Abstract
It has been discovered that the transient receptor potential ankyrin 1 (TRPA1) proteins of Boidae (boas), Pythonidae (pythons), and Crotalinae (pit vipers) are used to detect infrared radiation, but the molecular mechanism for detecting the infrared radiation is unknown. Here, relating the amino acid substitutions in their TRPA1 proteins and the functional differentiations, we propose that three parallel amino acid changes (L330M, Q391H, and S434T) are responsible for the development of infrared vision in the three groups of snakes. Protein modeling shows that the three amino acid changes alter the structures of the central region of their ankyrin repeats.
Keywords: infrared vision, transient receptor potential ankyrin 1 (TRPA1) proteins, ankyrin repeats, parallel evolution, snakes
The transient receptor potential (TRP) ion channels are involved in various biological processes, including calcium and magnesium homeostasis, neuronal growth, temperature sensation, and pain sensation (Venkatachalam and Montell 2007; Gaudet 2008). Based on sequence similarity, the TRP superfamily is divided into seven subfamilies: TRPC (canonical), TRPV (vanilloid), TRPM (melastatin), TRPA (ankyrin), TRPN (NOMPC), TRPP (polycystin), and TRPML (mucolipin). The total numbers of TRP channels in worm (Caenorhabditis elegans), fruit fly (Drosophila melanogaster), mouse (Mus musculus), and human vary between 13 and 28 (Venkatachalam and Montell 2007).
A fiery sensation caused by the pungent agents of wasabi and other mustard plants is generated by our transient receptor potential ankyrin 1 (TRPA1) channel (Venkatachalam and Montell 2007). Recently, it has been discovered that the orthologous receptors of the western diamondback rattlesnake (Crotalus atrox), ball python (Python regius), and garden tree boa (Corallus hortulanus) detect infrared radiation, whereas that of the Texas rat snake (Pantherophis obsoletus lindheimeri) does not (Gracheva et al. 2010). The genetic mechanism of infrared sensitivity of these snake-specific TRPA1 proteins is unknown. To solve the problem, we need to first identify amino acid changes that are responsible for the dramatic functional changes in the three groups of snakes. One effective way of doing this is to relate amino acid substitutions and functional changes in the phylogenetic tree of the orthologous receptors (Yokoyama R and Yokoyama S 1990; Yokoyama 1997; Yokoyama et al. 2000, 2008). This function-based method is entirely different from commonly used statistical methods of detecting positive selection such as ADAPTSITE (Suzuki et al. 2001), PAML (Yang 2007), and HyPhy (Pond et al. 2005).
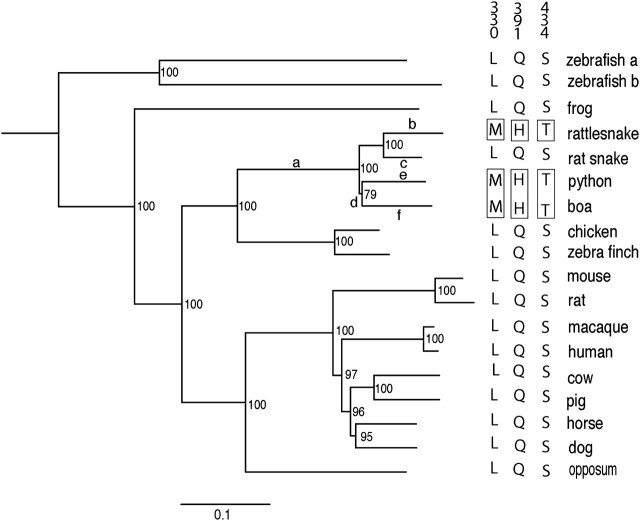
When we apply the neighbor joining (NJ) method (Saitou and Nei 1987) to the nucleotide sequences of a total of 18 representative TRPA1 genes in vertebrates, the phylogenetic tree obtained agrees with the species tree, where the TRPA1 proteins of snakes are most closely related to those of birds, and then mammals, amphibians, and fishes, in that order (fig. 1). The bootstrap analysis shows that many nodes are supported by high bootstrap values (>95%) and highly reliable. The clustering of the rattlesnake and rat snake TRPA1 proteins is supported by 100% bootstrap value but that of the python and boa proteins is supported by a bootstrap value of 79%. The latter value is rather low, but the phylogenetic relationships of the four snake groups are consistent with the species tree of snakes based on other molecular data set (Vidal et al. 2009).
FIG. 1.
Phylogenetic tree of 18 representative TRPA1 receptors constructed by applying the NJ method (Saitou and Nei 1987) to the nucleotide sequence of the corresponding genes. The numbers next to the different nodes indicate clustering percentage supports generated by 1,000 bootstrap analysis. Relevant branches are lettered (a–f) so that they can be referenced in the text. Three amino acids in TRPA1 that are suspected to enable infrared detection are boxed. The numerical column headings specify the amino acid positions of the rattlesnake TRPA1.
To identify critical amino acids that are involved in infrared vision, we assume that infrared vision is a derived character. This seems reasonable for two reasons: 1) only a limited number of snake lineages can detect infrared radiation and 2) there is no obvious reason why the infrared sensitivity had to be eliminated many times during snake evolution.
When we search for the amino acids that are common to the three infrared-sensitive snake TRPA1 proteins, a total of 19 amino acid sites are found (supplementary fig. 1, Supplementary Material online). For many proteins, a majority of amino acid changes are not involved in functional changes (King and Jukes 1969; Perutz 1983; Yokoyama 2008; Yokoyama et al. 2008). TRPA1 proteins should not be an exception. The elimination of nonessential amino acid substitutions was done in two steps. First, we inferred the ancestral sequences using PAML (Yang 2007). The results suggest that 12 out of 19 amino acid sites (positions 49, 179, 184, 198, 228, 342, 746, 828, 829, 913, 916, and 946; numbering follows the rattlesnake sequence) occurred in the rat snake TRPA1 receptor and are not involved in infrared vision (supplementary fig. 1, Supplementary Material online). Second, the ancestral sequences of four other amino acid sites (positions 1017, 1028, and 1074) have not been inferred (supplementary fig. 1, Supplementary Material online); however, the variable amino acids at these sites also suggest that they are not functionally important (supplementary fig. 2, Supplementary Material online).
After these considerations, 3 of the 19 amino acid sites (positions 330, 391, and 434) emerge: the infrared-sensitive TRPA1 proteins have M330, H391, and T434 (or M330/H391/T434), but all other TRPA1 proteins have L330/Q391/S434 (fig. 1). L330, Q391, and S434 of the TRPA1 receptor in the common ancestor of snakes and birds have posterior probabilities of >0.95. Hence, the amino acid differences in figure 1 can be explained by two alternative evolutionary changes: 1) L330M (L330 was replaced by M330), Q391H, and S434T (or L330M/Q391H/S434T) at “branch a” followed by M330L/H391Q/T434S at “branch c” or 2) L330M/Q391H/S434T at “branches b” and “d.” However, if the infrared vision is a derived form and these amino acid changes are involved in the development of infrared sensitivity, then the first possibility disappears.
To test the second hypothesis that the parallel evolution occurred by Darwinian natural selection, we examined the null hypothesis by determining the probabilities that the three changes occurred by random genetic drift. For that purpose, we first evaluated the number of amino acid substitutions at “branch b” (105 substitutions) and “branch d” (18 substitutions) using PAML. The ancestral snake TRPA1 had L330 (encoded by CTG), Q391 (CAA or CAG), and S434 (TCT). Therefore, if we assume that one nucleotide mutates to any one of the other three nucleotides with equal probabilities, then the respective amino acids mutate to M330, H391, and T434 with probabilities 1/5, 2/7, and 1/6. Then, at branch b, assuming that amino acid substitutions follow a Poisson process, L330M, Q391H, and S434T occur roughly with probabilities 0.0214, 0.0305, and 0.0178, respectively. At branch d, the respective substitutions occur roughly with probabilities 0.0038, 0.0055, and 0.0032. Hence, under neutral evolution, the chance that the three changes occurred by molecular convergence is about 0.78 × 10−12.
This result rejects the null hypothesis that L330M/Q391H/S434T were generated by random genetic drift alone, strongly suggesting that they are caused by Darwinian natural selection via the acquisition of infrared vision. In fact, the python–boa clade also includes some infrared-nonsensitive snakes (Lawson et al. 2004). Therefore, it is most likely that parallel evolution of L330M/Q391H/S434T have occurred at branches b, e, and f rather than at branches b and d.
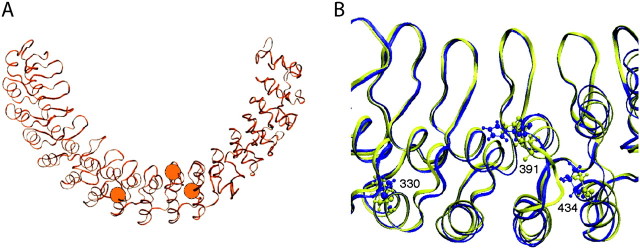
As its name suggests, TRPA1 is characterized by 17 functionally important ankyrin repeats (ANK 1–ANK 17) in the N-terminal segment (supplementary fig. 1, Supplementary Material online). Interestingly, amino acid sites 330, 391, and 434 are on ANK 8, ANK 10, and ANK 11, respectively (fig. 2). It is suspected that long ankyrin repeats function as springs when pulled from both ends and are directly involved in sensing mechanical stimuli (Sotomayor et al. 2005; Lee et al. 2006). Being located in the middle of the ankyrin repeats (fig. 3A), the three amino acid sites may play key roles in infrared vision. Moreover, amino acid sites 420 and 427 on ANK 11 are shown to be critical in detecting various pungent compounds (Macpherson et al. 2007).
FIG. 2.
Sequence of ANK 8–ANK 11 of the snake ancestor, rattlesnake, rat snake, python, and boa. The ancestral amino acids that have a posterior probability of <0.95 are underlined. Amino acid sites where L330M/Q391H/S434T occurred are shown by rectangles.
FIG. 3.
The molecular structures of ankyrin receptor. (A) The entire polyankyrin of the snake ancestor; the three orange balls illustrate amino acid sites 330, 391, and 434 (from left to right). (B) Superimposed ANK 8–ANK 11 of the snake ancestor (yellow ribbon) and its L330M/Q391H/S434T mutant (blue ribbon). The candidate amino acids at critical sites are shown with ball and stick models.
To evaluate the effects of L330M/Q391H/S434T on the structural change in the ankyrin repeats, we modeled the ancestral snake ankyrin with and without the three mutations (Fig. 3B). The mutations on the ANK 8 (L330M), ANK 10 (Q391H), and ANK 11 (S434T) cause the most significant change in the helicity of ANK 10. ANK 8 structure does not change much and the helicity of ANK 11 changes slightly. In particular, the distances between Cα atoms of sites 330 and 391 (23.2 Å), sites 330 and 434 (29.2 Å), and sites 391 and 434 (10.6 Å) in the ancestral snake ankyrin become 22.9, 28.1, and 11.3 Å in the mutant, respectively. These changes effectively increase the area of the triangle formed by the Cα atoms of these sites from 111.6 Å2 of the TRPA1 of the ancestral snake to 124.7 Å2 of those of the infrared-sensitive snakes.
These theoretical results strongly suggest that both M330/H391/T434 and the expanded space of the ANK 8–ANK 11 in the infrared-sensitive snake TRPA1 channels may be required for interacting with either a specific protein or ligand involved in infrared vision. Being located in the middle of the ankyrin repeats, M330/H391/T434 may also change the elastic properties of the snake ankyrins.
Supplementary Material
Supplementary figures 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
This work is supported by the National Institutes of Health (R01EY016400) and Emory University.
References
- Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Ingolia NT, Kelly YM, Cordero-Morales JF, Hollopeter G, Chesler AT, Sanchez EE, Perez JC, Weissman JS, Julius D. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- Lawson R, Slowinski JB, Burbrink FT. A molecular approach to discerning the phylogenetic placement of the enigmatic snake Xenophidion schaeferi among the Alethinophidia. J Zool. 2004;263:285–294. [Google Scholar]
- Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, Patapoutian A. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 2007;445:541–545. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Species adaptation in a protein molecule. Mol Biol Evol. 1983;1:1–28. doi: 10.1093/oxfordjournals.molbev.a040299. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Corey DP, Schulten K. In search of the hair-cell gating spring elastic properties of ankyrin and cadherin repeats. Structure. 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T, Nei M. ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics. 2001;17:660–661. doi: 10.1093/bioinformatics/17.7.660. [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal N, Rage J-C, Couloux A, Hedges SB. Snakes. (Serpentes) In: Hedeges SB, Kumar S, editors. The timetree of life. New York: Oxford University Press; 2009. pp. 390–397. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular genetic basis of adaptive selection: examples from color vision in vertebrates. Annu Rev Genet. 1997;31:315–336. doi: 10.1146/annurev.genet.31.1.315. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Evolution of dim-light and color visual pigments. Annu Rev Genom Hum Genet. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB, Blow NS. Ultraviolet pigments in birds evolved from violet pigments by a single amino acid change. Proc Natl Acad Sci U S A. 2000;97:7366–7371. doi: 10.1073/pnas.97.13.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Natl Acad Sci U S A. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.