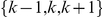 distribution: a node's degree is chosen randomly from
distribution: a node's degree is chosen randomly from 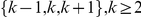 ,
,Poisson distribution (ER graph),
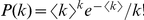
exponential distribution
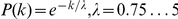 ,
,-
power-law distribution with exponential cutoff
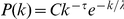 and parameters:
and parameters:
In all networks, the lengths of links between nodes were uniformly distributed in (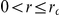 ). As one can see in Figure 8A, the wave speed profiles vary widely when plotted against network mean degree
). As one can see in Figure 8A, the wave speed profiles vary widely when plotted against network mean degree 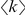 . However, they merge into nearly the same shape when plotted against
. However, they merge into nearly the same shape when plotted against 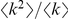 (Figure 8B, inset). The key role of
(Figure 8B, inset). The key role of 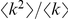 ratio is evident from general network theory. For a randomly chosen link, the degree of a node on its end follows the nearest-neighbor distribution
ratio is evident from general network theory. For a randomly chosen link, the degree of a node on its end follows the nearest-neighbor distribution 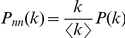 , where
, where 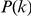 is the original network degree distribution [6]. The mean degree of a connected node
is the original network degree distribution [6]. The mean degree of a connected node 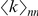 is therefore different from a randomly picked node:
is therefore different from a randomly picked node: 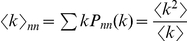 , so our ratio is merely the nearest-neighbor mean degree. In other words, when activation travels from one node to another, the degree of a node in the end of a link follows the nearest-neighbor distribution
, so our ratio is merely the nearest-neighbor mean degree. In other words, when activation travels from one node to another, the degree of a node in the end of a link follows the nearest-neighbor distribution 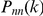 , which has a mean of
, which has a mean of 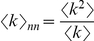 and gives the actual branching ratio of the activation in the new node.
and gives the actual branching ratio of the activation in the new node.
Figure 8. Wave speeds in networks with different degree distributions.
Wave speeds in networks of six different degree distributions (explained in legend) are plotted A. against mean degree 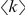 ; B. against ratio of network moments
; B. against ratio of network moments 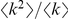 . Note the convergence of wave speeds in B. The mean field formulae are shown in both panels by broken lines (Eqn. 7 in A; Eqn. 11 in B). Inset. The
. Note the convergence of wave speeds in B. The mean field formulae are shown in both panels by broken lines (Eqn. 7 in A; Eqn. 11 in B). Inset. The 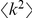 versus
versus 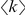 in the simulated networks. Line markers are consistent with legend in panel A. Errorbars are smaller than symbols, due to simulations on many networks of large size (1000×1000).
in the simulated networks. Line markers are consistent with legend in panel A. Errorbars are smaller than symbols, due to simulations on many networks of large size (1000×1000).
For an ER graph, 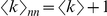 . Substitution of
. Substitution of 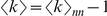 into wave speed (7) gives us a more general formula for fitting wave speeds
into wave speed (7) gives us a more general formula for fitting wave speeds
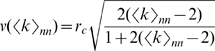 |
(11) |
where 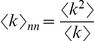 as before. As shown in (Figure 8, inset), this formula gives a good estimate of wave speeds in a variety of network degree distributions.
as before. As shown in (Figure 8, inset), this formula gives a good estimate of wave speeds in a variety of network degree distributions.
Discussion
In summary, we have analysed behavior of excitable random networks with spatially constrained connections (SCC), and derived a mean field theory of the activity propagation. We conclude that the hyperbolic PDE
| (12) |
is necessary and sufficient to capture the wave speed in random (uncorrelated) networks with spatially constrained connections.
The wave speed is mainly determined by the longest possible connection between the firing node and a node it can activate, so the mean maximum of 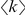 random radii
random radii 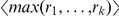 serves as a good predictor of wave speed in Erdös-Rényi SCC networks with various radii distributions.
serves as a good predictor of wave speed in Erdös-Rényi SCC networks with various radii distributions.
We have derived formula (7) for wave speed 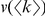 , which agrees with simulated behavior of CA on Erdös-Rényi SCC networks. Simulations of CA on networks with other (non-Poisson) degree distributions suggest a more general formula (11) for wave speed, which depends on
, which agrees with simulated behavior of CA on Erdös-Rényi SCC networks. Simulations of CA on networks with other (non-Poisson) degree distributions suggest a more general formula (11) for wave speed, which depends on 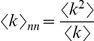 , the nearest-neighbor mean degree. So, our mean field theory extends to networks with various degree distributions, provided that
, the nearest-neighbor mean degree. So, our mean field theory extends to networks with various degree distributions, provided that 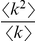 is used as a more universal measure of network's average branching, rather than simple
is used as a more universal measure of network's average branching, rather than simple 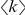 , which is explained by general network theory.
, which is explained by general network theory.
The original [8], [9], [13] cellular automaton model of very fast brain oscillations assumed that pyramidal neurons are connected via gap junctions into an Erdös-Rényi SCC network. Our results show that wave speed (hence network excitability) will scale with radius 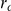 and degree
and degree 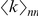 the same way even if the network topology is different. This has experimental implications: accounting for the wave speed does not require reconstruction of full gap junction network topology. Rather, determining the mean
the same way even if the network topology is different. This has experimental implications: accounting for the wave speed does not require reconstruction of full gap junction network topology. Rather, determining the mean 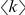 and variance
and variance 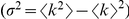 of the gap junctions per cell, and the maximum inter-neuronal distance
of the gap junctions per cell, and the maximum inter-neuronal distance 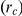 for coupled cells should suffice. Also, our simulations show that wave speed is primarily driven by the longest of
for coupled cells should suffice. Also, our simulations show that wave speed is primarily driven by the longest of 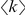 connections emanating from an average node. Therefore knowledge of the longest axoaxonal connections is more important than recovering their full range.
connections emanating from an average node. Therefore knowledge of the longest axoaxonal connections is more important than recovering their full range.
Our present knowledge of wave speed will facilitate a detailed analysis of waves interaction and coalescence, which generate complex oscillatory dynamics in large networks shown in [9].
In general perspective, the fact that wave speed depends on 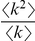 shows that variation in node degree plays an important role in wave propagation. High variation of node degrees provides high wave speed, presumably due to presence of highly-connected nodes (hubs).
shows that variation in node degree plays an important role in wave propagation. High variation of node degrees provides high wave speed, presumably due to presence of highly-connected nodes (hubs).
The hyperbolic PDE (12) should not be confused with similar-looking telegrapher's equation, because the 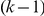 term in Eqn 12 is positive and gives a self-sustaining wave in time, in contrast to the decaying solution of telegrapher's equation.
term in Eqn 12 is positive and gives a self-sustaining wave in time, in contrast to the decaying solution of telegrapher's equation.
The special type of networks we study (with spatially constrained connections) should not be confused with small-world networks. The limited length of link is crucial for spatial phenomena, whereas in conventional small-world networks the “short-cuts” are unlimited in length. This makes wave speed infinite and destroys spatial coherence, but improves temporal coherence [25], [26]. However, small-world networks might be constructed with spatially constrained shortcuts, which could be an interesting system for analysis.
The CA model we use is similar to epidemiological SIRS model (susceptible-infected-recovery-susceptible). Therefore, our theory may help predict spatial spread of epidemics in large networks. For example, it may be applied to predict spatial spread of SIR type malware through a sufficiently large network of WiFi routers [27], where the length of connections is limited by the router's range.
We studied networks with degree distributions that cover a broad spectrum of possible 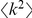 versus
versus 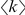 combinations (Figure 8B, inset), suggesting that our MFT holds for uncorrelated networks of arbitrary degree distribution (with spatially constrained connections). The degree distributions studied here appear in real-world networks, such as the neuronal network of C. elegans, the power grid network, acquaintance networks and the WWW (see [4] and references therein). Our MFT apply to these and other networks, provided there is a metric to measure connection length, and connections are limited by some constant
combinations (Figure 8B, inset), suggesting that our MFT holds for uncorrelated networks of arbitrary degree distribution (with spatially constrained connections). The degree distributions studied here appear in real-world networks, such as the neuronal network of C. elegans, the power grid network, acquaintance networks and the WWW (see [4] and references therein). Our MFT apply to these and other networks, provided there is a metric to measure connection length, and connections are limited by some constant 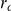 .
.
Methods
Cellular automaton model
A node in excitable state 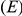 becomes firing
becomes firing 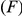 if one or more of its neighbours are
if one or more of its neighbours are 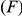 . After one time step
. After one time step 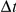 the firing
the firing 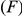 node becomes refractory
node becomes refractory  for a relatively long period
for a relatively long period 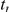 , after which it becomes excitable
, after which it becomes excitable 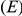 again. Thus each node rests in
again. Thus each node rests in 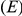 or undergoes a sequence of states
or undergoes a sequence of states 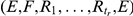 if activated by a neighbor. The formal rules of CA are as follows
if activated by a neighbor. The formal rules of CA are as follows
excitable (E)
 firing (F) if any neighbor is (F)
firing (F) if any neighbor is (F)F
 refractory (
refractory (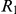 )
)
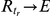
The states of all nodes are updated simultaneously every time step. Initially all nodes are in excitable 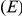 state, except a small number of firing nodes that initiate the wave.
state, except a small number of firing nodes that initiate the wave.
Network with spatially constrained connections
The network consists of excitable nodes, which are are set on a uniform 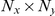 grid, with unit space between adjacent nodes. The network with defined degree distribution
grid, with unit space between adjacent nodes. The network with defined degree distribution 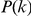 and connectivity radius
and connectivity radius 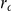 is constructed by a procedure similar to that for spatially homogeneous networks [28]. Initially, each node is assigned to a random number of ‘stubs’
is constructed by a procedure similar to that for spatially homogeneous networks [28]. Initially, each node is assigned to a random number of ‘stubs’ 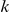 , which is picked from distribution
, which is picked from distribution 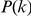 . Next, the program picks nodes from a randomized (shuffled) list of all nodes. The list of nodes must be randomized to avoid artificial correlations imposed by node order. For each picked node with nonzero number of stubs, the program randomly picks one of its neighbor within distance
. Next, the program picks nodes from a randomized (shuffled) list of all nodes. The list of nodes must be randomized to avoid artificial correlations imposed by node order. For each picked node with nonzero number of stubs, the program randomly picks one of its neighbor within distance 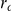 . If the neighbor has nonzero number of stubs, too, both nodes are linked. Their numbers of stubs are decremented by -1, and their numbers of links are incremented by +1. The procedure is repeated until all stubs of the chosen node become links. To avoid infinitely long search in a situation when all neighbors are already linked and they have no more free stubs, the search of potential neighbors is stopped after
. If the neighbor has nonzero number of stubs, too, both nodes are linked. Their numbers of stubs are decremented by -1, and their numbers of links are incremented by +1. The procedure is repeated until all stubs of the chosen node become links. To avoid infinitely long search in a situation when all neighbors are already linked and they have no more free stubs, the search of potential neighbors is stopped after 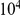 unsuccessful attempts. The procedure is repeated for each node.
unsuccessful attempts. The procedure is repeated for each node.
The 2D network differs from the quasi-1D network only by the way that the connectivity distance is measured. In 2D network, connectivity of node 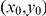 is
is 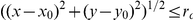 (round ‘footprint’) or
(round ‘footprint’) or 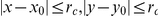 (square ‘footprint’). In quasi one-dimensional network,
(square ‘footprint’). In quasi one-dimensional network, 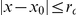 (interval footprint). In most simulations
(interval footprint). In most simulations 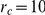 , if not specified otherwise.
, if not specified otherwise.
Single networks of size up to 400 400 were simulated in Matlab. Large-scale simulations of multiple (64 to 128) networks of size
400 were simulated in Matlab. Large-scale simulations of multiple (64 to 128) networks of size 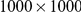 with
with 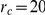 (used in Figure 8) were carried out in C/C++ MPI program on an IBM Blue Gene supercomputer.
(used in Figure 8) were carried out in C/C++ MPI program on an IBM Blue Gene supercomputer.
High-order analysis
The use of Taylor's approximation in space and time could potentially bring unwanted errors because 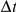 and
and  are in fact not small. Here we estimate wave speed in exact terms by looking for a traveling wave solution in the form
are in fact not small. Here we estimate wave speed in exact terms by looking for a traveling wave solution in the form 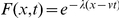 without using Taylor's expansion, thus taking all high-order terms into account. Both cases of links with maximum and random length are analyzed below.
without using Taylor's expansion, thus taking all high-order terms into account. Both cases of links with maximum and random length are analyzed below.
Maximum length
Substitution of 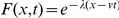 into Eqn. 2 gives
into Eqn. 2 gives
| (13) |
For convenience, we will change to new variables 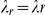 and
and 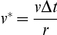 . The equation then reads
. The equation then reads 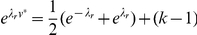 and can be solved numerically for marginal stability analysis.
and can be solved numerically for marginal stability analysis.
In order to find the minimal wave speed, we need to find 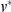 such that the function in the left part
such that the function in the left part 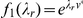 has a unique common point with the function in the right part
has a unique common point with the function in the right part 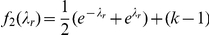 , for given
, for given 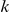 . This happens when
. This happens when 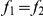 and
and 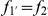 (plots touch each other). Parameter
(plots touch each other). Parameter 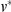 is the minimal wave speed, normalized to the radius
is the minimal wave speed, normalized to the radius  . Figure 5 shows that
. Figure 5 shows that 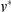 (upper dashed line) obtained by this method is close to
(upper dashed line) obtained by this method is close to 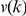 obtained from a hyperbolic PDE (upper solid line), so considering derivatives of order above 2 (in time and space) will not affect the wave speed qualitatively. Therefore, the hyperbolic PDE is both necessary and sufficient for qualitative wave speed prediction.
obtained from a hyperbolic PDE (upper solid line), so considering derivatives of order above 2 (in time and space) will not affect the wave speed qualitatively. Therefore, the hyperbolic PDE is both necessary and sufficient for qualitative wave speed prediction.
Random length
This case is analyzed in a similar way. Eqn. 2 linearized around the unstable steady state reads: 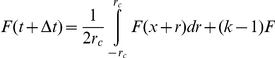 . Substitution of
. Substitution of 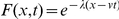 and changing to
and changing to 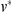 and
and 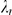 gives
gives
| (14) |
This equation is solved numerically in the same way as in the maximum-length case. As one can see in Figure 5, the obtained minimal wave speed 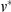 (lower dashed line) is close to
(lower dashed line) is close to 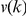 obtained in hyperbolic PDE (lower solid line). However, as shown in Results, the MFT for maximum link length provides better approximation of CA simulations, so this case is shown here only for completeness of analysis.
obtained in hyperbolic PDE (lower solid line). However, as shown in Results, the MFT for maximum link length provides better approximation of CA simulations, so this case is shown here only for completeness of analysis.
Radii distributions
We studied five different radii distributions: uniform in  , fixed-value (always 10), bell-shaped
, fixed-value (always 10), bell-shaped 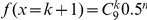 with binomial coefficients
with binomial coefficients 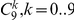 , and two exponentially shaped distributions
, and two exponentially shaped distributions 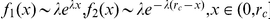 , both normalized to compensate the cutoff of exponential tails.
, both normalized to compensate the cutoff of exponential tails.
Numerical integration of PDE system
Analytic formulas for wave speeds are supported by numerical integration of corresponding PDE systems. In the hyperbolic case, the closed nonlinear system reads
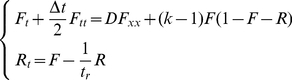 |
(15) |
By the normalization 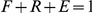 the equation for
the equation for 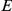 is not necessary. Parameters are
is not necessary. Parameters are 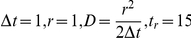 . This system was replaced by a system of three parabolic PDEs (by introducing
. This system was replaced by a system of three parabolic PDEs (by introducing 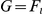 ), which was solved by a method of lines in Matlab, with parameters
), which was solved by a method of lines in Matlab, with parameters 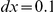 ,
, 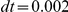 , mesh size 200
, mesh size 200 200, and explicit Euler scheme. Initial conditions were
200, and explicit Euler scheme. Initial conditions were 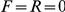 in all points except the center line where
in all points except the center line where 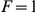 .
.
Simulations of traveling waves in the Fisher-Kolmogorov equation were carried out similarly, with the first equation changed to  .
.
Both systems demonstrate the effect of wave coalescence – two waves cancel each other at the areas where they meet, while their outer borders merge into one big circular wave. The main difference between the two PDE systems is the speed of wave, which grows infinitely with 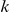 in the Fisher-Kolmogorov system, but saturates to unity for the hyperbolic PDE system, as discussed in the Results.
in the Fisher-Kolmogorov system, but saturates to unity for the hyperbolic PDE system, as discussed in the Results.
Acknowledgments
We thank Nancy Kopell for stimulating discussions.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts: All authors are employed by the IBM T. J. Watson Research Center. There are no patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Funding: The work was supported by grants NIH/NINDS (http://nih.gov) R01NS062955 to NV and RDT, R01NS44133 to RDT, R01GM081747 to YT, and Alexander von Humboldt Stiftung (www.humboldt-foundation.de) to RDT. These funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors are employed by the IBM T. J. Watson Research Center and contributed to all aspects of the study.
References
- 1.Erdős P, Rényi A. On random graphs. I. Publ Math Debrecen. 1959;6:290–297. [Google Scholar]
- 2.Watts D, Strogatz S. Collective dynamics of small-world networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 3.Barabási AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 4.Strogatz S. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 5.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Complex networks: Structure and dynamics. Physics Reports. 2006;424:175–308. [Google Scholar]
- 6.Dorogovtsev SN, Goltsev AV, Mendes JFF. Critical phenomena in complex networks. Rev Mod Phys. 2008;80:1275–1335. [Google Scholar]
- 7.Barthelemy M. Spatial networks. Physics Reports. 2011;499:1–101. [Google Scholar]
- 8.Lewis TJ, Rinzel J. Self-organized synchronous oscillations in a network of excitable cells coupled by gap junctions. Network: Computation in Neural Systems. 2000;11:299–320. [PubMed] [Google Scholar]
- 9.Traub R, Duncan R, Russell A, Baldeweg T, Tu Y, et al. Spatiotemporal patterns of electrocorticographic very fast oscillations (>80 Hz) consistent with a network model based on electrical coupling between principal neurons. Epilepsia. 2010;51:1587–1597. doi: 10.1111/j.1528-1167.2009.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waxman B. Routing of multipoint connections. IEEE Journal on Selected Areas in Communications. 1988;6:1617–1622. [Google Scholar]
- 11.Zegura E, Calvert K, Bhattacharjee S. How to model an internetwork. Proceedings of IEEE Infocom. 1996;volume 2:594–602. [Google Scholar]
- 12.Traub R, Whittington M. Oxford University Press; 2010. Cortical oscillations in health and disease. [Google Scholar]
- 13.Traub RD, Schmitz D, Jefferys JGR, Draguhn A. High-frequency population oscillations are predicted to occur in hippocampal pyramidal neuronal networks interconnected by axoaxonal gap junctions. Neuroscience. 1999;92:407–426. doi: 10.1016/s0306-4522(98)00755-6. [DOI] [PubMed] [Google Scholar]
- 14.Traub R, Contreras D, Cunningham M, Murray H, LeBeau F, et al. Single-column thalamocortical network model exhibiting gamma oscillations, sleep spindles, and epileptogenic bursts. Journal of Neurophysiology. 2005;93:2194. doi: 10.1152/jn.00983.2004. [DOI] [PubMed] [Google Scholar]
- 15.Roopun AK, Middleton SJ, Cunningham MO, LeBeau FEN, Bibbig A, et al. A beta2-frequency (20–30 Hz) oscillation in nonsynaptic networks of somatosensory cortex. Proc Natl Acad Sci U S A. 2006;103:15646–15650. doi: 10.1073/pnas.0607443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traub R, Middleton S, Knoepfel T, Whittington M. Model of very fast (>75 Hz) network oscillations generated by electrical coupling between the proximal axons of cerebellar Purkinje cells. European Journal of Neuroscience. 2008;28:1603–1616. doi: 10.1111/j.1460-9568.2008.06477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg JM, Hastings SP. Spatial patterns for discrete models of diffusion in excitable media. SIAM J Appl Math. 1978;34:515–523. [Google Scholar]
- 18.Munro E, Boergers C. Mechanisms of very fast oscillations in networks of axons coupled by gap junctions. Journal of Computational Neuroscience. 2010;28:539–555. doi: 10.1007/s10827-010-0235-6. [DOI] [PubMed] [Google Scholar]
- 19.Kinouchi O, Copelli M. Optimal dynamical range of excitable networks at criticality. Nature Physics. 2006;2:348–351. [Google Scholar]
- 20.Furtado L, Copelli M. Response of electrically coupled spiking neurons: a cellular automaton approach. Phys Rev E. 2006;73:11907. doi: 10.1103/PhysRevE.73.011907. [DOI] [PubMed] [Google Scholar]
- 21.Zorzenon dos Santos R, Coutinho S. Dynamics of HIV infection: A cellular automata approach. Phys Rev Lett. 2001;87:168102. doi: 10.1103/PhysRevLett.87.168102. [DOI] [PubMed] [Google Scholar]
- 22.Berg HC. Princeton University Press; 1993. Random Walks in Biology. [Google Scholar]
- 23.Murray JD. Springer; 2003. Mathematical Biology II. [Google Scholar]
- 24.Aronson DG, Weinberger HF. Multidimensional nonlinear diffusion arising in population genetics. Adv Math. 1978;30:33–76. [Google Scholar]
- 25.Perc M. Effects of small-world connectivity on noise-induced temporal and spatial order in neural media. Chaos, Solitons & Fractals. 2007;31:280–291. [Google Scholar]
- 26.Sun X, Perc M, Lu Q, Kurths J. Spatial coherence resonance on diffusive and small-world networks of Hodgkin–Huxley neurons. Chaos: An Interdisciplinary Journal of Nonlinear Science. 2008;18:023102. doi: 10.1063/1.2900402. [DOI] [PubMed] [Google Scholar]
- 27.Hu H, Myers S, Colizza V, Vespignani A. WiFi networks and malware epidemiology. Proc Natl Acad Sci U S A. 2009;106:1318. doi: 10.1073/pnas.0811973106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman MEJ, Strogatz SH, Watts DJ. Random graphs with arbitrary degree distributions and their applications. Phys Rev E. 2001;64:026118. doi: 10.1103/PhysRevE.64.026118. [DOI] [PubMed] [Google Scholar]
- 29.Roopun A, Traub R, Baldeweg T, Cunningham M, Whittaker R, et al. Detecting seizure origin using basic, multiscale population dynamic measures: Preliminary findings. Epilepsy and Behavior. 2009;14:39–46. doi: 10.1016/j.yebeh.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]