Abstract
Genetic screens in zebrafish (Danio rerio) have isolated mutations in hundreds of genes essential for vertebrate development, physiology, and behavior. We have constructed a genetic linkage map that will facilitate the identification of candidate genes for these mutations and allow comparisons among the genomes of zebrafish and other vertebrates. On this map, we have localized 771 zebrafish genes and expressed sequence tags (ESTs) by scoring single-stranded conformational polymorphisms (SSCPs) in a meiotic mapping panel. Of these sequences, 642 represent previously unmapped genes and ESTs. The mapping panel was comprised of 42 homozygous diploid individuals produced by heat shock treatment of haploid embryos at the one-cell stage (HS diploids). This “doubled haploid” strategy combines the advantages of mapping in haploid and standard diploid systems, because heat shock diploid individuals have only one allele at each locus and can survive to adulthood, enabling a relatively large quantity of genomic DNA to be prepared from each individual in the mapping panel. To integrate this map with others, we also scored 593 previously mapped simple-sequence length polymorphisms (SSLPs) in the mapping panel. This map will accelerate the molecular analysis of zebrafish mutations and facilitate comparative analysis of vertebrate genomes.
[A table of the mapped genes and ESTs is provided online at http://www.genome.org.]
Lethal mutations in zebrafish (Danio rerio) define the functions of hundreds of essential genes in the vertebrate genome (Driever et al. 1996; Haffter et al. 1996). Powerful methods for the analysis of mutant phenotypes can provide an understanding of the function of these genes at the cellular level (Kimmel 1989; Schier and Talbot 1998). More than 25 genes defined by mutations have been cloned, and these molecular studies demonstrate the ability of forward genetic approaches in zebrafish to reveal new genes and new gene functions that are conserved among vertebrates (Postlethwait and Talbot 1997; Talbot and Hopkins 2000). More than 1800 additional mutations have been identified in genetic screens, and developing genetic maps and other resources to accelerate the molecular analysis of these mutations is an important objective.
Beginning in 1994, several maps of the zebrafish genome have been produced to facilitate the identification of mutated genes by positional cloning and the candidate gene approach. The first map (Postlethwait et al. 1994) was constructed by scoring randomly amplified polymorphic DNA (RAPD) markers in a panel of haploid embryos. The use of haploid embryos simplified the analysis of RAPDs, which usually segregate as dominant markers for which the two alleles are scored as the presence or absence of an amplified fragment. Another mapping strategy employed simple-sequence length polymorphisms (SSLPs) and a standard diploid breeding protocol (Knapik et al. 1998; Shimoda et al. 1999). SSLPs segregate typically as codominant markers and they are more useful in diploid crosses than are dominant markers such as RAPDs. An advantage of the diploid protocol is that diploid zebrafish survive to adulthood, providing much more genomic DNA for mapping studies than haploids, which survive for only a few days after fertilization. These genetic maps benefit positional cloning projects by increasing the likelihood that a mutation will lie near a mapped DNA sequence.
To facilitate the identification of candidate genes for mutations, genes have been mapped within frameworks of RAPDs or SSLPs by scoring polymorphisms in haploid mapping panels (Postlethwait et al. 1998; Gates et al. 1999). These gene maps have also been useful in comparative studies, which have identified groups of genes that are syntenic in zebrafish and other vertebrates.
The recent advent of two radiation hybrid (RH) maps provides a complementary approach for mapping zebrafish genes (Geisler et al. 1999; Hukriede et al. 1999). RH maps assign positions to markers based on their retention in particular members of a panel of cell lines constructed by fusing irradiated zebrafish cells to rodent cells. As such, markers that tend to be present in the same hybrid cell lines are closer together than those that are coretained infrequently. One limitation of this approach is that RH maps tend to have more uncertainty in the order of closely spaced markers than genetic maps, but an important advantage is that mapped sequences need not be polymorphic.
To expand the zebrafish gene map, we have localized single-stranded conformational polymorphisms (SSCPs) corresponding to 642 previously unmapped genes and expressed sequence tags (ESTs) in a meiotic mapping panel comprised of 42 homozygous diploid individuals. To facilitate comparisons with other maps, we also scored in the mapping panel 593 previously localized SSLPs, which were used widely in genetic linkage and RH maps, as well as 129 previously mapped genes and ESTs. This map will facilitate the molecular analysis of zebrafish mutations and provide an important resource for comparative mapping analysis.
RESULTS
Construction of Homozygous Diploid Mapping Panel
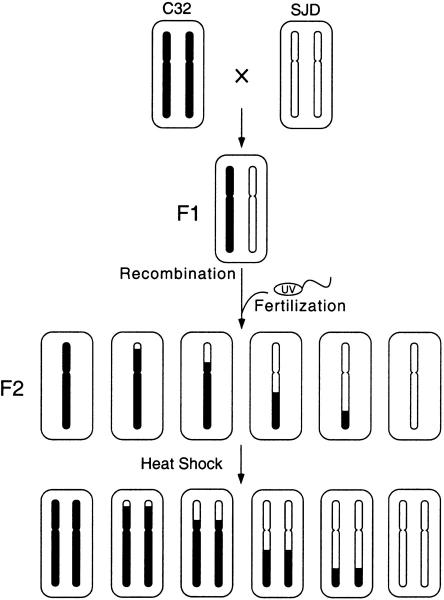
We have assembled a mapping panel comprised of gynogenetic homozygous diploid individuals (also called heat shock diploids), which are produced by heat shock treatment of haploid embryos during the one-cell stage (Fig. 1). The heat shock treatment is applied after the S-phase of the first zygotic cell cycle so that chromosomes have replicated, but because heat shock blocks the subsequent cytokinesis, the haploid chromosome set becomes doubled (Streisinger et al. 1981; Westerfield 1995). This produces diploid embryos that are homozygous at every locus. Many embryos subjected to heat shock treatment are inviable, but a substantial fraction develops normally and some individuals grow to fertile adults. Because heat shock diploids are doubled haploids, the use of this protocol in mapping studies provides the key advantages of haploid embryos, simplifying linkage analysis and facilitating the use of dominant markers (Young et al. 1998). An additional advantage is that heat shock diploids can survive beyond embryonic stages, allowing genomic DNA to be collected in relatively large quantities from juveniles and adults, whereas haploid embryos survive for only ∼4 days.
Figure 1.
Production of the homozygous diploid mapping panel. C32 and SJD fish were crossed to produce a heterozygous F1 generation. Eggs from two F1 females were fertilized with UV-irradiated sperm, which makes no genetic contribution to the progeny. The resulting haploid embryos were subjected to heat shock treatment, which doubles the haploid chromosome set. DNA for the mapping panel was prepared from homozygous diploid F2 adults.
The mapping panel (HS panel) was comprised of 42 heat shock diploid F2 progeny obtained from two C32 × SJD F1 females. C32 and SJD are inbred mapping strains (Streisinger et al. 1981; Nechiporuk et al. 1999) and F1 females were obtained from natural crosses of C32 and SJD adults (Fig. 1). Nineteen of the F2 individuals in the HS panel were progeny of one C32 × SJD female and 23 were progeny of the second C32 × SJD female. The HS panel contains products of 42 meioses and the panel therefore provides an average resolution of 2.4 cM. The C32 and SJD strains are highly polymorphic with respect to one another, with ∼90% of markers showing different alleles in a previous comparison of the two strains (Nechiporuk et al. 1999). Both strains are inbred, and only ∼10% of SSLP markers reveal heterogeneity among different individuals of the same strain (Nechiporuk et al. 1999). Thus, we expected most markers that were heterozygous in one F1 female also to be heterozygous in the other, such that F2 progeny of the two females should segregate identical alleles. Accordingly, ∼10% of SSLPs (21 of 200 for which we tracked allele differences) showed differences between the two families in the heat shock panel (HS panel). Most markers displaying allele differences were polymorphic in both families in the panel and only 14 of 1364 markers that we scored in the HS panel were polymorphic in one family but monomorphic in the other. The positions of these 14 markers were assigned according to the genotypes of the family in which the marker was informative.
Markers and Linkage Analysis
To construct a genetic linkage map, we scored 1364 polymorphic markers in the HS panel (Figs. 2 and 3; Table 1, available online at http://www.genome.org). These markers included 593 previously mapped SSLPs and 771 SSCPs derived from zebrafish ESTs and fully sequenced cDNAs. Locus names, accession numbers, and primer sequences for the genes and ESTs scored in the HS panel are shown in Table 1. The genes and ESTs represented 482 different UniGene clusters (four of which were mapped in duplicate, see below) and there were 285 sequences that were not assigned to UniGene clusters in the current release (UniGene Build 6; http://www.ncbi.nlm.nih.gov/UniGene/Dr.Home.html). Sequence comparisons with TIGR Assembler (Sutton et al. 1995) and the BLAST algorithm (Altschul et al. 1997) did not reveal overlap among these 285 sequences, suggesting that most correspond to different genes.
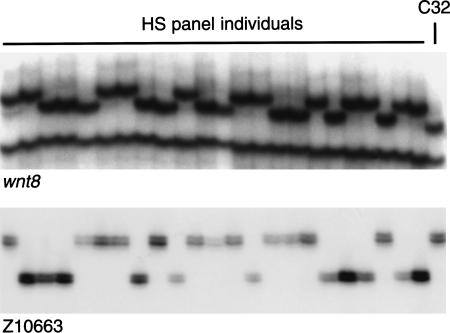
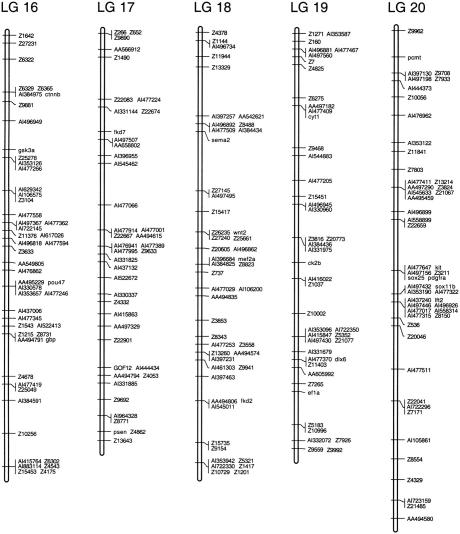
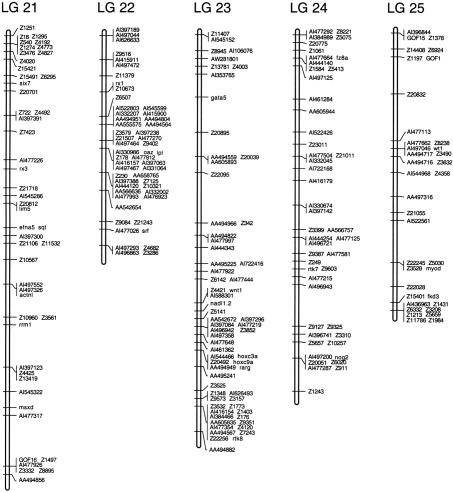
Figure 2.
Examples of polymorphisms scored in 23 individuals in the heat shock (HS) mapping panel and C32. An SSCP in the wnt8 gene and the SSLP Z10663 are shown.
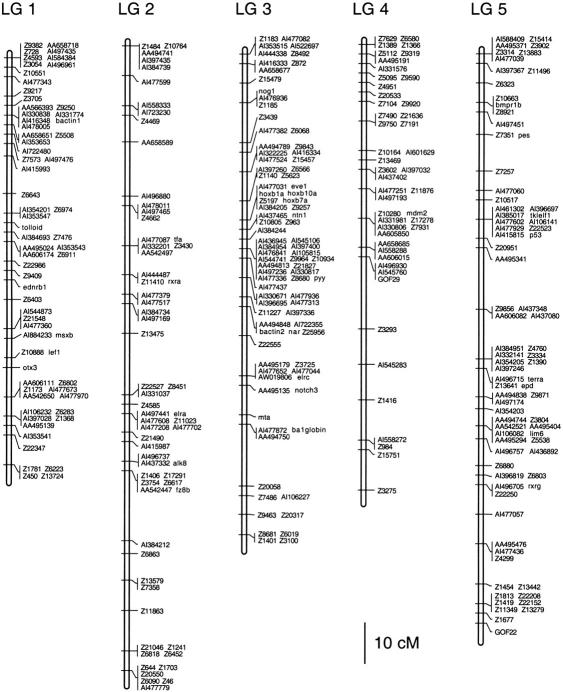
Figure 3.
(See pages 561–565.) Genetic linkage map of the zebrafish genome. Positions of 1364 polymorphic markers scored on the HS panel are shown. GenBank accession numbers, UniGene numbers, and primer sequences for genes and ESTs are shown in Table 1 (available online at http://www.genome.org). SSLP markers were described by Shimoda et al. (1999).
Of the SSCPs scored in the HS panel, 642 correspond to previously unmapped genes and ESTs (Table 1, available online at http://www.genome.org). In addition, we scored 59 SSCPs corresponding to genes localized in previous genetic maps (Postlethwait et al. 1998; Gates et al. 1999) and an additional 25 genes that were localized in RH maps (Geisler et al. 1999; Hukriede et al. 1999). Forty-five of the ESTs scored in the HS panel were localized in previous maps.
Linkage analysis assembled the 1364 polymorphisms into a map with 25 linkage groups, each representing one zebrafish chromosome (Fig. 3). The markers occupied 598 unique map positions. All map positions in the 25 linkage groups were supported by lod scores of ≥3 and there were no gaps >22 cM.
The 25 linkage groups in the HS panel map spanned 3011 cM, using the Kosambi mapping function to estimate the occurrence of double crossovers. Previous estimates put the total length of the female meiotic map at 2900 cM (Johnson et al. 1996), suggesting that the HS panel map covers nearly the entire genome. Accordingly, we were able to assign map positions to all of the polymorphic markers scored in the HS panel, because all markers in the data set showed significant linkage (lod ≥3) to at least one other marker.
As one measure of the error frequency in genotype data, we examined apparent double crossovers occurring in short intervals, which are uncommon and thus likely to reflect erroneous genotype assignments. Among the 53522 individual genotype assays in the data set (an average of 39.2 individuals scored per marker), there were only two double crossovers in intervals <20 cM. This suggests that the frequency of genotyping errors in the data set is low.
As another measure of the accuracy of the map, we compared the positions of markers mapped in the HS panel and in previous work. The HS panel map assigned 591 of 593 SSLPs to the same linkage groups as Shimoda et al. (1999). Of these 591 markers, there were eight cases where the order of closely spaced markers differed between the two maps. Because closely spaced markers are separated by few recombinants, these differences may reflect a small number of genotyping errors in one or both data sets. It is also possible that these discrepancies were caused by differences in the strains used to construct the two mapping panels. Two SSLPs (Z3054, Z9559) were assigned to different linkage groups; we repeated the genotype assays and confirmed the assignment of these two markers in the HS panel. In addition, the positions we assigned for these two SSLPs matched those obtained from RH mapping (M. Ekker, pers. comm.). Thirty-nine genes localized in our map were analyzed previously in a C32 × SJD haploid mapping panel (Postlethwait et al. 1998) and all of these were assigned to the same linkage group in both maps. There was one discrepancy among the 49 genes and ESTs common to our map and that of Gates et al. (1999). We mapped EST AI544741 to LG 3, and Gates et al. (1999) localized AA606157, another EST assigned to the same UniGene cluster (Dr.1344), to LG 1. One EST that we mapped (AA495459) was assigned to a different linkage group in the T51 RH panel. The assignments of the discrepant markers AI544741 and AA495459 in the HS panel were confirmed by a second set of genotype assays. In addition, there were four cases, all confirmed in duplicate genotype assays, in which sequences assigned to the same UniGene cluster were mapped to different linkage groups in the HS panel (Dr.261, Dr.274, Dr.345, Dr.1339). These may represent genes with pseudogenes or they may be pairs of duplicate genes generated by chromosomal duplications.
DISCUSSION
We have mapped 771 zebrafish genes and ESTs by scoring SSCPs in a meiotic mapping panel comprised of homozygous diploid individuals. In previous genetic linkage maps, ∼150 genes were analyzed in each of two haploid mapping panels (144 in Postlethwait et al. 1998; 157 in Gates et al. 1999). In RH mapping projects, Geisler et al. (1999) and Hukriede et al. (1999) mapped 355 and 148 genes and ESTs in the T51 and LN54 panels, respectively. In total, the sequences mapped in these previous studies define ∼500 different genes. Sequence comparisons and analysis of UniGene cluster assignments indicate that 642 of the 771 polymorphisms we localized correspond to previously unmapped sequences. Consequently, our map more than doubles the number of mapped zebrafish genes and ESTs.
The strategy we used in constructing the HS panel has a number of advantages for mapping experiments. First, the inbred mapping strains C32 and SJD are divergent and many markers exhibit polymorphisms in C32 × SJD crosses (Nechiporuk et al. 1999). Second, because homozygous diploid fish can survive to adulthood, we were able to collect enough genomic DNA to map >10,000 markers in future mapping studies with the HS panel. This is a substantial advantage over mapping panels comprised of haploid embryos, which survive only a few days and therefore yield much less genomic DNA. The ability to map many markers in a single cross diminishes the need to integrate maps produced in different mapping panels. Third, the use of homozygous diploids simplifies linkage analysis in comparison to sexual diploid protocols, because no heterozygotes are present in a population of homozygous diploid fish. This is useful particularly in the analysis of SSCPs, because these polymorphisms can be subtle, so that it can be difficult to distinguish heterozygotes from both classes of homozygotes.
The map presented here will accelerate the molecular analysis of zebrafish mutations by the positional cloning and candidate gene approaches. Positional cloning projects are accelerated as the density of mapped polymorphisms grows and the number of possible entry points for chromosomal walks increases. Gene maps augment the candidate approach because a comparison of map positions is a straightforward initial test to determine if a mutation might disrupt a particular gene. The polymorphisms corresponding to the genes we have mapped allow one to test candidate genes near a mutation of interest for direct linkage in a mutant mapping cross. Finally, this gene map will advance comparative mapping resources by allowing the identification of groups of syntenic genes that are conserved among vertebrates (Postlethwait et al. 1998).
METHODS
Production of Homozygous Diploids
Heat shock diploids were prepared from F1 females that were heterozygous for C32 and SJD genomes according to the procedures in Westerfield (1995) and raised to maturity. DNA was extracted from fully grown adult heat-shocked fish as described for the isolation of high molecular weight DNA from mammalian cells (Sambrook et al. 1989). Animals varied in mass between 0.19 g and 0.64 g and yielded ∼100–500 μg of genomic DNA.
Data Collection and Linkage Analysis
PCR was performed in 12.5-μl volumes as described previously (Gates et al. 1999). In typical PCR assays, 0.6 ng of genomic DNA was used as a template. SSLP markers were electrophoresed on denaturing acrylamide gels and SSCP markers were analyzed on nondenaturing acrylamide gels as described previously (Gates et al. 1999). Because SSCP analysis can generate complex banding patterns and because some primer pairs amplify multiple, unlinked fragments, polymorphisms were scored for major bands on SSCP gels.
Map data were collected and analyzed using Map Manager 2.6.5 (Manly 1993; http://mcbio.med.buffalo.edu/mapmgr.html). The complete genotype data set in Map Manager format can be obtained at http://zebrafish.stanford.edu. Each locus was placed initially at the position that maximized its lod score as indicated with the “Links” feature of Map Manager. Markers were then placed manually to minimize the number of double recombinants. In cases where double recombinants remained, autoradiographs were reexamined, and if an individual was again scored as a double recombinant, the genotype assay was repeated. Map graphics were drawn with MapMaker software (Lander et al. 1987) using the Kosambi mapping function and modified for clarity with Canvas 5.0.
Primer Design
D. rerio sequences from the NCBI nonredundant sequence database (NR) are housed in a MySQL relational database system (http://www.mysql.com), accessible through in-house-developed web query interfaces. The database was updated monthly with new ESTs (M. Clark and S. Johnson, Washington University Zebrafish Genome Resources Project; http://zfish.wustl.edu) deposited in the NR database. Genes and ESTs were clustered into groups of overlapping sequences using TIGR Assembler (Sutton et al. 1995). Primers used to develop SSCPs were designed from these contigs using Primer 3.0.6 (Rozen and Skaletsky 1997) and synthesized at the Stanford DNA Sequencing and Technology Center. SSLP primer sequences were obtained from Shimoda et al. (1999; http://zebrafish.mgh.harvard.edu/) and primers were synthesized at the Stanford DNA Sequencing and Technology Center or obtained from Research Genetics.
Acknowledgments
We thank Michael Gates and the members of our laboratories for helpful discussions, Michele Mittman and Lauren Jow for technical assistance, Marc Ekker for communicating unpublished data, and the Stanford DNA Sequencing and Technology Center for oligonucleotide synthesis. This work was supported by NIH grants R01DK55378 (W.S.T. and J.H.P.), R01RR12349 (W.S.T.), P01HD22486 (J.H.P. and S.L.J.), and R21HG01704 (A.F.S.). A.F.S. is a Scholar of the McKnight Endowment Fund for Neuroscience. W.S.T. is a Pew Scholar in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL talbot@cmgm.stanford.edu; FAX (650) 725-7739.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SCF, Malicki J, Stemple DL, Stainier DYR, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Gates MA, Kim L, Egan ES, Cardozo T, Sirotkin HI, Dougan ST, Laskari D, Abagyan R, Schier AF, Talbot WS. A genetic linkage map for zebrafish: Comparative analysis of genes and expressed sequences. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- Geisler R, Rauch G-J, Baier H, van Bebber F, Bross L, Dekens MPS, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJM, Jiang Y-J, Heisenberg C-P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Gates MA, Johnson M, Talbot WS, Horne S, Baik K, Rude S, Wong JR, Postlethwait JH. Centromere-linkage analysis and consolidation of the zebrafish genetic map. Genetics. 1996;142:1277–1288. doi: 10.1093/genetics/142.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB. Genetics and early development of zebrafish. Trends Genet. 1989;5:283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Ekket M, Chevrette M, Delgado J, Neuhauss S, Shimoda N, Driever W, Fishman MC, Jacob HJ. A microsatellite genetic linkage map for zebrafish. Nat Genet. 1998;18:338–343. doi: 10.1038/ng0498-338. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Manly KF. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Finney JE, Keating MT, Johnson SL. Assessment of polymorphism in zebrafish mapping strains. Genome Res. 1999;9:1231–1238. doi: 10.1101/gr.9.12.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Talbot WS. Zebrafish genomics: From mutants to genes. Trends Genet. 1997;13:183–190. doi: 10.1016/s0168-9525(97)01129-3. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Johnson S, Midson CN, Talbot WS, Gates M, Ballinger EW, Africa D, Andrews R, Carl T, Eisen JS, et al. A genetic map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan Y-L, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer 3.0. ( http://www.genome.wi.mit.edu) Cambridge, MA: Whitehead Institute; 1997. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schier AF, Talbot WS. The zebrafish organizer. Curr Opin Genet Dev. 1998;8:464–471. doi: 10.1016/s0959-437x(98)80119-6. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish genetic map with 2000 microsatellite markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- Sutton G, White O, Adams M, Kerlavage A. TIGR Assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci & Tech. 1995;1:9–19. [Google Scholar]
- Talbot WS, Hopkins N. Zebrafish mutations and functional analysis of the vertebrate genome. Genes & Dev. 2000;14:755–762. [PubMed] [Google Scholar]
- Young WP, Wheeler PA, Coryell VH, Keim P, Thorgaard GH. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics. 1998;148:839–850. doi: 10.1093/genetics/148.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; 1995. [Google Scholar]