Abstract
Human L1 retrotransposons can produce DNA transduction events in which unique DNA segments downstream of L1 elements are mobilized as part of aberrant retrotransposition events. That L1s are capable of carrying out such a reaction in tissue culture cells was elegantly demonstrated. Using bioinformatic approaches to analyze the structures of L1 element target site duplications and flanking sequence features, we provide evidence suggesting that ∼15% of full-length L1 elements bear evidence of flanking DNA segment transduction. Extrapolating these findings to the 600,000 copies of L1 in the genome, we predict that the amount of DNA transduced by L1 represents ∼1% of the genome, a fraction comparable with that occupied by exons.
The LINE-1 (L1) retrotransposon family is estimated to contain 600,000 copies, accounting for at least 15 % of the human genomic DNA (Smit 1996). L1's second ORF (ORF2) encodes endonuclease and reverse transcriptase activities (Mathias et al. 1991; Feng et al. 1996), and is the most abundant ORF in the genome. As a major source of reverse transcriptase, L1 is likely to be indirectly responsible for the spreading of other retrotranscripts, such as Alu sequences and processed pseudogenes (Maestre et al. 1995; Boeke and Stoye 1997; Dhellin et al. 1997; Jurka 1997). A novel feature of L1 propagation and function was described recently by Moran et al. (1999), who showed that L1 can efficiently comobilize a 3′- flanking segment of non-L1 DNA to new genomic locations in tissue culture cells. This makes L1 a potential player in such genomic events as exon shuffling and regulatory region combinatorics (Boeke and Pickeral 1999; Eickbush 1999). We have studied 129 full-length L1 elements with high similarity to L1.2 (an active element). Computational analysis shows that at least 10% of these L1s have an associated putative 3′-transduced segment, and on this basis, the total amount of DNA transduced by L1 can be extrapolated to represent at least 1% of the human genome. This finding demonstrates that L1s are often involved in shuffling genomic DNA, and are thus important contributors to genome plasticity.
Several examples of naturally occurring 3′-transduction events were identified previously as mutagenic L1 insertions, in which additional (non-L1) sequences were incorporated downstream from each newly transposed L1 (Miki et al. 1992; Holmes et al. 1994; McNaughton et al. 1997). Transduction of 3′-flanking sequence by engineered L1 elements also readily occurs in HeLa cells, and can be driven by either the cytomegalovirus promoter, or the native L1 promoter; notably, transposition efficiency is higher when a strong polyadenylation signal is introduced downstream from the L1 (Moran et al. 1999). An important open question remains—how efficiently does L1-driven 3′ transduction occur naturally in the human genome? Opportunities to observe abnormal human phenotypes caused by 3′-transducing L1 insertions may be extremely limited because only a small fraction of the human genome is currently attributed to genes and upstream regulatory regions. In this study, we took advantage of the tremendous sequence production by the Human Genome Project to computationally estimate the extent of naturally occurring L1-driven 3′ transduction (Fig.1).
Full-length L1 elements are ∼6000 bp long; the majority of L1s in the human genome, however, are severely 5′ truncated or rearranged, including 5′-inverted and deleted-inverted forms (Hutchison et al. 1989). Newly inserted L1 sequences are frequently flanked by short direct repeats, which have been shown to represent target site duplications (TSDs) created upon L1 integration (Kazazian et al. 1988; Holmes et al. 1994; Moran et al. 1996). With these sequence features in mind, we designed a TSD-based strategy to look for potential 3′-transduced segments associated with full-length L1s (see Methods). If a pair of TSDs is found immediately flanking the L1 and its poly(A) tail, this represents a standard L1 insertion, with no additional sequences transposed. In contrast, in cases of 3′ transduction the 3′ TSD is found further downstream from the L1 (Miki et al. 1992; Holmes et al. 1994; McNaughton et al. 1997).
RESULTS AND DISCUSSION
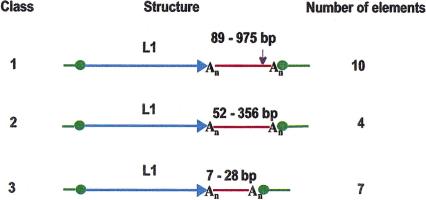
We limited our present study to full-size L1s with high similarity (>94% identity) to L1.2. Studying whole insertion events is critical for the TSD-based algorithm for 3′-transduction detection, and this allows us to avoid introducing the extra ambiguity of the precise 5′ boundaries of each L1, which is a prominent factor when truncated elements are analyzed. We studied 129 full-length L1 elements; of these, 16 were uninformative because of insufficient flanking sequence in the GenBank records. An additional 16 examples lacked TSDs >6 bp in length. Another 76 cases represented standard insertions. Finally, 21 qualified as 3′ transduction candidates. These 21 elements could be divided into three classes on the basis of sequence characteristics of the transduced DNA segment (Fig. 2; Table 1). Class 1 elements had downstream segments 89–975-bp long, and contained a consensus polyadenylation signal (AATAAA or ATTAAA) (Tabaska and Zhang 1999) 10–35 bp upstream from the poly(A) tail immediately preceding the 3′ TSD. The 10 elements in this class are the most likely candidates for 3′ transduction. Class 2 elements had 3′ segments 52–356-bp long, and lacked a consensus polyadenylation signal. Class 3 segments are shorter than the other two classes (7–26 bp), and could represent aberrant poly(A) tails. Even though class 3 segments may well have been formed by the same mechanism, further sequence analysis was not pursued because the results would not be statistically significant.
Figure 2.
Three classes of L1 elements with 3′ transduced segments. Transduced segments are shown in red. Range of transduced segment lengths is shown above each line. (Ovals) Target side duplications; (purple arrow) polyadenylation signal found in the transduced regions of class 1; (An) poly(A) tail.
Table 1.
Summary of L1-Associated 3′ Transduction Events
| L1 element (gi no. and coordinates) | L1 structurea | Class | TSDs | pA signal | Poly(A) tail (bp) | Transduced segment length (bp) | |||
|---|---|---|---|---|---|---|---|---|---|
| length (bp) | ORF1 | ORF2 | poly(A) tail (bp) | ||||||
| 1279499, 11400 . . 17418 | 6019 | — | — | 25 | 1 | AAGAAAGAGCCAA | ATTAAA | 27 | 479 |
| 1669380, (16606 . . 22620) | 6015 | — | — | 24 | 1 | AGAAATTGTC | ATTAAA | 17 | 89 |
| 2275172, (73720 . . 79740) | 6021 | + | — | 22 | 1 | AGAAATCTGATTTGA | AATAAA | 13 | 975 |
| 2588618, (59187 . . 65202) | 6016 | + | — | 27 | 1 | GAAATTTTTCATC | AATAAA | 12 | 337 |
| 2588627, 131175 . . 137191 | 6017 | — | + | 14 | 1 | AAAAATTTACTGTCTA | AATAAA | 10 | 455 |
| 2809267, (11716 . . 17735) | 6020 | + | + | 47 | 1 | AAAAAAAATCACCA | AATAAA | 15 | 137 |
| 2853183, 4261 . . 10276 | 6016 | + | + | 36 | 1 | AAAGAAACTGAAGAGG | AATAAA | 25 | 175 |
| 3319674, 61629 . . 67639 | 6011 | + | — | 16 | 1 | AAAGAAAAAATG | AATAAA | 17 | 272 |
| 3810569, 21041 . . 27057 | 6017 | + | — | 24 | 1 | AGAAGCA | ATTAAA | 14 | 821 |
| 3845408, (22572 . . 28582) | 6011 | — | — | 17 | 1 | AGAATCTGACCTTCC | AATAAA | 13 | 460 |
| 2076718, 29372 . . 35393 | 6022 | — | — | 18 | 2 | AAGAAAGAGAATA | — | 9 | 356 |
| 3288437, 65129 . . 71148 | 6020 | + | — | 16 | 2 | AAAGAACACCTGGG | — | 11 | 52 |
| 3522964, (24909 . . 30926) | 6018 | + | + | 61 | 2 | AAAAAACACAGTGAA | — | 14 | 119 |
| 4006838, 117572 . . 123596 | 6025 | + | — | 28 | 2 | AGAAGAAATG | — | 22 | 36 |
| 3421083, 114651 . . 120654 | 6004 | — | — | 37 | 3 | AATGTTTA | AATAAA | 28 | 16 |
| 3510243, (4788 . . 10794) | 6007 | — | — | 16 | 3 | AAATAGT[T/C]GAAGA | AATAAA | 21 | 7 |
| 3808075, 6601 . . 12628 | 6028 | + | + | 23 | 3 | AAAAAGGAGCCCGG | AATAAA | 18 | 11 |
| 3873180, 114223 . . 120240 | 6018 | + | — | 26 | 3 | AAGATTTTGTG | AATAAA | 26 | 10 |
| 3928116, 61076 . . 67082 | 6007 | — | — | 18 | 3 | AGAAACTTGGAACACA | AATAAA | 6 | 8 |
| 3953485, 108923 . . 114934 | 6012 | — | — | 31 | 3 | AAGCAATTTGT | — | 17 | 26 |
| 4107187, 91010 . . 97031 | 6022 | — | — | 9 | 3 | AAACAGTATTTTCCCT | AATAAA | 8 | 13 |
Table 1 is also available at http://www.bs.jhmi.edu/mbg/boekelab/3'tstable1links.html, where links to the corresponding GenBank records are provided for all gi numbers.
ORF1 and ORF2 are L1 open reading frames; (+) An ORF of predicted size is seen at the predicted location within the L1.
We then searched for a possible origin of the 14 elements of classes 1 and 2 and their 3′ transduced segments. Each of the potential transduced segments was masked by RepeatMasker (A. Smit and P. Green, unpubl.) to suppress highly repetitive matches, and the masked sequences were then used as queries in BLASTN and BLASTX similarity searches. A high-scoring matching segment elsewhere in the genome, if found immediately downstream from another L1 element, would represent a potential master element if no poly(A) tail followed the segment of interest, or a related 3′ transduction event if another poly(A) tail with target site duplications were found.
Of the fourteen 3′-transduced segments, three were completely masked by RepeatMasker, and four were partially masked. Three of the four L1s with partially masked 3′-transduced segments represent a previous integration into an L1 3′ UTR, and one represents integration into an Alu repeat. Interestingly, all of these L1 elements inserted in the same orientation as the target element (Fig. 3), which may be indicative of an L1 targeting preference. The TSD-based algorithm allows one to distinguish a transduction event with inclusion of repetitive sequence present at the master locus from a new L1 insertion into a pre-existing L1 3′ UTR: In each case in Figure 3, a full-length, uninterrupted L1 is followed by the poly(A) tail and the sequence of interest (including other transposon sequence), followed by another poly(A) tail. All of this inserted material is flanked by TSDs. This sequence signature is consistent with a 3′-transduction event in which the master L1 that produced each insertion had inserted previously into another transposon (see Fig. 3 legend for details). These events also suggest a simple mechanism for the rapid evolution of L1 (and other retrotransposon) 3′ ends, which are structurally quite diverse.
Figure 3.
L1 integration into other transposons. Numbers at left indicate gi numbers of GenBank records in which the L1 elements and their associated 3′-transduced segments are found (see Table 1 for exact coordinates). Repetitive elements recognized in the 3′-transduced segments by RepeatMasker are represented by filled gray arrows, with annotation shown at top (L1 or Alu subfamily name is followed by the coordinates projected onto the consensus sequence for that subfamily). Remarkably, in all four cases, the L1 that produced the observed new insertion depicted here must have inserted previously into another L1 or Alu in the same orientation. [A similar tendency has been noticed previously for newly inserted Alu elements (Jurka 1995)]. In two of the examples shown above, the transduced segment terminates at the end of the pre-existing L1. This suggests that once an L1 element inserts into such a region, subsequent transcription of the newly inserted element frequently reads through into the flanking DNA, and is then polyadenylated using a signal in the 3′ UTR of the pre-existing L1 element. By this means, L1 elements may acquire new, hybrid 3′ UTRs. This process likely contributes to the rapid evolution of L1 3′ UTR sequences (Smit 1996) and could explain why they are so rich in A residues, as well as provide a clue to the mechanism leading to the similarity of L1 and Alu 3′ UTR sequences (Boeke 1997).
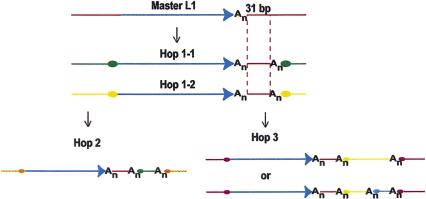
Several of the unique sequences present in 3′-transduced segments produced significant nucleotide matches, whereas no protein matches (representing potential coding exons) were found. Twelve of the fourteen segments did, however, contain at least one ORF >50 bp long: two were <80 bp long, four between 80 and 100 bp long, and six were >100 bp long. Six of the fourteen segments associated with L1s of classes 1 and 2 (identified in the text by the gi numbers of their GenBank records—see Table 1) produced significant non-self nucleotide matches in the human nonredundant database. In three cases, gi2076718, 2853183, and 3522964, no L1 was found upstream from the matches. Sequence gi2275172 represents an example of 3′ transduction we expected to find as a positive control—this case is a previously described L1-driven 3′-transduction event in the human dystrophin gene (McNaughton et al. 1997). The last two of the query segments, gi2588627 and 3288437, shared a unique 31-bp sequence immediately following L1. Further analysis of the DNA flanking each of these two elements suggests that they are several transposition events removed from the same master element, which acquired additional flanking sequences in several steps (Fig. 4).
Figure 4.
Model for the generation of L1 elements and their 3′-transduced regions in gi 3288437 and gi 2588627. Hop 2 (gi 3288437) and Hop3 (gi 2588627) are likely to be related descendants of the same Master L1 element. At least two, and possibly three (in case of gi 2588627) 3′-transducing intermediate transposition events would result in the structures seen at the DNA sequence level. The 31-bp sequence immediately downstream from the L1 is 100% identical between the two transduced segments, and in both cases is followed by a poly(A) tail. The proposed intermediates have not yet been found in the human genomic sequence available to date. The blue segment in Hop 3 reflects the possibility of two rather than one intermediate retrotransposition events between the master and Hop 3 elements, as there are two internal poly(A) sequences within the transduced segment.
Of 113 informative L1 elements in the data set studied here, 97 (86%) had TSDs consistent with either a standard insertion or a 3′-transduction event. Of these 97 L1s, 10% (class 1 elements) are excellent candidates for 3′ transduction, whereas 14%–22% have a 3′-transduced segment by more relaxed criteria (including classes 2 and 3). Importantly, known and new examples of related 3′-transduction events were identified. To estimate the total fraction of the human genome that can be accounted for by 3′ transduction, we assumed ∼600,000 copies of L1 in the human genome (Smit 1996) and the average length of transduced segments 420 bp for class 1 elements, 340 bp for classes 1 and 2 combined, and 231 bp for all 3 classes of transducing L1s. By use of these values, the total amount of 3′-transduced sequence shuffling extrapolated to the entire human genome is between 25.2 and 30.5 Mb, or ∼1% of the human genome, a fraction comparable with that occupied by exons.
The estimate of L1 numbers on which the above calculation is based, was extrapolated from the amount of repeat DNA found in various large DNA clones selected from the human genome for sequencing. There is some ambiguity in the number of L1s in the human genome, mainly due to the difficulty of recognizing old elements in the sequence. In addition, our current knowledge of highly heterochromatic and potentially difficult to clone regions is limited; these are likely to be enriched in transposon sequences. Balancing this, the L1s without TSDs may result from either 3′-transduction events of very large segments or from very short TSDs, thus the actual frequency of transduction events may be higher than our estimate. Finally, because L1s tend to localize to less gene-rich regions, the chances of carrying an exon may be decreased.
A final issue regarding the above estimate is the validity of extrapolating our numbers to the very large class of 5′-truncated L1 elements. We expect that they will have as high a number of associated transduced segments as full-length elements, if not higher. We carried out a pilot survey of 25 randomly selected 100–1000-bp long 5′-truncated L1 elements. These had characteristics similar to the full-length L1 elements in this study. The truncated elements analyzed were 85%–99% identical to L1.2 sequence, and of the 25 truncated L1 elements, 1 was uninformative (insufficient flanking sequence), 9 had no TSDs >7-bp long, and 15 had TSDs. Of the elements with TSDs, 11 were classified as standard insertion, and 4 represented a 3′-transduction signature. Of these four candidates, two were completely masked (L1 3′ UTR sequence, in the same orientation as the L1 of interest). This preliminary survey supports our expectation of finding an equivalent or slightly higher fraction of 3′ transduction in truncated elements as opposed to full-length L1s.
A possible expansion of this study would be the study of older L1 subfamilies. However, this would bring a higher ambiguity level to the signals seen at the DNA sequence level. Elements of the young subfamilies are more likely to have inserted relatively recently, and thus would have a higher chance of preserving intact TSDs. The older an insertion, the lower the likelihood of finding authentic TSDs, because of the accumulation of random mutations.
Thus, L1-driven transduction of flanking DNA is likely to be an important mechanism of genome evolution via increasing genome plasticity and facilitating new combinations of coding and regulatory sequences. Although most observed cases of 3′ transduction represent relatively small gene segments, it is possible that entire genes are included occasionally into transduced segments. This would lead to the introduction of a processed (no introns) copy of a gene into a new genomic locus. Thus, 3′ transduction is potentially a mechanism for outright gene duplication as well as gene scrambling. A 3′ transduction might represent a relatively noninvasive mechanism by which an organism can test novel sequence combinations, because transposon-plus-flanking-sequence integration could be significantly less disruptive to genome organization than larger-scale genome rearrangements such as inversions or translocations.
METHODS
Data set
A total of 129 L1 elements, at least 6000-bp long, were collected as high-scoring BLAST matches to L1.2 (accession no. M80343), one of the currently active LINE-1 transposons. The search was performed against GenBank release 113.0 plus daily updates as of September 1, 1999. All of these elements are at least 94% identical to L1.2.
Target Site Duplication Determination
One hundred base pairs upstream of each L1 element studied were compared with 3000 bp downstream of the same element, in search of short direct repeats that are the putative TSDs. BLAST2SEQUENCES (Tatusova and Madden 1999) and DOTTER (Sonnhammer and Durbin 1995) computer programs were used to find matching substrings at least 6-bp long. One mismatch was allowed for repeats >11-bp long. A pair of short direct repeats was considered a TSD pair if the 5′ TSD adjoined the 5′ end of the L1 element and the 3′ TSD immediately followed a poly(A) tail.
DNA Sequence Analysis
The BLASTN (Altschul et al. 1997) program was used to search human-specific nucleotide databases for sequences similar to the putative transduced regions; BLASTX (Altschul et al. 1997) program was used to search for protein matches. We used the e-value cutoff of 0.1 in both BLASTN and BLASTX searches. REPEATMASKER (A. Smit and P. Green, unpubl.) was used to mask and characterize the transduced segments.
Databases Used
The nonredundant (nr) protein database was used in BLASTX searches. In BLASTN searches, four human-specific databases were used, nr, EST, GSS, and HTGS.
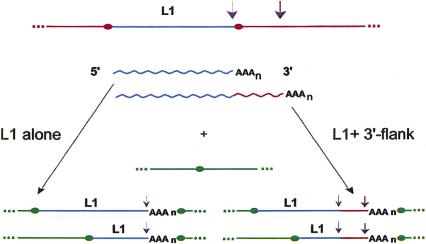
Figure 1.
L1-driven 3′ transduction. Incorporation of additional (3′ flanking) sequence into the transcript generated from the L1 promoter is followed by reverse transcription and integration of this longer cDNA into new genomic loci. (Ovals) Target site duplications; (purple arrows) polyadenylation signals (weak and strong); (AAAn) poly(A) tail.
Acknowledgments
We thank John Moran and Greg Cost for helpful discussions. Our work was supported in part by NIH grant CA16519 to J.D.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jboeke@jhmi.edu; FAX (410) 614-2987.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Pickeral OK. Retroshuffling the genomic deck. Nature. 1999;398:108–109. doi: 10.1038/18118. , 111. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Varmus H, Hughes S, Coffin J, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 343–435. [PubMed] [Google Scholar]
- Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush T. Exon shuffling in retrospect. Science. 1999;283:1465–1467. doi: 10.1126/science.283.5407.1465. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Hutchison CA, Hardies SC, Loeb DD, Shehee WR, Edgell MH. Long interspersed repeated sequences in the eukaryotic genome. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, DC.: American Society for Microbiology; 1989. pp. 593–617. [Google Scholar]
- Jurka J. Origin and evolution of Alu repetitive elements. In: Maraia R, editor. The impact of short interspersed elements (SINEs) on the host genome. Austin, TX: R.G. Landes; 1995. pp. 25–41. [Google Scholar]
- ————— Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: Processed pseudogene formation. EMBO J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- McNaughton JC, Hughes G, Jones WA, Stockwell PA, Klamut HJ, Petersen GB. The evolution of an intron: Analysis of a long, deletion-prone intron in the human dystrophin gene. Genomics. 1997;40:294–304. doi: 10.1006/geno.1996.4543. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Tabaska JE, Zhang MQ. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]