Abstract
Sporadic colon cancer is caused predominantly by dietary factors. We have selected bile acids as a focus of this review since high levels of hydrophobic bile acids accompany a Western-style diet, and play a key role in colon carcinogenesis. We describe how bile acid-induced stresses cause cell death in susceptible cells, contribute to genomic instability in surviving cells, impose Darwinian selection on survivors and enhance initiation and progression to colon cancer. The most likely major mechanisms by which hydrophobic bile acids induce stresses on cells (DNA damage, endoplasmic reticulum stress, mitochondrial damage) are described. Persistent exposure of colon epithelial cells to hydrophobic bile acids can result in the activation of pro-survival stress-response pathways, and the modulation of numerous genes/proteins associated with chromosome maintenance and mitosis. The multiple mechanisms by which hydrophobic bile acids contribute to genomic instability are discussed, and include oxidative DNA damage, p53 and other mutations, micronuclei formation and aneuploidy. Since bile acids and oxidative stress decrease DNA repair proteins, an increase in DNA damage and increased genomic instability through this mechanism is also described. This review provides a mechanistic explanation for the important link between a Western-style diet and associated increased levels of colon cancer.
Keywords: bile acids, genomic instability, colon cancer
Introduction
Sporadic colon cancer is caused predominantly by dietary factors, most notably a high-fat diet accompanied by low intake of micronutrients (folate, niacin, zinc), antioxidants and other plant phytochemicals. A high intake of fat induces the release of bile acids from the gall bladder into the small intestine. Bacteria in the colonic lumen then convert conjugated and hydrophilic bile acids to unconjugated, hydrophobic bile acids such as deoxycholic acid and lithocholic acid (Figure 1). These hydrophobic bile acids, deoxycholic acid in particular, have been reported to be multiple stress inducers and at high physiologic levels can create chaos within colon epithelial cells. These stresses include membrane perturbation, oxidative DNA damage, decrease in DNA repair proteins, mitotic stress, micronuclei formation, mitochondrial damage (metabolic stress) and endoplasmic reticulum (ER) stress (Figure 2).
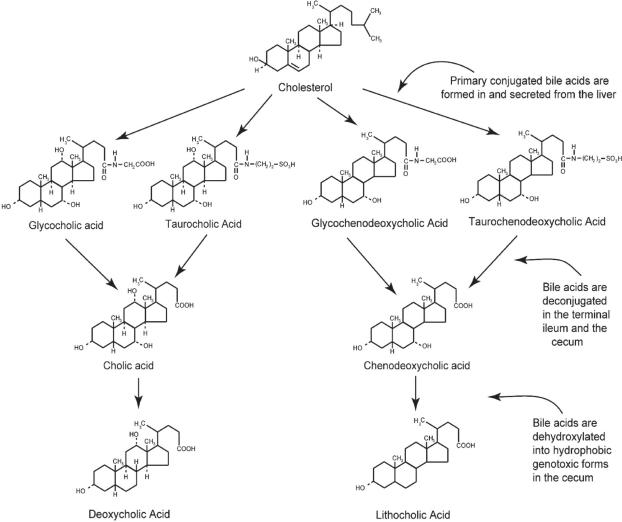
Figure 1.
The primary bile acids, cholic acid and chenodeoxycholic acid, are formed in the liver from cholesterol and conjugated, usually with glycine or taurine. They are then secreted into the gall bladder. From the gall bladder, glyco-and tauro- conjugated primary bile acids are released into the duodenum to aid in the digestion of fats. In the terminal ileum and in the cecum, the primary bile acids are deconjugated. Further, in the cecum, they are dehydroxylated through the action of bacterial 7-alpha dehydroxylases. By this reaction, cholic acid and chenodeoxycholic acid are converted to the more genotoxic-hydrophobic bile acids deoxycholic acid and lithocholic acid, respectively.
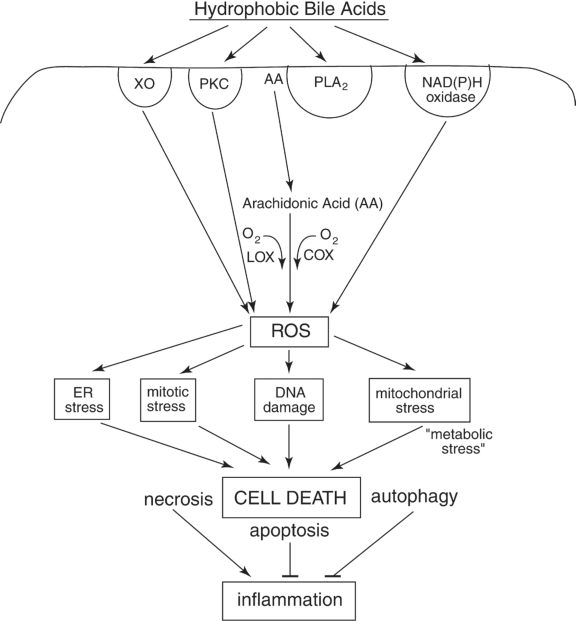
Figure 2.
Schematic diagram indicating some of the hydrophobic bile acid (HBA)-induced signaling pathways that begin with the activation of surface enzymes, the subsequent generation of ROS, and the resultant stresses/damages that, if excessive, result in cell death. The most well documented deleterious effects of HBAs in colon cells are DNA damage, mitochondrial stress and ER stress. DNA damage results in the activation of mitotic checkpoint proteins leading to growth arrest. Mitochondrial damage results in the activation of caspases and the cleavage of multiple substrates in the cell. ER stress activates several pro-apoptotic molecules that result in cell death. Repair processes responsive to DNA damage, mitochondrial stress and ER stress can deplete the energy reserves of the cell, resulting in “metabolic stress”. Too much stress on the cell results in cell death through mechanisms that involve apoptosis, necrosis, and/or autophagy. HBA-induced mitotic stress can lead to abnormal cell division. Excessive mitotic stress may lead to mitotic catastrophe and cell death, although the details of this mode of cell death have not been well described. Necrotic cells induced by HBA may, especially, elicit an inflammatory response in vivo, and this could explain, in part, the induction of colitis by chronic feeding of HBA in mouse models. See text for details.
Abbreviations: AA, arachidonic acid; COX, cyclooxygenase; PKC, protein kinase C; PLA2, phospholipase A2; LOX, lipoxygenase; ROS, reactive oxygen species; XO, xanthine oxidase.
Although excessive stress will lead to apoptosis, this altruistic act of removing DNA-damaged cells for the survival of the organism becomes largely circumvented in cells with activated survival pathways. In this “war zone”,1 stimulated by the presence of hydrophobic bile acids, mutated epithelial cells may act in a selfish manner,2 like bacterial cells, and Darwinian selection3–10 can come into play at the cellular level. The very stress-response pathways that result in cell survival (eg, activation of nuclear factor-kappa B [NF-κB], autophagy) (Figure 3) can become constitutively upregulated through clonal selection of mutated cells, and can serve to propagate cells even with unrepaired DNA damages and genomic instability. Although it has been known for decades (using animal models of carcinogenesis) that hydrophobic bile acids can promote colon cancer,11–14 the evidence is now convincing that bile acids are also carcinogens.15–17 Evidence that bile acids are carcinogens in human gastrointestinal (GI) cancers has been reviewed by Bernstein and colleagues.15 Proof that hydrophobic bile acids cause mutation in vitro in epithelial cells of the GI tract was provided by the elegant studies of Jenkins and colleagues.16,17 Even though proliferation is essential to cancer development, it is not the high level of proliferation itself that is responsible for cancer. For example, adenocarcinomas are infrequent in the small intestine, although the proliferation rate of epithelial cells at this site is similar to that of the epithelial cells of the colon. The colon may be particularly susceptible to cancer because it contains tissues that are often exposed to unique stresses (eg, high levels of hydrophobic bile acids resulting from anaerobic bacterial metabolism). Since there may be insufficient time for DNA repair in this “war zone”,18 the selection of mutated cells with a proliferative advantage may occur repeatedly. This proliferative advantage may take the form of apoptosis resistance and/or increased cell division.
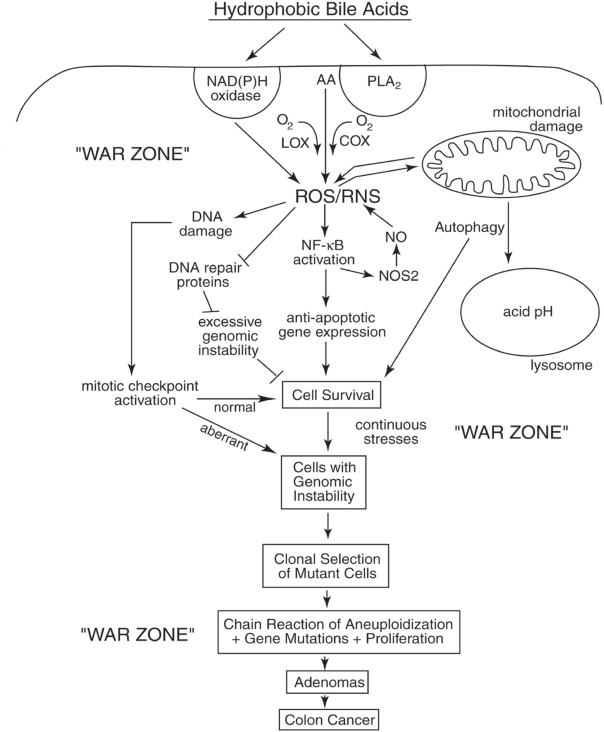
Figure 3.
Schematic diagram illustrating a probable sequence of events resulting from exposure to high levels of hydrophobic bile acids (HBAs) that accompany a high-fat diet. This exposure leads to HBA-induced generation of ROS/RNS, activation of survival pathways (eg, autophagy, NF-κB), the generation of cells with genomic instability (eg, mutations, aneuploidy) and clonal selection of mutant cells with survival and proliferative advantages. The end results are the production of adenomas that progress to colon cancer. The epithelial cells of the colon of a person on a Western-style (high-fat/low vegetable/low micronutrient) diet are probably in a persistant “war zone” (bombardment with HBA-induced ROS/RNS, presence of food carcinogens, toxins, etc.). Cells in different stages of progression to malignancy are thus persistently receiving damages to their genome, resulting in clones of cells that are selected for survival in the adverse environment of the colon. While cells in the previous population, if receiving excessive DNA damage, underwent cell death altruistically for the overall benefit of the organism, the new clones of cells may behave selfishly. The new clones may acquire resistance to apoptosis and undergo clonal selection on the basis of their survival advantage, even when their DNA is damaged and after their genomes become unstable. This allows progression to adenomas and colon cancer.
Abbreviations: AA, arachidonic acid; COX, cyclooxygenase; LOX, lipoxygenase; PLA2, phospholipiase A2; NO, nitric oxide; ROS, reactive oxygen species; RNS, reactive nitrogen species.
How, in particular, do bile acids contribute to colon carcinogenesis? Since somatic mutation rates are normally too low to account for the high incidence of colon cancers, it has been proposed that genomic instability (ie, a mutator phenotype) plays a major role in producing the plethora of random genomic changes needed to produce a malignancy.19–21 A mutator phenotype can explain the complex histopathology and behavior of tumors,22–24 including different tumor subsets,25 divergence of clonal populations,26 and different molecular pathways27,28 that may be selected during colon cancer progression and lead to different clinical outcomes.29,30 Genomic instability in cancer appears in three major forms: (i) aneuploidy, in which entire chromosomes are gained or lost, (ii) intrachromosomal instability, characterized by insertions, deletions, translocations, amplifications, and other forms all sharing the feature of utilizing DNA breakage as an early step, and (iii) point or oligobase mutations.31 Genomic instability is a dynamic process in which the three major forms of genetic instability may co-exist at different stages of colon carcinogenesis, providing support for the chaotic roots of neoplastic development and continuous growth of aberrant clones.32,33
Mutations, aneuploidy or other chromosomal aberrations affecting genes that govern genetic stability5,34–37 could be the cause of a mutator phenotype.38,39 The occurrence of a mutator phenotype would be made more likely by conditions that cause DNA damage, such as increased oxidative stress, due to persistent exposure to high levels of bile acids that accompanies a high-fat diet. Increased levels of oxidative stress caused by defective mitochondria have been proposed as a possible mechanism by which a “mutator phenotype” could arise.40 As discussed below, bile acids cause oxidative DNA damage, double-strand DNA breaks (evidenced by activation of phosphorylated H2AX41), aneuploidy,16,17 broken chromosomes16,17 and aberrant mitoses (unpublished data from our laboratory). In addition, high concentrations of bile acids in the lumen provide a persistent stressful environment that may select for mutated cells that can better survive in that environment (Figure 3). Hydrophobic bile acids thus may serve to both initiate and propagate the mutator phenotype. Cycles of aneuploidization42 coupled with specific mutations (eg, APC, ras, p53) and large-scale structural alterations to chromosomes (eg, nonreciprocal translocations, inversions, deletions and insertions resulting in “genome scrambling”43) may then give rise to a population of cells with multiple karyotypes and genotypes. The clonal selection of cells with particular karyotypes and/or mutations may then produce a “field defect” or “field cancerization” associated with gene expression changes. Frequently, one of the selected genotypes will be apoptosis resistance as found in the normal-appearing flat colonic mucosa of humans44,45 which is by definition a “survival genotype”. Within such a field defect, further mutations may then be selected for increased proliferation, altered cell-cell interactions and/or migratory behavior. On this basis a neoplastic clone can emerge (eg, as found in flat adenomas and polyps). These neoplastic survival karyotypes may undergo further mutation and selection to produce a cancer.
This review emphasizes the plethora of cellular stresses imposed by hydrophobic bile acids. Our review provides possible mechanisms by which a high-fat diet can lead to the development of sporadic colon cancers. The roles of mutations in critical genes (eg, APC, ras, p53) during colon carcinogenesis was previously addressed in the insightful discussions by the Vogelstein group46–48 and will not be discussed in this review.
Hydrophobic bile acids generate cellular reactive oxygen species (ROS) and reactive nitrogen species (RNS) that damage DNA and proteins. These damages can lead to genomic instability (eg, point mutations and aneuploidy).49 Point mutations, aneuploidy, and other gross structural alterations of chromosomes (eg, balanced or unbalanced translocations, dicentric chromosomes) appear to interact to produce the unstable and/or aberrant genomes associated with colon carcinogenesis. Aneuploidy is observed in most cancers, although it is not ordinarily found in colonic tumors that have a mismatch repair deficiency and display microsatellite instability.50 A small subset of colorectal tumors display microsatellite instability, whereas the rest of colorectal tumors display numerical and/or structural chromosomal alterations as the most prominent outcome of genetic disruption.51
Loss of heterozygosity (LOH) of a portion of a chromosome or a whole chromosome is already evident at the adenoma stage of colon carcinogenesis52–65 and is prominent at the colon cancer stage.59,60,62,63,66–68 A review of the literature indicates that at least 19 of the 22 human somatic chromosomes exhibit aneuploidy during the adenoma to colon cancer stage. The loss or gain of whole or portions of chromosomes involves thousands of genes.69 This chromosomal imbalance could have dramatic effects on cellular homeostasis, including an increase in oxidative/redox state of the cell and the loss of key DNA repair and pro-apoptotic proteins. Approximately 11,000 genomic events were identified in sporadic colonic polyps using the technique of inter-(simple sequence repeat) polymerase chain reaction (PCR), indicating that genomic destabilization is an early event in sporadic tumor development.31
Pre-neoplastic lesions smaller than adenomas are referred to as aberrant crypt foci (ACF).70,71 The ACF are not identified during routine colonoscopies since vital stains are often not used and biopsies are not usually taken of the flat colonic mucosa. However, human ACF have unequivocally been shown to exhibit genomic instability, including LOH.72–76 Since numerous genetic alterations and aberrant mitotic changes are also observed in the nonneoplastic colon of mice77 and humans78–82 at high risk for colon cancer, biomarkers based on aberrant karyotypes have promise for identifying high-risk individuals. Figures 2 and 3 are based on data from our laboratory and those of others and emphasize the role of hydrophobic bile acids in the induction of multiple stresses and genomic instability. We have selected bile acids as a focus of this review since bile acids play a key role in colon carcinogenesis. We describe how bile acid-induced stresses cause cell death in susceptible cells, contribute to genomic instability in surviving cells, impose Darwinian selection on survivors in a “war zone” and enhance initiation and progression to colon cancer. This review provides a mechanistic explanation for the important link between a Western-style diet and associated increased levels of colon cancer. However, other factors such as smoking, food-related carcinogens, long-chain fatty acids (eg, palmitic acid), excess iron, arsenic, viruses, bacteria, inflammatory cells, low dietary micronutrients, low omega-3 fatty acids, etc., may also contribute to genomic instability-based carcinogenesis.
Bile acids cause membrane perturbation
It has been known for over thirty years that bile salts perturb plasma membranes, and that deoxycholate is more membrane-damaging than cholate and its conjugates.83 Binding of bile acids with rat colon and the resultant perturbation of membrane organization was later measured using 31P nuclear resonance spectroscopy.84 Deoxycholate alters membrane composition, as evidenced by a significant upregulation in membrane cholesterol and phospholipids.85 Bile acids also induce the modification and upregulation of caveolin-1 in a hydrophobicity-dependent manner, implying widespread receptor dysregulation.85 These findings were mimicked by other hydrophobic molecules unrelated to bile acids, such as sodium lauryl sarcosine and cholesteryl hemisuccinate, strongly implicating hydrophobicity as an important determinant of bile acid effects on signaling.85 Deoxycholate also perturbs membrane structure by alterating membrane microdomains.86 Depletion of membrane cholesterol by treating cells with methyl-β-cyclodextrin suppressed deoxycholate-induced apoptosis, and staining for cholesterol with filipin showed that deoxycholate caused a marked rearrangement of this lipid in the membrane.86 Additionally, fluorescence anisotropy studies indicated that deoxycholate caused a decrease in membrane fluidity consistent with the increase in membrane cholesterol content measured after deoxycholate treatment.86
Bile acids activate surface enzymes, such as phospholipase A2 (PLA2),87,88 epidermal growth factor receptor (EGFR),86,89–94 Fas receptor,89 protein kinase C (PKC),90 bile acid receptor M-BAR/TGR5 (G-protein-coupled receptor),93 phospholipase C (PLC),95 nicotinamide adenine dinucleotide phosphate (NAD(P)H) oxidase96,97 and the Na+/K+ -ATPase.97 We found that the surface enzymes, Na+/K+ -ATPase and NAD(P)H oxidase are involved in activating NF-κB,97 a redox-sensitive transcription factor upregulated during the development of apoptosis resistance in colon cancer cells,98 and associated with colon carcinogenesis. The activation of surface and other enzymes through membrane perturbation is, therefore, responsible for modulating signal-transduction pathways in colon cells.85,86,89,99
The fact that hydrophobic bile acids cause secretory diarrhea (diarrhea is the most common and debilitating symptom of patients with ulcerative colitis [a pre-neoplastic condition of the colon]), warrants a greater effort to understand how these bile acids induce changes in ion transport.100 Taurodeoxycholic acid, for example, increases electrogenic Cl− secretion into the proximal colonic lumen.101 This stimulated secretion is believed to occur via an increase in intracellular Ca++ concentration. Deoxycholic acid reduces transepithelial Na+ absorption by inhibiting amiloride-sensitive Na+ channels, and increasing secretion of Na+, K+, and Cl− ions in the distal colon.102 Hydrophobic bile acids have been reported to act as calcium ionophores,103,104 most probably by partitioning bile acid/Ca++ complexes into the hydrophobic core of the membrane bilayer104 and/or Ca++-transporting channels.105 We have recently reported that high cytosolic Ca++ levels are a major factor in the activation of NF-κB in HCT-116 colon cells by bile acids.97 Two agents (EGTA and Ruthenium Red (RuR)) used in that study prevented NF-κB activation by bile acids through at least two different mechanisms. EGTA does not cross cell membranes and chelates Ca++ ions in the extracellular milieu, thereby preventing Ca++ entry into the cell. RuR is an inorganic polycationic dye that inhibits Ca++ influx through voltage-sensitive calcium channels and blocks the release of Ca++ from the ER.
Bile acids cause oxidative and nitrosative stress in colon cells
Bile acids induce production of ROS and RNS in colon epithelial cells.7,106–117 Bile acids generate ROS and RNS by several different pathways, as reviewed by Bernstein and colleagues15 and summarized here. Bile acids damage mitochondria and damaged mitochondria can “leak” electrons from the electron transport chain to form superoxide (O2−). ROS can also be generated by bile acid activation of PLA2, which releases arachidonic acid from the cell membrane. Arachidonic acid may then be acted on by cyclooxygenase and lipooxygenase to release ROS through partial reduction of O2, during the conversion of arachidonic acid to prostaglandins and leukotrienes.
Deoxycholate activates the redox-sensitive transcription factor NF-κB in colon epithelial cells.97–99,110,118–124 When NF-κB is activated it can induce an increase in nitric oxide synthase 2 (NOS2), an enzyme that can generate micromolar quantities of NO. We have found that repeated exposure of HCT-116 colon epithelial cells to increasing concentrations of deoxycholate results in the consistent upregulation of S-nitrosylated proteins. Some of these post-translationally modified proteins were identified by mass fingerprinting analysis and found to include cytoskeletal proteins, metabolic enzymes, signaling proteins, chaperones, redox-related proteins and differentiation-related proteins.114 We also found that feeding mice deoxycholate can induce inflammation in a mouse model,115 and that the inflammation is mediated in large part by RNS, since colitis is markedly attenuated in NOS2 knockout mice.116
DNA repair proteins can be modulated by oxidative stress and deoxycholate
The promoting and mutagenic effects of bile acids can be explained, in part, by a decrease in DNA repair proteins. Since oxidative stress is known to reduce expression of DNA repair proteins, bile acid-induced decreases in DNA repair proteins may also be through an oxidative mechanism. The cell types and experimental conditions in which oxidative stress decreases DNA repair proteins, and bile acids decrease proteins involved in 6 different DNA repair pathways are described below. Since DNA repair proteins often have multiple functions in the cell in addition to DNA repair, such as participation in apoptosis125 and regulation of mitosis126,127 and checkpoint pathways,128 a deleterious effect of bile acids on DNA repair protein expression can have important and varied implications for inducing genomic instability.
Oxidative stress causes a decrease in DNA repair proteins
Langie and colleagues,129 working with human pulmonary epithelial cells A549, showed that oxidative stress caused a reduction in mRNA for the nucleotide excision repair (NER) enzyme ERCC1 as well as a reduction in NER capacity that correlated with level of ERCC1 mRNA. Feng and colleagues,130 using the human colon epithelial cell line HCT-116 and the human hepatic fetal epithelial line CL-48, showed by in vitro DNA repair synthesis and host cell reactivation assays, that ROS also inhibit NER through production of malondialdehyde. This effect is likely mediated by malondialdehye-induced inactivation of DNA repair proteins. Furthermore, as shown in the human erythroleukemia HEL cell line, oxidative stress inactivates the mismatch DNA repair proteins MSH6 and Pms2 (but not MSH2 or MLH1).131 In HeLa cells and the human Boleth lymphoblastoid cell line, the base excision repair enzyme OGG1 is degraded or reversibly inactivated following oxidative stress.132,133
In mitochondria, DNA polymerase γ is responsible for both mtDNA replication and repair. In an SV40-transformed human fibroblast cell line, the functional ability of the catalytic p140 subunit of DNA polymerase γ is reduced by oxidative stress.134
Bile acids decrease DNA repair proteins
Short term exposure to deoxycholate decreases the expresson of the DNA repair proteins p53 and BRCA1
The enzyme p53 has multiple roles in the cell. In particular, it has roles in 6 different DNA repair pathways: (1) homologous recombinational repair, (2) nonhomologous end joining, (3) base excision repair, (4) nucleotide excision repair, (5) mismatch repair, and (6) transactivation of expression of O6-methyl-guanine-DNA-methyl-transferase for direct repair of alkyl adducts at the O6-methyl-guanine position.125,135 While treatment of colon HCT-116 cells with non-cytotoxic (200 μM) deoxycholate concentrations increases mRNA expression of the p53 gene, the level of p53 protein is decreased by 40% through proteasome-mediated degradation.136 Although mutations in p53 are a late event in colon carcinogenesis, the persistent decrease in p53 function by deoxycholate may have similar consequences as that of a mutation. Work from our group has also shown that deoxycholate decreases BRCA1 at the protein and mRNA levels.137 BRCA1 has a key role in recombinational repair of DNA damages, and also regulates the function of the centrosomes which carry out mitotic chromosome segregation through spindle organization.127 Failure to adequately repair DNA damages can lead to replication errors and thus mutation; and failure of mitotic segregation can lead to aneuploidy. Since BRCA1 expression is also reduced in colon adenocarcinomas,137 it is possible that the modulation of expression of DNA repair enzymes by dietary-related factors may, in part, be responsible.
Long term exposure of colon cells to deoxycholate selects for decreased protein or mRNA expression of DNA repair proteins
Three HCT-116 colon cell lines were developed in our laboratory for stable resistance to bile acids by repeated exposure to increasing concentrations of deoxycholate over a period of approximately 40 weeks.98 These cell lines were found to have constitutively decreased protein expression of the DNA repair enzymes MSH3 (mismatch repair), Ku80 (non-homologous end joining), P36/MAT1 (nucleotide excision repair) and XPA (nucleotide excision repair).138 These cell lines were also found to have decreased expression at the mRNA level for ATM (homologous recombinational repair), MSH3 (mismatch repair) and XRCC4 (non-homologous end joining).98
Exposure of cholangiocytes to glycochenodeoxycholate (GCDA) decreases DNA repair enzymes
Komichi and colleagues139 exposed immortalized mouse cholangiocytes to 200 μM glycochenodeoxycholate (GCDC) for 4 weeks and then performed a microarray gene expression analysis. mRNA expression of OGG1 and MUTYH was downregulated 0.8-fold after exposure to CGDC compared to control cells.
Bile acids cause DNA damage in colon cells
Bile acids induce DNA damage in cells of the colon.41,90,99,108,109,115,118,140–143 One type of induced damage in colon cells is oxidative DNA damage,90,108,115,143 suggesting that oxidative stress is a significant cause of the overall bile acid-induced DNA damage in the colon. Exposure of rat colon ex vivo with high physiologic concentrations of bile acids resulted in the activation of poly(ADP-ribose) polymerase (PARP).99 PARP is activated by DNA strand breaks and attaches a polymer of ADP-ribose units to proteins which modulates their functional activity and assists in the opening up of the chromatin to allow for more efficient DNA repair.144 It is probable that the increase in DNA strand breaks is mediated, in part, through the bile acid-induced increase in ROS and/or RNS. Treatment of human colonic biopsies ex vivo with deoxycholate also resulted in the activation of phosphorylated H2AX,41 implying the generation of double-strand breaks in DNA.145 Double-strand breaks can cause chromosome breakage with the generation of micronuclei upon exit from mitosis.16,17 Increased DNA damage may be caused by the bile acid-induced decrease in DNA repair enzymes (see Bile acids decrease DNA repair proteins, above).
Bile acids cause mutation
The repeated observation that bile acids cause DNA damage in colon cells suggests that bile acids also increase the incidence of mutation, since replication of a damaged DNA template strand frequently results in a replication mistake, and thus a mutation. Bile acids have been shown to induce mutation in esophageal cells in culture and in vivo in rat esophageal cells. When cultured esophageal cells were treated with deoxycholate, an increase in the frequency of GC to AT mutations in the p53 gene was observed.16 In other experiments, Big Blue F1lacI transgenic rats were subjected to esophagoduodenostomy, a surgical procedure that increases duodenogastroesophageal reflux.146 In these surgically altered rats, the frequency of mutant lacI cells of the esophageal mucosa was significantly greater than in the control rats that were not subjected to surgery. Thus, components of refluxate, such as bile acids, appear to cause mutation. Forty-six percent of the mutant esophageal cells were altered at CpG dinucleotide sites, and most of these mutations (61%) were C to T or G to A transitions. This pattern of mutation seen in the surgical model approximates that seen in human esophageal adenocarcinoma, suggesting that duodenogastroesophageal reflux is carcinogenic. In surgically treated Big Blue mice (rather than rats) that were altered to increase duodenogastroesophageal reflux, increased mutagenesis was also observed.147 In another rat model of colon carcinogenesis based on exposure to azoxy-methane, deoxycholate increased both the incidence of colon tumors and the incidence of tumors with K-ras mutations.148 This finding suggests that deoxycholate may induce K-ras mutations. In a sensitive bacterial flucturation test based on the Ames Salmonella test system, bile acids were also found to be mutagenic.149
Hydrophobic bile acids induce aneuploidy and micronuclei formation
The induction of aneuploidy by deoxycholate was first reported 26 years ago by Assinder and Uphall150 in a heterozygous diploid strain of Aspergillus nidulans. The detection of aneuploidy was made possible by the unique orientation of mutations on the chromosomes. Each pair of homologues carried mutations, in repulsion, in at least 2 gene loci, those in linkage groups I, III, IV, and V being on opposite sides of the centromere. This orientation allowed a distinction between cross-over and nondisjunctional segregants (aneuploids). It was suggested that aneuploidy was a result of interference with the normal functioning of the mitotic apparatus through disruption of the nuclear membrane. Ferguson and Parry151 confirmed that bile acids, including deoxycholate, were potent inducers of aneuploidy using the diploid yeast, Saccharomyces cerevisiae. The assay used to detect mitotic chromosome aneuploidy involved scoring of white cycloheximide resistant monosomic colonies. Albertini and colleagues152 determined that lithocholic acid induces mitotic chromosome loss in the diploid yeast strain, S. cerevisiae D61.M. The assay to detect mitotic chromosome loss involved scoring of segregants expressing three linked recessive markers, two of which were located close to the centromere on opposite arms of chromosome VII.
The evaluation of the induction of aneuploidy by bile salts in eukaryotic cells of higher organisms was neglected for almost 20 years, even though a role of bile acids in colon carcinogenesis was established in animal models in the 1970s. The first study of bile acid-induced aneuploidy in cells of multicellular organisms in vitro was reported by Jenkins and colleagues16 using human esophageal cell lines. In this elegant study, the frequency of deoxycholate-induced kinetocore-positive and -negative micronuclei was assessed as a measure of the level of aneuploidy, using the cytokinesis-block micro-nucleus assay.153,154 The rationale for this assay is based on data which indicate that aneuploid cells and cells with broken chromosomes will often produce micronuclei after exit from mitosis. The presence of kinetochore-negative micronuclei indicated that chromosome fragmentation occurred, probably as a result of deoxycholate-induced double strand breaks. The deoxycholate-induced increase in micronuclei was significantly reduced by the antioxidant, vitamin C, indicating the importance of diet in the possible prevention of genotoxicity. Unpublished data from our laboratory also indicate that deoxycholate can induce micronuclei in colon epithelial cells in vitro in addition to inducing aberrant mitoses (implying perturbation of the mitotic machinery). These unpublished results are consistent with published data from our laboratory that persistent exposure of cells to deoxycholate in vitro98,138 or in vivo in an animal model,115 overall, results in the modulation of genes/proteins involved in chromosome maintenance and cell cycle progression (see Persistent exposure of colon cells to deoxycholate results in the modulation of expression of chromosomal maintenance/mitosis-related genes, below). These findings indicate that deoxycholate is a true carcinogen,15 with important implications for dietary intervention strategies to prevent the initiation and/or progression of GI cancer.
A major stress induced by hydrophobic bile acids is the generation of ROS/RNS. Chronic exposure of cells to oxidative stress has been reported to result in increased genomic instability.155,156 An important source of ROS is damaged/mutated mitochondria.40,157 The mechanisms by which oxidative stress can lead to genomic instability are probably multifactorial, and include induction of double-strand breaks (resulting in broken chromosomes) and overriding the spindle checkpoint during cell cycle progression,158 which can result in aneuploidy.
Persistent exposure of colon cells to deoxycholate results in the modulation of expression of chromosomal maintenance/mitosis-related genes
The induction of aneuploidy with the formation of micronuclei after cells exit mitosis may be caused by a number of defects during cell division. Defects such as insufficient or excessive sister chromatid cohesion,159,160 kinetochore-microtubule attachment defects,161,162 failure of DNA damage checkpoints,163 failure of spindle checkpoints,164–166 telomere defects167,168 and aberrant centrosomal organization and multiplicity169,170 may result in an incorrect number of chromosomes in daughter cells after mitosis. These defects coupled with chromosome breaks can produce an aberrant karyotype that may be carcinogenic. We, therefore, searched for the aberrant expression of mitosis-related genes in colon epithelial cells and colonic tissues that were modulated by deoxycholate. cDNA microarray (Table 1) and proteomic analyses (Table 2) of HCT-116 cell lines persistently exposed to increasing concentrations of deoxycholate in vitro98,138 and cDNA microarray analysis of colonic tissue of wild-type mice receiving dietary supplementation with deoxycholate in vivo115 (Table 3) indicate that deoxycholate modulates the expression of numerous genes associated with mitosis and chromosome maintenance. The list includes 71 genes/proteins associated with cohesion and segregation of chromosomes, telomeres, kinetochore structure, spindle assemby and function, centrosome regulation, various aspects of the cell cycle (metaphase, anaphase), cell cycle progression (kinases, phosphatases, ubiquitin-conjugating enzymes), cell cycle checkpoints and cytokinesis (Tables 1–3).
Table 1.
Fold reduction/induction of mRNA levels of chromosomal maintenance/mitosis related genes in HCT-116 colon cancer cells persistently exposed to deoxycholate
| Systematic gene notation | Fold reduction/induction | Gene and function |
|---|---|---|
| AA479781 | 0.32 | Radixin (highly concentrated in the cleavage furrow during the late stages of mitosis) |
| H24707 | 0.35 | DLG1 (Discs, large (Drosophila) homolog 1 found in the midzone during mitosis); multidomain scaffolding protein; interacts through its guanylate kinase-like domain with KIF13B (kinesin family member 13B) |
| N69204 | 0.36 | CSE1L (chromosome segregation 1 (yeast homolog)-like); microtubule-associated protein that functions in the mitotic spindle checkpoint; also associates with chromatin and increases the transcription of select p53 target genes, including the PIG3 gene; Ran-binding protein implicated in the nuclear to cytoplasmic reshuffling of importin α, which is necessary for the nuclear transport of several proliferation activating proteins, transcription factors, oncogene and tumor suppressor genes |
| H23021 | 0.50 | RBBP6 (retinoblastoma binding protein 6); phosphorylated upon DNA damage; co-localizes with mitotic chromosomes and localizes to nucleoli in interpase cells; binds p53 and Rb1; overexpression restricts mitotic progression at prometaphase and promotes mitotic apoptosis |
| H84048 | 0.50 | RBL1 (retinoblastoma-like 1 (p107); key regulator of entry into cell division; directly involved in heterochromatin formation by maintaining overall chromatin structure; controls histone H4 ‘lys-20’ trimethylation; interacts with transcription factor E2F-4 and with cyclin E/A-CDK2; inhibits cell cycle progression in response to DNA damage in S phase cells; inhibits G1 to S phase progression by downregulating expression of the F-box protein SKp2; reduced expression in colorectal tumors may indicate a poor prognosis |
| AA490213 | 0.51 | TOB1 (Transducer of p185ERBB2 1 receptor tyrosine kinase); antiproliferative protein; acts as a transcriptional corepressor and suppresses the promoter activity of the cyclin D1 gene through an interaction with histone deacetylase; mice lacking TOB are predisposed to cancer indicating that TOB is a tumor suppressor |
| R02820 | 0.52 | PDS5A [regulator of cohesion maintenance, homolog A (S. cerevisiae)]; component of the molecular glue along with cohesin that maintains sister chromatid cohesion during S phase and maintains it during metaphase; sumoylated at anaphase to promote chromatid separation |
| R92435 | 0.54 | NUF2R (NDC80 subunit of the outer kinetochore complex component, homolog); localizes to kinetochores from late prophase to anaphase; the NDC80 complex is required for chromosome segregation, spindle checkpoint activity, kinetochore integrity and the organization of stable microtubule binding sites in the outer plate of the kinetochore |
| AA428749 | 0.56 | PPP1R2 [protein phosphatase 1, regulatory (inhibitor) subunit 2]; alias: PNUTS; serine/threonine phosphatase involved in exit from mitosis and targeted to the reforming nuclei in telophase following the assembly of nuclear membranes and enhances chromosome decondensation as cells re-enter interphase |
| AA448676 | 0.59 | UBE2V2 (Ubiquitin-conjugating enzyme E2 variant 2; facilitates progress through the cell cycle but has no ubiquitin ligase activity on its own); plays a role in error-free DNA repair pathway and contributes to the survival of cells after DNA damage |
| R66447 | 0.60 | MYCN (V-myc avian myelocytomatosis viral related oncogene, neuroblastoma-derived); involved in centrosome amplification |
| N54344 | 0.60 | NCAPH (non-SMC condensin 1 complex, subunit H); regulatory subunit of the condensin complex which is required for the conversion of interphase chromatin into mitotic-like condensed chromosomes; the condensin complex introduces positive supercoils into relaxed DNA in the presence of type 1 topoisomerases and converts nicked DNA into positive knotted forms in the presence of type II topoisomerases; the condensin 1 complex is required not only to promote sister chromatid resolution but also to maintain the structural integrity of centromeric heterochromatin during mitosis; in NCAPH-depleted cells the pericentromeric and centromeric heterochromatin cannot withstand the forces exerted by the mitotic spindle and undergoes irreversible distortion; highest expression occurs during the G2 phase of the cell cycle |
| AA489007 | 0.60 | Aurora borealis (BORA) (binding partner to Aurora A); in interphase cells, Bora is located in the nucleus, but upon entry into mitosis, bora translocates to the cytoplasm (in a cdc2-dependent manner) where it binds to and activates the protein kinase Aurora A. The activation of Aurora A is necessary for centrosome maturation, spindle assembly, and asymmetric protein localization during mitosis |
| AA608568 | 0.61 | CCNA2 (Cyclin A2); major regulator of cell cycle progression; its synthesis is required for progression to S phase; regulates nuclear envelope breakdown and the nuclear accumulation of cyclin B1; also synthesized during G2/M transition; associated with cyclin-dependent protein kinases 1 and 2; cyclin A2 levels decrease as a result of p53-dependent G2 arrest |
| AA262211 | 0.63 | DLG7 discs, large homolog 7; HURP (Hepatoma Up-Regulated Protein); mitotic phosphoprotein mediates Ran-GTP-dependent assembly of the bipolar spindle, allows for efficient kinetochore capture at prometaphase, promotes chromosome congression to the metaphase plate, chromosome alignment at the metaphase plate and proper interkinetochore tension for anaphase initiation during mitosis; binds to microtubules through its N-terminal domain which hyperstabilizes spindle microtubules in the vicinity of chromosomes by forming specialized tubulin configurations to form a sheet that wraps microtubule ends; interacts with CDC2 which localizes to the spindle poles in mitotic cells; co-localizes with CDH1 at sites of cell-cell contact in intestinal epithelial cells; phosphorylated by Aurora A which provides a regulatory mechanism for the control of spindle assembly and function; increased expression in the G2/M phase of the cell cycle |
| R88741 | 0.67 | RBBP8 (retinoblastoma-binding protein 8); tumor suppressor that interacts with CTBP, with the terminal (BRCT) domain of BRCA1, and with the retinoblastoma protein to regulate the G1/S-phase transition of the cell cycle; confers resistance to double-strand break-inducing agents and is recruited to double strand breaks exclusively in S and G2 cell cycle phases |
| AA448194 | 1.25 | SMN2 (Survival of motor neuron 2, centromeric); function not known at the present time |
| T90375 | 1.26 | KIF3B (microtubule plus end-directed kinesin motor family member 3B); forms a heterodimer with KIF3A which interacts with SMC3 subunit of the cohesin complex; involved in tethering the chromosomes to the spindle pole and in chromosome movement through plus end-directed microtubule sliding activity in vitro; prevents aneuploidy and abnormal spindle formation |
| AA598887 | 1.27 | SMC1L1 [SMC1 (structural maintenance of chromosomes 1, yeast)-like 1]; a cohesin subunit that is a central component of the cohesin complex required for the cohesion of sister chromatids after DNA replication; forms a heterodimer with SMC3 in cohesin complexes consisting of CDCA5, RAD21, PDS5A/APRIN and PDS5B/SCC-112; before prophase it is scattered along chromosome arms; during prophase, most of the cohesin complexes dissociate from chromosomes except at centromeres where cohesin complexes remain forming part of the kinetochore; phosphorylation of Ser-957 and Ser-966 activates it and is required for S-phase checkpoint activation; role in spindle pole assembly during mitosis; SMC1L1 localizes to centrosomes throughout the cell cycle where it is involved in the organization of dynamic arrays of microtubules |
| R74078 | 1.27 | TNKS1BP1 [182 kDa tankyrase 1 binding protein 1 that binds to the ankyrin domain (comprises 24 ankyrin repeats) of TNKS1]; serves as an acceptor of poly(ADP-ribosylation) by tankyrase 1 (telomeric PARP); tankyrase 1-mediated polymerization of poly(ADP-ribose) is required for spindle structure and function; TNKS1BP1 co-localizes with chromosomes during mitosis, is phosphorylated upon DNA damage and binds to cytosolic actin in interphase |
| N68492 | 1.29 | Anaphase-promoting complex (APC) 1; alias is MCPR (meiotic checkpoint regulator); one of the 12 subunits that make up the APC/C, a 20S ubiquitin-ligase complex that targets proteins for proteasomal destruction, an essential step in chromatid separation that is necessary for the metaphase to anaphase transition; phosphorylation of Ser-355 occurs specifically during mitosis; |
| T75522 | 1.32 | PPP1R10 (Protein Phosphatase 1, regulatory subunit 10); serine/threonine – protein phosphatase involved in chromosome decondensation in a PP1-dependent manner; alias PNUTS; inhibits PPP1CA and PPP1CC phosphatase activities; phosphorylated on Ser-398 and Thr-400 by PKA within the region necessary for interaction with PPP1CA; found in discrete nucleoplasmic bodies and within nucleoli |
| W95346 | 1.34 | SAC3D1 (Saccharomyces cerevisiae 3 domain-containing 1 protein); involved in centrosome duplication and mitotic progression (by similarity); localizes to centrosomes in interphase cells and at spindle poles and mitotic spindles at M phase, similar to α-tubulin; functions in the spindle assembly checkpoint; RNA interference suppression of endogenous SAC3D1 causes defects in centrosome duplication and spindle formation resulting in cells with a single centrosome and downregulated Mad2 expression, generating increased micronuclei; increased expression of SAC3D1 by DNA transfection resulted in cells with multiple centrosomes and deregulated spindle assembly with upregulated Mad2 expression until anaphase, generating polyploid cells |
| AA486312 | 1.34 | CDK4 (cyclin-dependent kinase 4); forms a stable complex with D-type G1 cyclins involved in regulating G1 to S transition; inhibition of CDK4 results in a mitotic delay associated with elevated Wee1 (mitotic delay results from failure of chromosomes to migrate to the metaphase plate; however, cells eventually exit from mitosis, resulting in an increase in cells with multiple or micronuclei); shows aberrant cytological localization in colorectal epithelia in the usual adenoma carcinoma sequence |
| H18838 | 1.34 | MAB21l1 [Mab-21 (C. elegans)-like 1]; meiotic instability associated with the CAGR1 trinucleotide repeat at 13q13 |
| W02403 | 1.36 | CLSPN (Claspin homolog of Xenopus laevis); required for checkpoint-mediated cell cycle arrest in response to inhibition of DNA relication or to DNA damage; acts as a sensor which monitors the integrity of DNA relication forks; expression peaks at S and G2 phases of the cell cycle; phosphorylated in response to replication stress and this phosphorylation is required for its association with Chk1 |
| R21614 | 1.36 | SUPT5H [Suppressor of Ty (S. cerevisiae) 5 homolog]; DS1F large subunit; human chromatin structural protein that is reversibly phosphorylated in mitosis |
| H45967 | 1.37 | CDK9 (Cyclin-Dependent Kinase 9); cell division cycle 2-like protein kinase 4 which regulates progression through the cell cycle; serine/threonine protein kinase PITALRE; member of the cyclin-dependent kinase pair (CDK9/cyclin-T) complex, also called positive transcription elongation factor b (TERb), which facilitates the transition from abortive to productive elongation by phosphorylating the C-terminal domain of the large subunit of RNA polymerase II and SUPT5H and RDBP, thereby increasing gene expression; forms a CDK9/cyclin-K complex which has kinase activity toward RNA polymerase II; phosphorylates retinoblastoma protein in vitro; phosphorylates p53 on serine 392 independently of CKII; also phosphorylates p53 on serine residues 33, 315 and 392 in a feedback loop between p53 and CDK9, pinpointing a novel mechanism by which p53 regulates the basal transcriptional machinery; upregulated upon exposure to various stresses |
| R06313 | 1.38 | CROCCL1 (ciliary rootlet coiled-coiled, rootletin-like 1); a major structural component of the ciliary rootlet; recombinant rootletin forms detergent-insoluble filaments radiating from the centrioles; the homopolymeric rootletin protofilaments bundle into variably shaped thick filaments; interacts with C-Nap1 and may function in centrosome cohesion by acting as a physical linker between the pair of centrioles/basal bodies; ciliary rootlet interacts with kinesin light chains and may provide a scaffold for kinesin-1 vesicular cargos; rootletin is phosphorylated by Nek2 kinase and is displaced from the centrosomes at the onset of mitosis; overexpression of rootletin in cells results in the formation of extensive fibers resulting in multinucleation, micronucleation and irregularity of nuclear shape and size, indicative of defects in chromosome separation |
| AA479771 | 1.40 | CUL7 (Cullin 7); an E3 ubiquitin ligase and a member of the Cullin Ring Ligase family involved in post-translational modifications that are important in the regulation of cell cycle progression; induced by DNA damage and promotes cell growth by antagonizing p53 function; functions as a novel antiapoptotic oncogene |
| AA455786 | 1.40 | MCM3 [minichromosome maintenance deficient (S. cerevisiae) 3]; required for cell proliferation; acts as a replication licensing factor that acts in early S phase and allows the DNA to undergo a single round of replication per cell cycle; ATM phosphorylates MCM3 on S535 in response to ionizing radiation; overexpressed in various cancers including those of the colon |
| AA032090 | 1.41 | DDX11 [(DEAD/H (Asp-Glu-Ala-Asp/His)] box polypeptide 11); S. cerevisiae CHL1-like DNA helicase involved in proliferation; required for sister chromatid cohesion and maintaining chromosome segregation; functions during S, G2, or M phase of cell cycle and is essential for prevention of aneuploidy |
| N76587 | 1.41 | CDC42BPB (Cell Division Cycle 42 Binding Protein Kinase beta [DMPK-like (dystrophy myotonic protein kinase)]; serine/threonine protein kinase involved in cytoskeletal organization and biogenesis in cell division by acting as a Cdc42 effector |
| H77797 | 1.41 | KIF12 (Kinesin Family Member 12) contains one kinesin-motor domain involved in microtubule motor activity during cell division |
| N91750 | 1.41 | E2F2 (E2F transcription factor 2); transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5′-TTTC[CG]CGC-′, found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA recognition; phosphorylated by CDK2 and Cyclin A-CDK2 in the S-phase of the cell cycle; component of the DRTF1/E2F transcription factor complex that functions in the control of cell cycle progression from G1 to S phase; the E2F-2 complex binds specifically hypophosphorylated retinoblastoma protein RB1; during the cell cycle, RB1 becomes phosphorylated in mid-to-late G1 phase, detaches from the DRTF1/E2F complex, thereby rendering the E2F transcriptionally active |
| AA488526 | 1.41 | NOLC1 (Nucleolar and coiled-body phosphoprotein 1); shuttles between nucleus, nucleolus and cytoplasm at telophase; begins to assemble into granular-like pre-nucleolar bodies which are subsequently relocated to nucleoli at early G1 phase |
| T77840 | 1.42 | DLG5 [Discs, large (Drosophila) homolog 5]; peripheral membrane protein involved in the negative regulation of cell proliferation; genetic variation associated with inflammatory bowel disease |
| R70925 | 1.44 | MKS1 (Meckel syndrome, type 1); FABB proteome-like protein required for primary cilium formation; function in mitosis not known at the present time |
| T67474 | 1.45 | APC7 (anaphase-promoting complex, subunit 7); one of 12 subunits that comprise the anaphase promoting complex, a ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle; The APC7 protein is located in the nucleus during interphase and the centrosome during metaphase/anaphase; functions with other members of the APC complex to regulate sister chromatid separation by degrading securins and targets cyclin B and other destruction box containing proteins for proteolysis; may recruit Cdh1 into the APC complex |
| AA489602 | 1.45 | TRAP1 (heat shock protein 75); refolds denatured retinoblastoma protein 1 into its native conformation during mitosis; mitochondrial Hsp90 analog; antagonizes ROS generation; protects mitochondria against damaging stimuli; protects cells from granzyme M-mediated apoptosis |
| AA026709 | 1.51 | DOCK6 (Dedicator of Cytokinesis 6); a Dock-C subfamily guanine nucleotide exchanger, has dual specificity for Rac1 and Cdc42 and regulates changes in the actin cytoskeleton during cell proliferation |
| H79234 | 1.51 | ACD [Adrenocortical dysplasia homolog (Drosophila)]; telomeric regulator and component of the TRF1 complex controlling telomere length; controls POT1 telomeric recruitment and telomere elongation by inhibition of telomerase activity |
| AA448755 | 1.54 | CDC25B (cell division cycle 25B); tyrosine phosphatase 2 that acts as a M-phase inducer; directly dephosphorylates CDC2 and stimulates its kinase activity; activates centrosomal Cdk1 in late prophase; activity controlled by centrosome-associated Chk1 which phosphorylates CDC25B in the absence of DNA damage; cooperates with Cdc25A to induce mitosis, but has a unique role in activating cyclin B1-Cdk1 at the centrosome resulting in centrosome separation |
| R93686 | 1.61 | MPHOSPH9 (M-phase phosphoprotein 9); involved in regulation of progression through the cell cycle |
| R93719 | 1.65 | GSPT1 (G1 to S Phase Transition 1 protein); GTP-binding protein 1 essential for G1 to S transition of the cell cycle; alias is eRF3 (eukaryotic release factor 3), which is a GTPase associated with eRF1 in a complex that mediates translation termination |
| AA459292 | 1.67 | CKS1B (CDC28 protein kinase regulatory subunit 1B); binds to and activates cyclin-dependent kinases and also interacts with SKP2 to promote the ubiquitination and proteasomal degradation of p27(Kip1); cyclin D1 regulates CKS1B-mediated degradation of p27(Kip1); exhibits altered expression in colorectal carcinoma |
| AA076063 | 1.71 | CALD1 (Caldesmon 1); actin-binding protein; during mitosis caldesmon dissociates from microfilaments |
| R40850 | 1.74 | ARP1 [Actin-Related Protein 1, yeast homolog A (centractin alpha)]; ACTR1A; major subunit of dynactin, a multiprotein complex known to contain 8 or 9 Arp1 monomers in a 37 nm filament involved in microtubule-based vesicle motility; attachment site for cargo directed to the dynein/dynactin complex; centrosome-associated actin homolog; overexpression results in cell cycle delay at prometaphase with appearance of supernumerary microtubule asters |
| AA488221 | 1.83 | DCTN1 [Dynactin 1 (p150, Glued) Drosophila homolog]; largest subunit of the 10 components that make up the large macromolecular dynactin complex that is involved in “search-and-capture” mechanisms that include the attachment of microtubules to kinetochores during mitosis, the maintenance of the spindle, formation of astral microtubules, chromosome motion and chromosome segregation; requires Aurora kinase B activity to be maintained at kinetochores; binds directly to microtubules and to cytoplasmic dynein; co-localizes with Arp1 to spindle microtubules; depletion in cells results in a metaphase delay and the poor connection of the centrosomes to the mitotic spindle poles; p150(Glued) is cleaved during apoptosis |
| T99336 | 1.88 | CEP164 (centrosomal protein of 164 kDa localized to the outer appendage); mediator protein required for the maintenance of genomic stability through the modulation of MDC1, RPA and CHK1; key player in the DNA damage-activated signaling cascade; phosphorylated upon replication stress; DNA damage-induced phosphorylation of CHK1 and activation of the G2/M checkpoint requires Cep164; plays a role in chromosome segregation, in addition to its function in checkpoint signaling; persists at centrioles throughout mitosis |
| R31831 | 2.00 | SHROOM3 (F-actin binding protein); induces cell elongation by redistributing γ-tubulin (associated with centrosomes) and directing microtubule arrays |
Notes: Fold reduction/induction is the ratio of mRNA levels in the deoxycholate-treated cells to the level in the untreated control cells. All fold reduction/induction ratios are the mean of all resistant cell lines HCT-116RB, HCT-116RC, and HCT-116RD compared to long-passage untreated cells and were statistically significant at the 95% probability level (p < 0.05).
Table 2.
Statistically significant increases/decreases in protein levels of chromosomal maintenance/mitosis related genes in HCT-116R colon cancer cells persistently exposed to deoxycholate
| Change in expression(1) in resistant cell lines | Gene and function | |||
|---|---|---|---|---|
| B | C | D | ||
| ↓ | ↓ | ↓ | KIF1A (Kinesin Family Member 1A); transports membrane-bound organelles toward the plus end of microtubules and important in cell division; functions in monomeric and dimeric states as a kinesin motor | |
| ↓ | ↓ | ↓ | Cyclin A (required during S phase and passage through G2); activates Cdk2 near the start of S phase and is necessary for the initiation of DNA replication; also binds to cdc2 (Cdk1); mutation or disruption of normal Cyclin A in cells results in a G2 arrest | |
| ↓ | ↓ | ↓ | EB1 [strong binding to the C-terminal domain of APC (adenomatous polyposis coli)]; interacts with the plus end of microtubules and targets APC to microtubule tips]; also required for the plus-end localization of CLIP-170 which is then required to localize p150(Glued) to plus-ends; targeted disruption of the interaction between EB1 and p150(Glued) suppresses anaphase astral microtubule elongation and a delay of cytokinesis | |
| ↓ | O | ↓ | SGT1 (G1/S and G2/M cell cycle transitions); important component in association with Skp1, Cul-I, F-box protein and CDC34/Ubc3 of the SCF ubiquitination ligase complex responsible for cell cycle transitions | |
| ↓ | O | ↓ | Chk1 (checkpoint kinase 1); activated after DNA damage; leads to G2/M arrest; regulates the S phase checkpoint by increasing the proteolysis of Cdc25A; cooperates with p21 to prevent apoptosis during DNA replication fork stress; inhibition causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage | |
| O | ↓ | ↓ | MAD2 (mitotic arrest-deficient 2); required for spindle assembly during mitosis and mitotic checkpoint control; localizes to the kinetochore of condensed chromosomes during mitosis; participates in checkpoint inhibition of the APC/C through a complex of BUBR1, BUB3 and CDC20; prevents premature proteolysis of cyclin B and securin; reduced cellular levels result in defective mitotic checkpoint control; averts aneuploidy by delaying anaphase onset until chromosomes align; functions as a tumor suppressor | |
| O | ↓ | ↓ | Eg5 (member 11 of the kinesin-5 family of microtubule-based motor proteins involved in mitosis and cytokinesis); phosphorylated exclusively on serine during S phase, but on both serine and Thr-926 during mitosis, thereby controlling the association of Eg5 with the spindle apparatus (probably during early prophase); important for bipolar spindle assembly and spindle function during mitosis; the rate of bipolar spindle assembly depends on the microtubule-gliding velocity of Eg5; Eg5 is also phosphorylated during mitosis at Thr-297, an evolutionarily conserved cdc2 phosphorylation site, by p34[cdc2]/cyclin B; phosphorylation by p34cdc2 regulates binding of Eg5 to the dynactin subunit 150(Glued); inhibition of phosphorylation blocks the interaction of Eg5 with centrosomes arresting cells in mitosis with monoastral microtubule arrays | |
| O | ↓ | ↓ | P140mDia (protein of 140mkDa); mammalian homolog of Drosophila’s diaphanous essential for cytokinesis | |
| ↑ | ↓ | ↑ | PTP1D/SHP2 (cytosolic Protein Tyrosine Phosphatase 1D/Src homology-2 (SH2) domain-containing phosphatase 2); helps maintain intracellular protein phosphotyrosine homeostasis and cell cycle progression | |
| ↑ | ↑ | ↑ | hRad9 (phosphorylated by ATM in response to DNA damage); checkpoint control; required for phosphorylation of Chk1; plays a role in cell cycle arrest during the G2 phase before entry into mitosis; phosphorylated hRad9 is found in the nucleus after DNA damage, and forms DNA damage-responsive complexes with other putative checkpoint control proteins, such as hRAD1 and hHUS1; binds the anti-apoptotic proteins Bcl-2 and Bcl-xL | |
| ↑ | ↑ | ↑ | Ran (Ras-related nuclear protein); highly conserved GTPase implicated in DNA replication and entry and exit from mitosis; at steady-state, 80%–90% of cellular Ran is located in the nucleus, with the remainder in the cytoplasm | |
| O | ↑ | ↑ | Cyclin D3 (regulatory subunit for cyclin-dependent kinase Cdk5); interacts with Cdk4 which is required for G2 phase cell cycle progression; regulator of progression through G1 phase during the cell cycle; promoter of cyclin D3 is regulated by E2F1; interacts with p58(PITSLRE), a G2/M-specific protein kinase; overexpression is associated with the accumulation of p27(kip1); interaction partner of lamin A/C; downregulated by rapamycin; when bound to cdks, the D-type cyclins also associate with the polymerase-delta subunit, PCNA; also binds pRb in vitro, suggesting that pRb may be an in vivo substrate of D-type cyclins; activates caspase 2, connecting cell proliferation with cell death; involved in liver metastasis of colorectal cancer | |
| O | ↑ | ↑ | Lap2 (Lamina-associated polypeptide 2); integral protein of the inner nuclear membrane; binding to lamins contributes to the attachment of the nuclear lamina to the inner nuclear membrane; organization of the nuclear envelope during cell cycle progression; also binds to chromatin, implying its role in chromosomal organization during mitosis; mitotic phosphorylation of LAP2 regulates its binding to lamins and chromosomes during the disassembly and reassembly of mitosis | |
Notes: Increase (↑), Decrease (↓), No Change (O).
Table 3.
Fold reduction/induction of mRNA Levels of mitosis-related genes in colon of mice fed a diet supplemented with deoxycholate
| Fold reduction/induction | Gene and function |
|---|---|
| 0.50 | Wee1 homolog (Schizosaccharomyces pombe); protein kinase that inhibits Cdc2 activity, thereby preventing cells from proceeding through mitosis; causes G2 arrest |
| 0.55 | APC (adenomatous polyposis coli); tumor suppressor protein that regulates free β-catenin levels and participates in Wnt signaling; binds to microtubules and increases microtubule ability; APC binds to microtubule plus ends and promotes microtubule net growth with or without EB1; haploinsufficiency accounts for a fraction of FAP patients without APC truncating mutations; decrease in APC expression after one week of supplemental deoxycholate feeding is dependent on the presence of NOS2 (inducible nitric oxide synthase) (unpublished data from the Bernstein et al 2006 study115) |
| 1.24 | CDC2L5 (cell division cycle 2-like 5); serine/threonine kinase; controller of the mitotic cell cycle; |
| 1.32 | Ubiquitin protein ligase E3C; E3 ubiquitin ligases degrade proteins via the ubiquitin-proteasomal pathway during mitosis |
| 1.33 | Cdc34 (cell division cycle 34); homolog of S. cerevisiae; Ubc3 ubiquitin-conjugating enzyme that controls proliferation through the regulation of p27 kip1 protein levels; mediates the degradation of wee1; inhibits the association of CENP-E with kinetochores and blocks the metaphase alignment of chromosomes |
| 1.34 | Tsg101 (Tumor susceptibility gene 101); necessary for cell proliferation and cell survival; may function as a dominant-negative inhibitor of ubiquination in pathways where protein expression is tightly regulated; interferes with MDM2 ubiquination leading to a decrease in MDM2 decay and downregulation of p53 protein |
| 2.10 | CDK5 (cell division protein kinase 5); involved in control of G1 to S phase transition of the cell cycle; one of the two main tau-kinases, complexing with cyclin D (D1, D2, D3); hyperphosphorylation of tau reduces tau’s ability to associate with microtubules; phosphorylates the tubulin-binding protein, stathmin; phosphorylates p53 and regulates its activity |
Since the microarray study was published six years ago,98 recently identified gene sequences related to mitosis and chromosome maintenance have been added to Table 1 to bring the 2002 study up to date.
In summary, persistent upregulation and/or downregulation of key proteins of the division process by deoxycholate, coupled with decreased DNA repair and increased DNA damage, may lead to genomic instability, including mutator phenotypes. Recent studies have indicated that 1) the spindle proteins, Aurora A and BUB1B, are aberrantly expressed in dysplastic mucosa of patients with longstanding ulcerative colitis,171 a pre-neoplastic condition), 2) both Aurora A and BUB1B undergo a shift in subcellular localization during malignant transformation,172 3) a reduced level of the spindle checkpoint protein, BUB1B, is associated with aneuploidy in colorectal cancers,173 and 4) chromatid cohesion defects may underlie chromosome instability in human colorectal cancers.174 It is an intriguing possibility that these defects occur very early during colon carcinogenesis, and may be caused, in part, by dietary-related factors, such as high levels of endogenous bile acids, coupled with polymorphisms in mitotic checkpoint genes.175
Oxidative stress as an important cause of genomic instability
The induction of kinetochore-positive and -negative micronuclei by bile acids may result from aneuploidy (involving the loss and gain of whole chromosomes) or portions of chromosomes, resulting from unrepaired double-strand breaks. This bile acid-induced induction of genomic instability is most probably caused by oxidative stress directed at parts of the mitotic machinery, checkpoint proteins and/or direct oxidative damage to DNA that results in double-strand breaks. The only studies that addressed this issue in the gastrointestinal tract are those of Jenkins and colleagues,16,17 who evaluated several antioxidants (eg, vitamin C, resveratrol, EGCG) to determine their effectiveness at reducing deoxycholate-induced micronuclei formation in esophageal cells. Of the antioxidants tested, only vitamin C had a significant effect on reducing micronuclei formation. Since bile acids damage mitochondria, resulting in an increase in mitochondrial ROS, and ROS derived from damaged mitochondria are known to induce genomic instability,157 various antioxidants specifically directed at mitochondria should be tested for their effectiveness at reducing bile acid-induced genomic instability.
Since this area of bile acid research is just beginning to be explored, we have reviewed the literature involving other cell types (beyond those of the GI tract) and found that oxidative stress plays a definite role in the induction of genomic instability.40,156,157,176–178 Aneuploidy may often reflect defects in mitotic segregation in cancer cells.50 The faithful cohesion and separation of chromosomes are controlled by a large number of proteins, some of which have a checkpoint function.180–185 Increased production of ROS has been reported to induce aneuploidy by impairing the spindle check-point function.158 The spindle checkpoint is the safeguard mechanism that halts anaphase onset until the mitotic spindle has been assembled and metaphase chromosomes properly attached. At the metaphase-anaphase transition, the APC (anaphase-promoting complex) initiates the separation of sister chromatids by catalyzing the ubiquitination of securin, a protein that prevents the proteolysis of the cohesin complex and the subsequent segregation of the chromosomes, in the securin-separase complex.186 The APC11 subunit functions as the catalytic core of the APC complex and contains a RING-H2-finger domain, which includes one histidine and seven cysteines residues that coordinate two Zn2+ ions. The RING-H2-finger domain is a target of hydrogen peroxide, which induces the release of of bound zinc as a result of the oxidation of cysteine residues.187 The oxidation of APC11 reduces the normal functions of the unoxidized APC11 protein, resulting in the inhibition of ubiquitination and degradation of cyclin B1, and the degradation of securin. Other chromosomal aberrations, such as translocations and dicentric chromosomes, may be caused by accumulation of double-strand breaks induced by oxidative damage, as in Werner syndrome cells.188 Based on the data from our laboratory that shows the modulation of 71 genes involved in chromosome maintenance, chromatid cohesion, kinetochore-microtubule attachments, chromatid separation, cytokinesis, etc. by deoxycholate (see Tables 1–3), some of these proteins may be oxidatively damaged and/or targeted for destruction by deoxycholate. However, no studies have yet addressed this important mechanism of bile acid-induced damage to dividing cells. Thus, in colon carcinogenesis, the persistent increase in oxidative stress in epithelial cells caused by excessive exposure to bile acids may induce mitotic defects very early in the sequence of events that lead to colon cancer. Hydrophobic bile acids damage mitochondria, and increased levels of oxidative stress caused by defective mitochondria have been proposed as a possible mechanism by which a “mutator phenotype” could arise.40
Bile acids induce ER stress
The induction of ER stress by hydrophobic bile acids is evidenced by an increase in Gadd153/CHOP,142,189,190 a pro-apoptotic transcription factor,191–197 and an increase in GRP78,98,190 a molecular chaperone that protects against ER stress.197,198 Furthermore, hydrophobic bile acids dilate the cisternae of the endoplasmic reticulum (identified using transmission electron microscopy199). The mechanisms by which bile acids cause ER stress probably include the generation of ROS that may damage the ER membrane and/or cause protein unfolding, and the release of Ca++ from the ER through the activation of inositol triphosphate and ryanodine receptors.200 Excessive ER stress may lead to cell death in the form of apoptosis and/or necrosis. Since deoxycholate induces the formation of nitrotyrosine residues in proteins of colon cells,112 ER stress may also be mediated through the generation of RNS.195,201 Persistent ER stress may then select for cells that exhibit activated ER stress-responsive survival pathways important in colon carcinogenesis. We have shown that ER stress-related survival proteins are consitutively upregulated in colon cancer cells that are resistant to deoxycholate-induced apoptosis, and include Grp78,98 S-nitrosylated Grp78114 and S-nitrosylated ORP150,114 another inducible ER stress-related chaperone protein.202 The observed constitutive increase in autophagic proteins98 observed in these resistant cells may also serve to protect against ER stress, although this area of research has not been fully explored.
Consequences of bile acid-induced mitochondrial damage: metabolic stress, autophagy, apoptosis, and necrosis
Bile acid-induced DNA damage, ER stress, mitotic stress, and mitochondrial damage may all lead to cell death through various processes including apoptosis, necrosis, autophagic cell death and/or mitotic catastrophe, depending on the level of combined stresses. Of all organelles, mitochondria are most central to the three main cell death pathways, apoptosis, necrosis, and autophagic cell death.203 Apoptosis as a mechanism of mitochondrially-mediated bile acid-induced cell death has been studied the most in cells of the GI tract, as discussed below.
Mitochondria generate most of the ATP used for energy-consuming processes within the cell. Moreover, mitochondria play an important role in the regulation of other cellular processes such as autophagy and apoptosis. Autophagy is a catabolic process involving the degradation of a cell’s own components through the activation of lysosomes. In response to stresses induced by hydrophobic bile acids, autophagy is initiated (unpublished data from our laboratory) as the first mechanism to cope with damage when mitochondrial injury is limited. The damaged mitochondria are sequestered within lysosomes, thereby preventing the release of apoptosis-inducing factors, such as cytochrome c, apoptosis-inducing factor (AIF), and Smac/DIABLO. As more mitochondria are damaged and undergo the mitochondrial membrane permeability transition in response to bile acids, the pro-apoptotic proteins are released from the mitochondria.204 These releases result in the formation of the apoptosome, caspase activation, degradation of vital proteins in the cell and, finally, apoptosis. Apoptosis can be triggered by two distinct signaling cascades, the extrinsic and intrinsic pathways. The extrinsic pathway involves the activation of specific cell surface receptors such as Fas/CD95 and the tumor necrosis factor receptor (TNFR), while the intrinsic pathway is primarily regulated by mitochondria. Importantly, in contrast to hepatocytes and hepatic cancer cell lines, in which hydrophobic bile acids induce apoptosis by an extrinsic pathway, deoxycholate induces apoptosis in colonic epithelial cells in a CD95-independent pathway.205 Bile acid-induced mitochondrial damage may initially increase autophagy to rid cells of damaged organelles. As stress continues, apoptosis or autophagic cell death may follow. If, however, the stresses on mitochondria are initially severe, oxidative phosphorylation fails, NAD+ levels drop, ATP levels are rapidly depleted, resulting in loss of ionic control,206 followed by cell swelling and lysis, characteristic features of necrosis.207–211 Apoptotic cell death induced by deoxycholate has been frequently reported in colon epithelial cells.41,44,45,98,99,110,112,118,199,205,212–224 Therefore, an understanding of bile acid-induced alterations in mitochondrial function are of great interest. At least two major alterations occur during bile acid-induced mitochondrial dysfunction. These are (1) disruption of electron transport leading to generation of ROS, loss of mitochondrial membrane potential (MMP), decrease in oxidative phosphorylation, and decrease in ATP production, and (2) release of mitochondrial proteins that trigger activation of caspases. Bile acids were shown to be involved in all three processes.
The exact mechanism by which bile acids disrupt electron transport and decrease oxidative phosphorylation has yet to be defined.225–227 Experiments performed with mitochondria isolated from hepatocytes clearly demonstrate that hydrophobic bile acids induced perturbations in mitochondrial bioenergetics.225,226 Specifically, complex I and complex III of the mitochondrial electron transport chain were inhibited by treatment with10 μM bile acid. Complex IV was inhibited at higher concentrations of bile acids.225 The authors speculated that the inhibition of the electron transport complexes may be due to bile acid incorporation into the mitochondrial membranes resulting either in direct toxicity or alteration in lipid composition. When they tested this hypothesis, they found that, indeed, bile acids were incorporated into the mitochondrial membranes. Furthermore, they found that the hydrophobic bile acid, chenodeoxycholic acid, induces alteration in the lipid composition of the mitochondrial membrane, while no alterations were found after treatment with lithocholic acid.225 However, it is possible that the changes in lipid composition induced by lithocholic acid were below the limit of detection used in the study.225 Membrane structural changes observed using spin-labelling techniques and electron paramagnetic resonance (EPR) spectroscopy analysis also support the involvement of mitochondria in bile acid-induced apoptosis.228 These mitochondrial structural changes include modified lipid polarity and fluidity, altered protein order and increased oxidative injury.228
Hydrophobic bile acids cause mitochondrial oxidative stress97,110,117,228,229 leading to mitochondrial swelling199 and formation of megamitochondria.110 The excessive generation of ROS that overwhelms the cells’ antioxidant defenses is most probably responsible for the death of colonic epithelial cells,230 and is consistent with the role of oxidative stress in mediating apoptosis in many other types of cells.231 Importantly, bile acid-induced apoptosis in HT-29 colon cells is dramatically reduced if mitochondria are protected against ROS by inhibiting complexes I and II of the electron transport chain.110 Later studies indicated, moreover, that perturbation of the mitochondrial electron transport chain, in general, and uncoupling of oxidative phosphorylation, attenuates deoxycholate-induced apoptosis.199 The mechanism by which this perturbation of mitochondrial function results in apoptotis resistance is not clearly understood. Another effect of hydrophobic bile acids on mitochondria is rapid loss of mitochondrial membrane potential (MMP).110,199,205 For example, treatment of SW480 cancer cell line with 0.5 mM deoxycholic acid induced loss of MMP in 5 minutes.205 This loss of MMP correlates with the induction of ROS.
The second major effect of hydrophobic bile acids is to induce rapid release of pro-apoptotic proteins in colon cancer cells, such as cytochrome c, which leads to the activation of caspases.218 Incubation of HT-29 and HCT-116 colon cells with deoxycholate resulted in the cleavage of procaspases and the activation of effector caspases 2, 3, 7 and 8,110,222 while caspase-1 proinflammatory caspase was not cleaved.222 Caspases 9 and 3 are rapidly activated by deoxycholate, while caspase 8 activation is a later event.110,218,222 Caspase-6 is of particular importance to deoxycholate-induced apoptosis of colon epithelial cells, since the antiapoptotic nitric oxide-targeted enzyme, guanylate cyclase-α1, is specifically cleaved by caspase-6, resulting in apoptosis.224 Apoptosis in HCT116 cells is not induced simply by a detergent effect, since sodium dodecyl sulfate (SDS), a detergent that is structurally distinct from bile acids, does not induce apoptosis.218 Although protein synthesis inhibitors have been shown to protect against programmed cell death, de novo synthesis of proteins does not appear to be necessary for bile acid-induced apoptosis.218 Altogether, these data suggest that in contrast to the extrinsic induction of apoptosis, as seen in hepatocytes, hydrophobic bile acids primarily activate the intrinsic mitochondrial pathway of apoptosis in colon cancer cell lines.
How exactly do bile acids interact with mitochondria to perturb their function? Hydrophobic bile acids may directly affect mitochondria, leading to disruption of the electron transport chain and increased production of mitochondrial ROS. Alternatively, mitochondria can be damaged indirectly by extramitochondrial ROS produced by plasma membrane bound enzymes targeted by bile acids, such as NAD(P)H oxidase96 (Figure 2). The importance of NAD(P)H to deoxycholate-induced apoptosis was ascertained by the use of an inhibitor of NAD(P)H oxidase, diphenyleneiodonium (DPI).232 DPI attenuated overall ROS production and prevented the occurrence of deoxycholate-induced apoptosis, assessed using translocated membrane phospholipid and nuclei containing condensed chromatin.232 Interestingly, NADPH oxidase levels are elevated in colon cancer cell lines as well as in the tissues from patients with colon cancer.233–236 Another extramitochondrial source of ROS occurs when molecular oxygen is partially reduced during the metabolism of arachidonic acid by the cyclooxygenase (COX) and lipoxy-genase (LOX) pathways (Figure 2). Another indirect effect of bile acids on mitochondria is the release of arachidonic acid from phospholipids following activation of PLA2 by bile acids.87 Arachidonic acid induces inhibition of mitochondrial complex I and III and causes an increase in mitochondrial ROS production.237 Hydrophobic bile acids may also trigger the formation of ceramide,238 which can damage mitochondria and induce apoptosis. The ceramide-induced disruption of mitochondrial function and induction of apoptosis is caused, in part, by modulation of gene expression of members of the Bcl-2 family of apoptosis-related proteins.239
Persistent exposure to deoxycholate selects for apoptosis resistance
Repeated long-term exposure of colon epithelial cells to high physiologic concentrations of bile acids may allow preferential survival of cells that are resistant to induction of apoptosis by bile acids. Such apoptosis resistant cells could arise and proliferate by the processes of mutation and Darwinian selection (Figure 3). Apoptosis-resistance in the colon appears to arise in settings where the colonic mucosa is chronically exposed to high physiologic levels of bile acids. Magnuson and colleagues240 reported that chronic feeding of 0.2% cholic acid (added to the AIN-76 diet) to azoxymethane (AOM)-treated Sprague-Dawley rats for 18 weeks resulted in a statistically significant reduction in the number of apoptotic bodies in ACF after an acute low dose of AOM, compared with normal-appearing crypts. Both normal and ACF crypts had fewer apoptotic bodies per 100 cells than crypts from rats fed the control diet.
Work from our laboratory indicated that in vitro exposure of apoptosis-sensitive colon epithelial cells (HCT-116) for approximately 40 weeks resulted in the generation of stable apoptosis-resistant cell populations.98 We also evaluated the nonneoplastic flat colonic mucosa of patients in different colon cancer risk groups for apoptosis competence using an ex vivo live cell bioassay.44,45,212,213,241 The apoptosis-inducing agent used in this bioassay was deoxycholate. The nonneo-plastic colonic mucosa of patients with colon cancer had a significantly reduced apoptotic index compared to that of low-risk and normal patients. The exact etiologic agents that were responsible for the development of apoptosis resistance are not known, but hydrophobic bile acids, other agents that generate ROS/RNS, and actual components of a high-fat diet may have been key players.
Bile acids, inflammation and colon cancer
Numerous epidemiological studies have shown that fecal bile acid concentrations are increased in populations with a high incidence of colon cancer, suggesting an etiological linkage.242–251 We have shown that supplemental feeding of deoxycholate in a mouse model can cause colitis in association with markers of oxidative stress.115 An important link between high bile acid concentrations and colon cancer may, therefore, be the induction of inflammation, a condition that is associated with many cancers of the GI tract. Bile acids may induce inflammation through a disruption of tight junctions between colonic epithelial cells115 and increase in transepithelial permeability,88 providing a means for entry of pro-inflammatory bacterial species.252 In addition, bile acids may cause dysregulation of cytokine expression through the constitutive activation of NF-κB,98 a redox-associated transcription factor, that contributes to apoptosis resistance and a persistent increase in pro-inflammatory proteins.
Field defects in the colon
A “field defect” or “field of cancerization” is a region of tissue that precedes and predisposes to the development of cancer.253 Field defects are of interest because they provide insight into the early events of progression to cancer and may lead to clinically useful biomarkers of cancer risk. It is generally considered that a solid tumor is initiated by a series of somatically inherited changes (ie, aneuploidy, mutations or transmissible epigenetic events (epimutations), such as methylation of CpG islands254). Aneuploidy, mutations and epimutations are considered to contribute to early progression, usually by causing a somatically inherited proliferative advantage relative to surrounding cells. Furthermore, a mutator phenotype or genetic instability may accelerate the process of mutation followed by Darwinian selection.19–21 In a normal population of dividing cells within a tissue, a cell may acquire a proliferative advantage through aneuploidy, mutation or epimutation. Such a cell will tend to undergo clonal expansion and replace neighboring cells, thus giving rise to a patch of abnormal cells. Within this patch, a cell might acquire a second aneuploidy, mutation or epimutation that provides a proliferative advantage compared to other cells within the established patch. This cell may then expand clonally forming a secondary patch within the first patch. Within this new patch, the process of mutation and Darwinian selection may be repeated several more times over a long period, perhaps decades, until a malignant cell arises that clonally expands into a cancer. If solid tumors generally arise by such a process, then tumors should ordinarily be accompanied by a field defect in associated tissue that may appear histologically normal.
The most common type of colorectal cancer, sporadic adenocarcinoma, develops by a pathway that appears to proceed through the following stages: normal flat mucosa, development of fields of defective flat mucosa, aberrant crypt foci, microadenomatous fields, adenoma with low grade dysplasia, adenoma with high grade dysplasia, and adenocarcinoma. However, currently, it is unclear whether all sporadic adenocarcinomas progress through each of these stages.
The morphologically normal appearing colonic mucosa of patients with colorectal neoplasia has been found by proteomic analysis to have numerous changes in protein expression.255 Alterations of gene expression that occur in the normal appearing colonic mucosa of human colon cancer patients correspond to alterations observed in the normal appearing colonic mucosa of cancer prone APCmin mice.256 The promoter of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) is methylated in 46% of colonic tumors. Within this group of tumors with MGMT methylation, 94% of tissue samples from apparently normal mucosa associated with these tumors also had MGMT promoter methylation.257 Often MGMT methylation was detected as far away as 10 cm from the tumor, indicating a field defect. Also patients with resected adenomas of the colon or rectum had altered expression of O-acetylated sialic acids in their “uninvolved” colon and rectal mucosa compared to sialic acids of colon or rectal mucosa of patients without colon tumors.258 These findings indicate a field defect of sialic acid expression in the patients with colorectal cancer. Since a decrease in the level of O-acetylated sialic acids on the surface of colon epithelial cells is found to be associated with the early stages of colorectal cancer,259 it is considered a risk factor. This decrease in the level of O-acetylated sialic acids may lead to an increase in the expression of sialyl LewisX, a tumor-associated antigen related to the progression of colorectal cancer cells to metastasis.260
An imbalance of proliferation and apoptosis in the left colon and sigmoid/rectum was found in patients who had a resected large adenoma (≥1.5 cm) compared to patients who never had a tumor.261 Several lines of evidence indicate that colorectal cancers often arise in a field of apoptosis-resistant cells. The Bcl-xL protein inhibits apoptosis, in part, by inhibiting release of cytochrome c from the mitochondria. The expression of Bcl-xL was found to be increased at 1 cm and 10 cm away from colorectal adenocarcinomas in the non-neoplastic colorectal mucosa, suggesting the presence of an apoptosis-resistant field.262 The prototypic anti-apoptotic protein, Bcl-2, also behaves similarly.263 Expression of the antiapoptotic survivin mRNA in colon tumors predicts poor patient survival due to subsequently recurrent colorectal tumors.264 The normal appearing mucosa of about half of patients with survivin-positive tumors also express this mRNA. This finding suggests that survivin- positive tumors often develop in a normal appearing, but survivin- positive, apoptosis-resistant field. When rats are fed a diet containing 0.2% cholic acid, their colon crypt cells develop increased resistance to apoptosis.240 Apoptosis-resistant crypts may then spread through the flat colonic mucosa by crypt fission.265 These various observations suggest that repeated exposure of the colon to high levels of bile acids as a result of a Western style diet cause Darwinian selection for cells resistant to induction of apoptosis, and give rise to fields of apoptosis-resistant epithelium that may further evolve to malignancy.
Work from our laboratory on biomarkers associated with colon cancer risk indicate that, in addition to loss of apoptosis competence, lectin staining using DBA (Dolichos biflorus agglutinin) (a measure of epithelial cell differentiation), cytochrome c oxidase subunit I (CcOI) (a mitochondrial DNA-encoded subunit of CcO) and Pms2 (a DNA repair protein) were all significantly decreased in the normal-appearing flat mucosa of patients with colon cancer compared to that of control subjects.213,241,266
Scalmati and Lipkin 267 reviewed numerous studies on cell proliferation pattern in patients with colon adenomas or cancer. Expansion of the proliferative compartment (a lumenward displacement of the zone of active cell proliferation in the crypts) occurred along the entire colon of individuals, indicating that there was increased risk throughout the colon, no matter where the actual lesion was located. We also determined that apoptosis resistance in the nonneoplastic flat mucosa of patients with colon cancer was present in several regions of the colon far removed from the tumor site,45 indicating a generalized field defect of apoptosis resistance, albeit “patchy” in nature,213 throughout the colon.
A recent study indicated that the consumption of a Western style diet plays a crucial role in the progression of a field defect to colon cancer.268 Among 1009 patients with resected stage III colon cancer, colon cancer recurred in 324 patients in a median of 5.3 years. This recurrence suggests the frequent presence of a field defect. The patients had all undergone fluorouracil-based adjuvant chemotherapy after surgery, and were evaluated for diet during and 6 months after adjuvant chemotherapy. The rate of cancer recurrence was almost threefold greater among patients in the highest quintile of Western style diet consumption compared to those in the lowest quintile of Western style diet consumption.
Prevention of bile acid-induced stresses and genomic instability: importance of butyrate
We have reviewed evidence that deoxycholate induces cellular stresses in epithelial cells of the GI tract and that bile acids play a role in colon carcinogenesis. Although hydrophobic bile acids were once believed to be mainly tumor promoters, the bile acid induction of DNA damage and aneuploidy indicates that they are also genotoxins relevant to the initiation and progression of the neoplastic process. It is well documented that phytosterols and other types of antioxidants, micronutrients and dietary fiber have antineoplastic effects. An overall discussion of dietary-related colon cancer chemopreventive agents is beyond the scope of this review. However, we will focus on the short-chain fatty acid, butyrate, since previous studies have shown that butyrate interferes with many deleterious effects of hydrophobic bile acids such as increased proliferation,269–272 and has been shown to have anti-genotoxic effects on deoxycholate143 and H2O2-induced273,274 genotoxicity in colon epithelial cells. Furthermore, butyrate affects many signal-transduction pathways and histological parameters associated with colon carcinogenesis.272,275–286
Butyrate is a 4-carbon fatty acid that is an end product of bacterial fermentation of dietary fiber and occurs at concentrations of 2–10 mM in the colon.287 Certain intestinal bacterial species (eg, Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia, Coprococcus) are butyrate producers288–290 and beneficial to colon cancer prevention, whereas others, such as Enterococcus faecalis (extracellular superoxide and H2O2-producing bacterial species291,292) and Desulfovibrio (sulfate-reducing bacteria which produces hydrogen sulfide) are deleterious in that they cause DNA damage and chromosomal instability.291–295 In a recent study that quantified specific bacterial species using real-time PCR in the feces of healthy control volunteers and patients with colon cancer, it was found that the butyrate-producing bacteria species were significantly decreased and the superoxide-producing bacterial species were significantly increased in the feces of colon cancer patients compared to control subjects.290 It is significant that the feces of patients with ulcerative colitis, a pre-neoplastic inflammatory condition of the colon, uniformly contain sulfate-reducing bacteria. Sulfide substantially increases the proliferative index of the crypts in mucosal biopsies, and this increase is prevented by the presence of butyrate.296 A general consequence of the fermentation of fiber is to reduce the pH of the gut lumen, thereby affecting bacterial metabolism and competition297 and reducing the bacteria-mediated formation of hydrophobic bile acids.298 These shifts in bacterial species may be important determinants of the risk of colon cancer.
It is significant that butyrate can protect against oxidative DNA damage induced by deoxycholate ex vivo in colono-cytes derived from human biopsies.143 The processes in the colon that are affected by butyrate include a suppression of proliferation,271,277,299 induction of apoptosis,214,215 induction of differentiation,275,276,278 a reduction in inflammation,280,286 suppression of NF-κB activation,300 increase in xenobiotic metabolizing enzymes301,302 and a decrease in DNA damage and genomic instability.143,273,274 All of these processes affect colon carcinogenesis in a profound way.280,303
In our studies of field defects in the human colon, we observed that in the nonneoplastic flat mucosa of patients with colon cancer there is an increase in apoptosis resistance,44,45,212,213 loss of differentiation,213 decrease in mitochondrial gene expression (eg, CcOI)266 and a decrease in PMS2, a mismatch repair protein.241 It is significant that butyrate has opposite effects. It can increase differentiation, induce apoptosis, increase CcOI levels,276 and has more potent antineoplastic effects on colon cancer cells defective in mismatch repair.304
Future directions
We have just begun to understand the role of hydrophobic bile acids in inducing DNA damage and genomic instability in colon epithelial cells, and potential strategies to reduce these deleterious effects. Some key areas that require further experimentation include: 1) the role of oxidative stress as mediators of the effect of bile acids on the reduction of DNA repair proteins (eg, Ku80, MSH3), tumor suppressors (eg, p53) and spindle checkpoint proteins (eg, Mad2); 2) the role of oxidative stress in the deoxycholate-induced induction of micronuclei and aberrant mitoses; 3) the ability of deoxycholate to induce aberrant mitoses and aneuploidy in vivo using mouse models; 4) the possible synergistic effect of bile acids and environmental agents (eg, nicotine from tobacco smoking, arsenic, food carcinogens) in inducing DNA damage and genomic instability; and 5) the effectiveness of dietary-related nutrients and co-factors (eg, zinc, niacin, selenium compounds, plant polyphenols, butyrate) in preventing the genotoxic effects of hydrophobic bile acids both in vitro using cell culture models and in vivo using mouse models. The results from these types of experiments will have relevance to other cancers of the GI tract (esophagus, stomach, liver, pancreas) in which hydrophobic bile acids have a potential etiologic role.
Summary
In this review, we have described the likely major mechanisms by which hydrophobic bile acids can induce stresses on cells, which can then result in colon carcinogenesis. Persistent exposure of colon epithelial cells to hydrophobic bile acids can result in the development of apoptosis resistance and the modulation of numerous genes/proteins associated with chromosome maintenance and mitosis. Hydrophobic bile acids also damage the genome through multiple mechanisms, including oxidative DNA damage, p53 and other mutations, micronuclei formation and aneuploidy, all major components of genomic instability. These findings link dietary-related factors to the development of genomic instability, extend our understanding of the underlying causes of colon cancer, and may open up new avenues for hypothesis-driven biomarker development to assess colon cancer risk.
Acknowledgments
This work was supported in part by NIH 5 R01 CA119087, Arizona Biomedical Research Commission Grant #0803, VA Merit Review Grant 0142 of the Southern Arizona Veterans Affairs Health Care System and Biomedical Diagnostics and Research, Inc., Tucson, Arizona, USA.
References
- 1.Breivik J. Don’t stop for repairs in a war zone: Darwinian evolution unites genes and environment in cancer development. Proc Natl Acad Sci U S A. 2001;98:5379–5381. doi: 10.1073/pnas.101137698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Israel L. Tumor progression: Random mutations or an integrated survival response to cellular stress conserved from unicellular organisms? J Theoret Biol. 1996;178:375–380. doi: 10.1006/jtbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- 3.Breivik J, Gaudernack G. Carcinogenesis and natural selection: a new perspective to the genetics and epigenetics of colorectal cancer. Adv Cancer Res. 1999;76:187–212. doi: 10.1016/s0065-230x(08)60777-0. [DOI] [PubMed] [Google Scholar]
- 4.Breivik J, Gaudernack G. Genomic instability, DNA methylation, and natural selection in colorectal carcinogenesis. Semin Cancer Biol. 1999;9:245–254. doi: 10.1006/scbi.1999.0123. [DOI] [PubMed] [Google Scholar]
- 5.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 6.Breivik J. The evolutionary origin of genetic instability in cancer development. Semin Cancer Biol. 2005;15:51–60. doi: 10.1016/j.semcancer.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Gorringe KL, Chin S-F, Pharaoah P, et al. Evidence that both genetic instability and selection contribute to the accumulation of chromosome alterations in cancer. Carcinogenesis. 2005;26:923–930. doi: 10.1093/carcin/bgi032. [DOI] [PubMed] [Google Scholar]
- 8.Tsafrir D, Bacolod M, Selvanayagam Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 9.Breivik J. Cancer–evolution within. Int J Epidemiol. 2006;35:1161–1162. doi: 10.1093/ije/dyl187. [DOI] [PubMed] [Google Scholar]
- 10.Vineis P, Berwick M. The population dynamics of cancer: a Darwinian perspective. Int J Epidemiol. 2006;35:1151–1159. doi: 10.1093/ije/dyl185. [DOI] [PubMed] [Google Scholar]
- 11.Narisawa T, Magadia NE, Weisburger JH, Wynder EL. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N-methyl-N’-nitro-N-nitrosoguanidine in rats. J Natl Cancer Inst. 1974;53:1093–1097. doi: 10.1093/jnci/53.4.1093. [DOI] [PubMed] [Google Scholar]
- 12.Reddy BS, Narasawa T, Weisburger JH, Wynder EL. Promoting effect of sodium deoxycholate on colon adenocarcinomas in germ-free rats. J Natl Cancer Inst. 1976;56:441–442. doi: 10.1093/jnci/56.2.441. [DOI] [PubMed] [Google Scholar]
- 13.Reddy BS, Watanabe K, Weisburger JH, Wynder EL. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res. 1977;37:3238–3242. [PubMed] [Google Scholar]
- 14.McSherry CK, Cohen BI, Bokkenheuser VD, Mosbach EH, Winter J. Effects of calcium and bile acid feeding on colon tumors in the rat. Cancer Res. 1989;49:6039–6043. [PubMed] [Google Scholar]
- 15.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins GJ, D’Souza FR, Suzen SH, et al. Deoxycholic acid at neutral and acid pH, is genotoxic to oesophageal cells through the induction of ROS: The potential role of anti-oxidants in Barrett’s oesophagus. Carcinogenesis. 2007;28:136–142. doi: 10.1093/carcin/bgl147. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins GJ, Cronin J, Alhamdani A, et al. The bile acid deoxycholic acid has a non-linear dose response for DNA damage and possibly NF-{kappa}B activation in oesophageal cells, with a mechanism of action involving ROS. Mutagenesis. 2008;23:399–405. doi: 10.1093/mutage/gen029. [DOI] [PubMed] [Google Scholar]
- 18.Breivik J, Gaudernack G. Resolving the evolutionary paradox of genetic instability: a cost-benefit analysis of DNA repair in changing environments. FEBS Lett. 2004;563:7–12. doi: 10.1016/S0014-5793(04)00282-0. [DOI] [PubMed] [Google Scholar]
- 19.Loeb KR, Loeb LA. Genetic instability and the mutator phenotype. Studies in ulcerative colitis. Am J Pathol. 1999;154:1621–1626. doi: 10.1016/S0002-9440(10)65415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3531–3557. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- 22.Bignold LP. The mutator phenotype theory can explain the complex morphology and behavior of cancers. Cell Mol Life Sci. 2002;59:950–958. doi: 10.1007/s00018-002-8482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bignold LP. The mutator phenotype theory of carcinogenesis and the complex histopathology of tumours: support for the theory from the independent occurrence of nuclear abnormality, loss of specialisation and invasiveness among occasional neoplastic lesions. Cell Mol Life Sci. 2003;60:883–891. doi: 10.1007/s00018-003-2226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bignold LP. Variation, “evolution”, immortality and genetic instabilities in tumour cells. Cancer Lett. 2007;253:155–169. doi: 10.1016/j.canlet.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Rahman WM, Katsura K, Rens W, et al. Spectral karyotyping suggests additional subsets of colorectal cancers by pattern of chromosome rearrangement. Proc Natl Acad Sci U S A. 2001;98:2538–2543. doi: 10.1073/pnas.041603298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masramon L, Vendrell, Trafa G, et al. Genetic instability and divergence of clonal populations in colon cancer cells in vitro. J Cell Sci. 2006;119:1477–1482. doi: 10.1242/jcs.02871. [DOI] [PubMed] [Google Scholar]
- 27.Hermsen M, Postma C, Baak J, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 28.Rowan A, Halford S, Gaasenbeek M, et al. Refining molecular analysis in the pathways of colorectal carcinogenesis. Clin Gastroenterol Hepatol. 2005;3:1115–1123. doi: 10.1016/s1542-3565(05)00618-x. [DOI] [PubMed] [Google Scholar]
- 29.Risques R-A, Moreno V, Marcuello E, et al. Redefining the significance of aneuploidy in the prognostic assessment of colorectal cancer. Lab Invest. 2001;81:307–315. doi: 10.1038/labinvest.3780239. [DOI] [PubMed] [Google Scholar]
- 30.Risques R-A, Moreno V, Ribas M, et al. Genetic pathways and genome-wide determinants of clinical outcome in colorectal cancer. Cancer Res. 2003;63:7206–7214. [PubMed] [Google Scholar]
- 31.Stoler DL, Chen N, Basik M, et al. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci U S A. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis. 2004;25:2033–2044. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- 33.Gagos S, Irminger-Finger I. Chromosome instability in neoplasia: chaotic roots to continuous growth. Int J Biochem Cell Biol. 2005;37:1014–1033. doi: 10.1016/j.biocel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Bignold LP. Initiation of genetic instability and tumour formation: a review and hypothesis of a nongenotoxic mechanism. Cell Mol Life Sci. 2003;60:1107–1117. doi: 10.1007/s00018-003-2314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieber OM, Heinimann K, Tomlinson IPM. Genomic instability – the engine of tumorigenesis? Nature Rev. 2003;3:701–708. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 36.Hanks S, Coleman K, Reid S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 37.Woo RA, Poon RYC. Gene mutations and aneuploidy. The instability that causes cancer. Cell Cycle. 2004;3:1101–1103. [PubMed] [Google Scholar]
- 38.Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. Aneuploidy vs gene mutation hypothesis of cancer: Recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci U S A. 2000;97:3236–3241. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nature Rev. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 40.Slane BG, Aykin-Burns N, Smith BJ, et al. Mutation of succinate dehydrogenase subunit C results in increased O2.−, oxidative stress and genomic instability. Cancer Res. 2006;66:7615–7620. doi: 10.1158/0008-5472.CAN-06-0833. [DOI] [PubMed] [Google Scholar]
- 41.Holubec H, Payne CM, Bernstein H, et al. Assessment of apoptosis by immunohistochemical markers compared to cellular morphology in ex vivo-stressed colonic mucosa. J Histochem Cytochem. 2005;53:229–235. doi: 10.1369/jhc.4A6386.2005. [DOI] [PubMed] [Google Scholar]
- 42.Duesberg P, Li R. Multistep carcinogenesis. A chain reaction of aneuploidizations. Cell Cycle. 2003;2:202–210. [PubMed] [Google Scholar]
- 43.Pihan G, Doxsey S. Mutations and aneuploidy: Co-conspirators in cancer? Cancer Cell. 2003;4:89–94. doi: 10.1016/s1535-6108(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 44.Payne CM, Bernstein H, Bernstein C, Garewal H. Role of apoptosis in biology and pathology: resistance to apoptosis in colon carcinogenesis. Ultrastruct Pathol. 1995;19:221–248. doi: 10.3109/01913129509064227. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein C, Bernstein H, Garewal H, et al. A bile acid-induced apoptosis assay for colon cancer risk and associated quality control studies. Cancer Res. 1999;59:2353–57. [PubMed] [Google Scholar]
- 46.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 47.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 48.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 49.Geigl JB, Obenauf AC, Schwarzbraun T, et al. Defining ‘chromosomal instability.’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 51.Ribas M, Masramon L, Aiza G, Capella G, Miro R, Peinado MA. The structural nature of chromosomal instability in colon cancer cells. FASEB J. 2003;17:289–291. doi: 10.1096/fj.02-0425fje. [DOI] [PubMed] [Google Scholar]
- 52.Paraskeva C, Corfield AP, Harper S, Hague A, Audcent K, Williams AC. Colorectal carcinogenesis: sequential steps in the in vitro immortalization and transformation of human colonic epithelial cells (review) Anticancer Res. 1990;10:1189–1200. [PubMed] [Google Scholar]
- 53.Griffioen G, Cornelisse CJ, Verspaget HW, et al. Association of aneuploidy in index adenomas with metachronous colorectal adenoma development and a comparison. Cancer. 1992;70:2035–2043. doi: 10.1002/1097-0142(19921015)70:8<2035::aid-cncr2820700804>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 54.Bomme L, Bardi G, Pandis N, Fenger C, Kronberg O, Heim S. Cytogenetic analysis of colorectal adenomas: Karyotypic comparisons of synchronous tumors. Cancer Genet Cytogenet. 1998;106:66–71. doi: 10.1016/s0165-4608(98)00047-8. [DOI] [PubMed] [Google Scholar]
- 55.Bomme L, Lothe RA, Bardi G, Fenger C, Kronberg O, Heim S. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13, 20. Int J Cancer. 2001;92:816–823. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- 56.Shih IM, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–12. [PubMed] [Google Scholar]
- 57.Nowak MA, Komarova NL, Sengupta A, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci U S A. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He QJ, Zeng WF, Sham JS, et al. Recurrent genetic alterations in 26 colorectal carcinomas and 21 adenomas from Chinese patients. Cancer Genet Cytogenet. 2003;144:112–118. doi: 10.1016/s0165-4608(02)00959-7. [DOI] [PubMed] [Google Scholar]
- 59.Richter H, Slezak P, Walch A, et al. Distinct chromosomal imbalances in nonpolypoid and polypoid colorectal adenomas indicate different genetic pathways in the development of colorectal neoplasms. Amer J Pathol. 2003;163:287–294. doi: 10.1016/S0002-9440(10)63652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugai T, Takahashi H, Haban W, et al. Analysis of genetic alterations, classified according to their DNA ploidy pattern, in the progression of colorectal adenomas and early colorectal carcinomas. J Pathol. 2003;200:168–176. doi: 10.1002/path.1340. [DOI] [PubMed] [Google Scholar]
- 61.Giaretti W, Molinu S, Ceccarelli J, Prevosto C. Chromosomal instability, aneuploidy, and gene mutations in human sporadic colorectal adenomas. Cell Oncol. 2004;26:301–305. doi: 10.1155/2004/816591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postma C, Hermsen MAJA, Coffa J, et al. Chromosomal instability in flat adenomas and carcinomas of the colon. J Pathol. 2005;205:514–521. doi: 10.1002/path.1733. [DOI] [PubMed] [Google Scholar]
- 63.Grade M, Becker H, Liersch T, Ried T, Ghadimi BM. Molecular cytogenetics: Genomic imbalances in colorectal cancer and their clinical impact. Cellular Oncol. 2006;28:71–84. doi: 10.1155/2006/173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishiguro K, Yoshida T, Yagishita H, Numata Y, Okayasu T. Epithelial and stromal genetic instability contributes to genesis of colorectal adenomas. Gut. 2006;55:695–702. doi: 10.1136/gut.2005.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin ES, Tonon G, Sinha R, et al. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res. 2007;67:10736–10743. doi: 10.1158/0008-5472.CAN-07-2742. [DOI] [PubMed] [Google Scholar]
- 66.Oliva MR, Ripoli F, Muniz P, et al. Genetic alterations and oxidative metabolism in sporadic colorectal tumors from a Spanish community. Mol Carcinogen. 1997;18:232–243. [PubMed] [Google Scholar]
- 67.Blaker H, Graf M, Rieker RJ, Otto HF. Comparison of losses of heterozygosity and replication errors in primary colorectal carcinomas and corresponding liver metastases. J Pathol. 1999;188:258–262. doi: 10.1002/(SICI)1096-9896(199907)188:3<258::AID-PATH350>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 68.Thiagalingam S, Kaken S, Willson JKV, et al. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc Natl Acad Sci U S A. 2001;98:2698–2702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasnick D, Duesberg PH. How aneuploidy affects metabolic control and causes cancer. Biochem J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
- 70.Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer Lett. 1987;37:147–151. doi: 10.1016/0304-3835(87)90157-1. [DOI] [PubMed] [Google Scholar]
- 71.Pretlow TP, Barrow BJ, Ashton WS, et al. Aberrant crypts: putative preneoplastic foci in human colonic mucosa. Cancer Res. 1991;51:1564–1567. [PubMed] [Google Scholar]
- 72.Augenlicht LH, Richards C, Corner G, Pretlow TP. Evidence for genomic instability in human colonic aberrant crypt foci. Oncogene. 1996;12:1767–1772. [PubMed] [Google Scholar]
- 73.Siu IM, Robinson DR, Schwartz S, et al. The identification of monoclonality in human aberrant crypt foci. Cancer Res. 59:63–66. [PubMed] [Google Scholar]
- 74.Luo L, Li B, Pretlow TP. DNA alterations in human aberrant crypt foci and colon cancers by random primed polymerase chain reaction. Cancer Res. 2003;63:6166–6169. [PubMed] [Google Scholar]
- 75.Pretlow TP, Pretlow TG. Mutant KRAS in aberrant crypt foci (ACF): Initiation of colorectal cancer? Biochim Biophys Acta. 2005;1756:83–96. doi: 10.1016/j.bbcan.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 76.Luo L, Shen GQ, Stiffler KA, Wang QK, Pretlow TG, Pretlow TP. Loss of heterozygosity in human aberrant crypt foci (ACF), a putative precursor of colon cancer. Carcinogenesis. 2006;27:1153–1159. doi: 10.1093/carcin/bgi354. [DOI] [PubMed] [Google Scholar]
- 77.Nambiar PR, Nakanishi M, Gupta R, et al. Genetic signatures of high- and low-risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res. 2004;64:6394–6401. doi: 10.1158/0008-5472.CAN-04-0933. [DOI] [PubMed] [Google Scholar]
- 78.Steinbeck RG. Imbalance of morphologically addressed telophases reflects interphase DNA aneuploidy in tumorigenesis. Eur J Histochem. 1997;41:243–254. 1997. [PubMed] [Google Scholar]
- 79.Steinbeck RG. The DNA content of chromosome division figures and interphase nuclei classifies ulcerative colitis. Eur J Cancer. 1998;34:175–181. doi: 10.1016/s0959-8049(97)00371-7. [DOI] [PubMed] [Google Scholar]
- 80.Rabinovitch PS, Dziadon S, Brentnall TA, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59:5148–5153. [PubMed] [Google Scholar]
- 81.Willenbucher RF, Aust DE, Chang CG, et al. Genomic instability is an early event during the progression pathway of ulcerative-colitis-related neoplasia. Amer J Pathol. 1999;154:1825–1830. doi: 10.1016/S0002-9440(10)65438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takayama T, Miyanishi K, Hayashi T, et al. Aberrant crypt foci: detection, gene abnormalities, and clinical usefulness. Clin Gastroenterol Hepatol. 2005;3(7 Suppl 1):S42–S45. doi: 10.1016/s1542-3565(05)00257-0. [DOI] [PubMed] [Google Scholar]
- 83.Vyvoda OS, Coleman R, Holdsworth G. Effects of different bile salts upon the composition and morphology of a liver plasma membrane preparation. Deoxycholate is more membrane damaging than cholate and its conjugates. Biochim Biophys Acta. 1977;465:68–76. doi: 10.1016/0005-2736(77)90356-x. [DOI] [PubMed] [Google Scholar]
- 84.Sugimoto Y, Saito H, Tabeta R, et al. Binding of bile acids with rat colon and resultant perturbation of membrane organization as studied by uptake measurement and 31P nuclear magnetic resonance spectroscopy. Gann. 1984;75:798–808. [PubMed] [Google Scholar]
- 85.Akare S, Martinez JD. Bile acid induces hydrophobicity-dependent membrane alterations. Biochim Biophys Acta. 2005;1735:59–67. doi: 10.1016/j.bbalip.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 86.Jean-Louis S, Akare S, Ali MA, Mash EA, Jr, Meuillet E, Martinez JD. Deoxycholic acid induces intracellular signaling through membrane perturbations. J Biol Chem. 2006;281:14948–14960. doi: 10.1074/jbc.M506710200. [DOI] [PubMed] [Google Scholar]
- 87.DeRubertis FR, Craven PA, Saito R. Bile salt stimulation of colonic epithelial proliferation. Evidence for involvement of lipoxygenase products. J Clin Invest. 1984;74:1614–1624. doi: 10.1172/JCI111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Araki Y, Katoh T, Ogawa A, et al. Bile acid modulates transepithelial permeability via the generation of reactive oxygen species in the Caco-2 cell line. Free Radic Biol Med. 2005;39:769–780. doi: 10.1016/j.freeradbiomed.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 89.Qiao L, Studer E, Leach K, et al. Deoxycholic acid (DCA) causes ligand-independent activation of epidermal growth factor receptor (EGFR) and Fas receptor in primary hepatocytes: inhibition of EGFR/mitogen-activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol Biol Cell. 2001;12:2629–2645. doi: 10.1091/mbc.12.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Booth LA, Bilton RF. DNA and Free Radicals: Techniques, Mechanisms and Applications. London: OICA International; 1998. Genotoxic potential of the secondary bile acids: a role for reactive oxygen species, In: Arouma OI, Halliwell B (editors) pp. 161–177. [Google Scholar]
- 91.Werneburg NW, Yoon JH, Higuchi H, Gores GJ. Bile acids activate EGF receptor via a TGF-alpha-dependent mechanism in human cholangiocyte cell lines. Amer J Physiol Gastrointest Liver Physiol. 2003;285:G31–G36. doi: 10.1152/ajpgi.00536.2002. [DOI] [PubMed] [Google Scholar]
- 92.Merchant NB, Rogers CM, Trivedi B, Morrow J, Coffey RJ. Ligand-dependent activation of the epidermal growth factor receptor by secondary bile acids in polarizing colon cancer cells. Surgery. 2005;138:415–421. doi: 10.1016/j.surg.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 93.Yasuda H, Hirata S, Inoue K, Mashima H, Ohnishi H, Yoshiba M. Involvement of membrane-type bile acid receptor M-BAR/TGR5 in bile acid-induced activation of epidermal growth factor receptor and mitogen-activated protein kinases in gastric carcinoma cells. Biochem Biophys Res Comm. 2007;354:154–159. doi: 10.1016/j.bbrc.2006.12.168. [DOI] [PubMed] [Google Scholar]
- 94.Raimondi F, Santoro P, Barone MV, et al. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Amer J Physiol Gastrointest Liver Physiol. 2008;294:G906–G913. doi: 10.1152/ajpgi.00043.2007. [DOI] [PubMed] [Google Scholar]
- 95.Nakanishi H, Takeyama Y, Ohyanagi H, Saitoh Y, Takai Y. Mode of stimulatory action of deoxycholate in signal transduction system of isolated rat pancreatic acini. Biochem Biophys Res Comm. 1990;170:111–118. doi: 10.1016/0006-291x(90)91247-p. [DOI] [PubMed] [Google Scholar]
- 96.Reinehr R, Becker S, Keitel V, Eberle A, Grether-Beck S, Haussinger D. Bile salt-induced apoptosis involves NADPH oxidase isoform activation. Gastroenterol. 2005;129:2009–31. doi: 10.1053/j.gastro.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 97.Payne CM, Weber C, Crowley-Skillicorn C, et al. Deoxycholate induces mitochondrial oxidative stress and activates NF-κB through multiple mechanisms in HCT-116 colon epithelial cells. Carcinogenesis. 2007;28:215–222. doi: 10.1093/carcin/bgl139. [DOI] [PubMed] [Google Scholar]
- 98.Crowley-Weber CL, Payne CM, Gleason-Guzman M, et al. Development and molecular characterization of HCT-116 cell lines resistant to the tumor promoter and multiple stress-inducer, deoxycholate. Carcinogenesis. 2002;23:2063–2080. doi: 10.1093/carcin/23.12.2063. [DOI] [PubMed] [Google Scholar]
- 99.Payne CM, Crowley C, Washo-Stultz D, et al. The stress-response proteins poly(ADP-ribose) polymerase and NF-κB protect against bile salt-induced apoptosis. Cell Death Differ. 1998;5:623–36. doi: 10.1038/sj.cdd.4400395. [DOI] [PubMed] [Google Scholar]
- 100.Payne CM, Fass R, Bernstein H, et al. Pathogenesis of diarrhea in the adult: diagnostic challenges and life-threatening conditions. Eur J Gastroenterol Hepatol. 2006;18:1047–1051. doi: 10.1097/01.meg.0000231748.60889.be. [DOI] [PubMed] [Google Scholar]
- 101.Moschetta A, Portincasa P, Debellis L, et al. Basolateral Ca2+-dependent K+-channels play a key role in Cl− secretion induced by taurodeoxy-cholate from colon mucosa. Biol Cell. 2003;95:115–122. doi: 10.1016/s0248-4900(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 102.Mauricio AC, Slawik M, Heitzmann D, et al. Deoxycholic acid (DOC) affects the transport properties of distal colon. Pflugers Arch. 2000;439:532–540. doi: 10.1007/s004249900226. [DOI] [PubMed] [Google Scholar]
- 103.Oelberg DG, Wang LB, Sackman JW, Adcock EW, Lester R, Dubinsky WP. Bile salt-induced calcium fluxes in artificial phospholipid vesicles. Biochim Biophys Acta. 1988;937:289–299. doi: 10.1016/0005-2736(88)90251-9. [DOI] [PubMed] [Google Scholar]
- 104.Zimniak P, Little JM, Radominska A, Oelberg DG, Anwer MS, Lester R. Taurine-conjugated bile acids act as Ca2+ ionophores. Biochemistry. 1991;30:8598–8604. doi: 10.1021/bi00099a015. [DOI] [PubMed] [Google Scholar]
- 105.Tapia R, Velasco I. Ruthenium red as a tool to study calcium channels, neuronal death and the function of neural pathways. Neurochem Int. 1997;30:137–147. doi: 10.1016/s0197-0186(96)00056-3. [DOI] [PubMed] [Google Scholar]
- 106.Craven PA, Pfanstiel J, DeRubertis FR. Role of reactive oxygen in bile salt stimulation of colonic epithelial proliferation. J Clin Invest. 1986;77:850–859. doi: 10.1172/JCI112382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Casellas F, Mourelle M, Papo M, et al. Bile acid induced colonic irritation stimulates intracolonic nitric oxide release in humans. Gut. 1996;38:719–723. doi: 10.1136/gut.38.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venturi M, Hambly RJ, Glinghammer B, Rafter JJ, Rowland IR. Genotoxic activity in human faecal water and the role of bile acids: a study using the alkaline comet assay. Carcinogenesis. 1997;18:2353–2359. doi: 10.1093/carcin/18.12.2353. [DOI] [PubMed] [Google Scholar]
- 109.Booth LA, Gilmore IT, Bilton RF. Secondary bile acid induced DNA damage in HT29 cells: are free radicals involved? Free Rad Res. 1997;26:135–144. doi: 10.3109/10715769709097792. [DOI] [PubMed] [Google Scholar]
- 110.Washo-Stultz D, Crowley-Weber CL, Dvorakova K, et al. Role of mitochondrial complexes I and II, reactive oxygen species and arachidonic acid metabolism in deoxycholate-induced apoptosis. Cancer Lett. 2002;177:129–144. doi: 10.1016/s0304-3835(01)00786-8. [DOI] [PubMed] [Google Scholar]
- 111.Lechner S, Muller-Ladner U, Schlottmann K, et al. Bile acids mimic oxidative stress induced upregulation ot thioredoxin reductase in colon cancer cell lines. Carcinogenesis. 2002;23:1281–1288. doi: 10.1093/carcin/23.8.1281. [DOI] [PubMed] [Google Scholar]
- 112.Washo-Stultz D, Hoglen N, Bernstein H, Bernstein C, Payne CM. Role of nitric oxide and peroxynitrite in bile salt-induced apoptosis: Relevance to colon carcinogenesis. Nutr Cancer. 1999;33:180–188. doi: 10.1207/S15327914NC352_13. [DOI] [PubMed] [Google Scholar]
- 113.Crowley-Weber CL, Dvorakova K, Crowley C, et al. Nicotine increases oxidative stress, activates NF-κB and GRP78, induces apoptosis and sensitizes cells to genotoxic/xenobiotic stresses by a multiple stress inducer, deoxycholate: relevance to colon carcinogenesis. Chemico-Biological Interact. 2003;145:53–66. doi: 10.1016/s0009-2797(02)00162-x. [DOI] [PubMed] [Google Scholar]
- 114.Dall’Agnol M, Bernstein C, Bernstein H, Garewal H, Payne CM. Identification of S-nitrosylated proteins after chronic exposure of colonic epithelial cells to deoxycholate. Proteomics. 2006;6:1654–1662. doi: 10.1002/pmic.200500240. [DOI] [PubMed] [Google Scholar]
- 115.Bernstein H, Holubec H, Bernstein C, et al. Unique dietary-related mouse model of colitis. Inflamm Bowel Dis. 2006;12:278–93. doi: 10.1097/01.MIB.0000209789.14114.63. [DOI] [PubMed] [Google Scholar]
- 116.Bernstein H, Holubec H, Bernstein C, et al. Deoxycholate-induced colitis is markedly attenuated in Nos2 knock-out mice in association with modulation of gene expression profiles. Dig Dis Sci. 2007;52:628–642. doi: 10.1007/s10620-006-9608-0. [DOI] [PubMed] [Google Scholar]
- 117.Fang Y, Han SI, Mitchell C, et al. Bile acids induce mitochondrial ROS, which promote activation of receptor tyrosine kinases and signaling pathways in rat hepatocytes. Hepatology. 2004;40:961–971. doi: 10.1002/hep.20385. [DOI] [PubMed] [Google Scholar]
- 118.Glinghammar B, Inoue H, Rafter JJ. Deoxycholic acid causes DNA damage in colonic cells with subsequent induction of caspases, COX-2 promoter activity and transcription factors NF-κB and AP-1. Carcinogenesis. 2002;23:839–845. doi: 10.1093/carcin/23.5.839. [DOI] [PubMed] [Google Scholar]
- 119.Muhlbauer M, Allard B, Bosserhoff AK, et al. Differential effects of deoxycholic acid and taurodeoxycholic acid on NF-κB signal transduction and IL-8 gene expression in colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:G1000–G1008. doi: 10.1152/ajpgi.00338.2003. [DOI] [PubMed] [Google Scholar]
- 120.Lee DK, Park SY, Baik SK, et al. Deoxycholic acid-induced signal transduction in HT-29 cells: role of NF-kappa B and interleukin 8. Korean J Gastroenterol. 2004;43:176–185. [PubMed] [Google Scholar]
- 121.Shah SA, Volkov Y, Arfin Q, Abdel-Latif MM, Kelleher D. Urso-deoxycholic acid inhibits interleukin beta 1 and deoxycholic acid-induced activation of NF-kappaB and AP-1 in human colon cancer cells. Int J Cancer. 2006;118:532–539. doi: 10.1002/ijc.21365. [DOI] [PubMed] [Google Scholar]
- 122.Shah SA, Mahmud N, Mftah M, Roche HM, Kelleher D. Chronic but not acute linoleic acid treatment inhibits deoxycholic acid-induced protein kinase C and nuclear factor-kappaB activation in human colorectal cancer cells. Eur J Cancer Prev. 2006;15:125–133. doi: 10.1097/01.cej.0000195708.72072.42. [DOI] [PubMed] [Google Scholar]
- 123.Fang C, Dean J, Smith JW. A novel variant of ileal bile acid binding protein is up-regulated through nuclear factor-kappaB activation in colorectal adenocarcinoma. Cancer Res. 2007;67:9039–9046. doi: 10.1158/0008-5472.CAN-06-3690. [DOI] [PubMed] [Google Scholar]
- 124.Raufman JP, Shant J, Guo CY, Roy S, Cheng K. Deoxycholyltaurine rescues human colon cancer cells from apoptosis by activating EGFR-dependent PI3K/Akt signaling. J Cell Physiol. 2008;215:538–549. doi: 10.1002/jcp.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 126.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J Cell Biochem. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 127.Sankaran S, Crone DE, Palazzo RE, Parvin JD. Aurora-A kinase regulates breast cancer – associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 2007;67:11186–11194. doi: 10.1158/0008-5472.CAN-07-2578. [DOI] [PubMed] [Google Scholar]
- 128.Barzilai A, Yamamoto K-I. DNA damage responses to oxidative stress. DNA Repair. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 129.Langie SA, Knaapen AM, Houben JM, et al. The role of glutathione in the regulation of nucleotide excision repair during oxidative stress. Toxicol Lett. 2007;168:302–309. doi: 10.1016/j.toxlet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 130.Feng Z, Hu W, Marnett LJ, Tang MS. Malondialdehyde, a major endogenous lipid peroxidation product, sensitizes human cells to UV- and BPDE-induced killing and mutagenesis through inhibition of nucleotide excision repair. Mutat Res. 2006;601:125–136. doi: 10.1016/j.mrfmmm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 131.Chang CL, Marra G, Chauhan DP, et al. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol. 2002;283:C148–C154. doi: 10.1152/ajpcell.00422.2001. [DOI] [PubMed] [Google Scholar]
- 132.Hill JW, Hu JJ, Evans MK. OGG1 is degraded by calpain following oxidative stress and cisplatin exposure. DNA Repair (Amst) 2008;7:648–654. doi: 10.1016/j.dnarep.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bravard A, Vacher M, Gouget B, et al. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Graziewicz MA, Day BJ, Copeland WC. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res. 2002;30:2817–2824. doi: 10.1093/nar/gkf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gatz SA, Wiesmüller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 136.Qiao D, Gaitonde SV, Qi W, Martinez JD. Deoxycholic acid suppresses p53 by stimulating proteasome-mediated p53 protein degradation. Carcinogenesis. 2001;22:957–964. doi: 10.1093/carcin/22.6.957. [DOI] [PubMed] [Google Scholar]
- 137.Romagnolo DF, Chirnomas RB, Ku J, et al. Deoxycholate, an endogenous tumor promoter and DNA damaging agent, modulates BRCA-1 expression in apoptosis-sensitive epithelial cells: loss of BRCA-1 expression in colonic adenocarcinomas. Nutr Cancer. 2003;46:82–92. doi: 10.1207/S15327914NC4601_11. [DOI] [PubMed] [Google Scholar]
- 138.Bernstein H, Payne CM, Kunke K, et al. A proteomic study of resistance to deoxycholate-induced apoptosis. Carcinogenesis. 2004;25:681–692. doi: 10.1093/carcin/bgh072. [DOI] [PubMed] [Google Scholar]
- 139.Komichi D, Tazuma S, Nishioka T, Hyogo H, Chayama K. Glyco-chenodeoxycholate plays a carcinogenic role in immortalized mouse cholangiocytes via oxidative DNA damage. Free Rad Biol Med. 2005;39:1418–1427. doi: 10.1016/j.freeradbiomed.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 140.Kulkarni MS, Yielding KL. DNA damage and repair in epithelial (mucous) cells and crypt cells from isolated colon. Chem Biol Interact. 1985;52:311–318. doi: 10.1016/0009-2797(85)90026-2. [DOI] [PubMed] [Google Scholar]
- 141.Pool-Zobel BL, Leucht U. Induction of DNA damage by risk factors of colon cancer in human colon cells derived from biopsies. Mutat Res. 1997;375:105–115. doi: 10.1016/s0027-5107(97)00006-7. [DOI] [PubMed] [Google Scholar]
- 142.Scott DW, Mutamba S, Hopkins RG, Loo G. Increased GADD gene expression in human colon epithelial cells exposed to deoxycholate. J Cell Physiol. 2005;202:295–303. doi: 10.1002/jcp.20135. [DOI] [PubMed] [Google Scholar]
- 143.Rosignoli P, Fabiani R, De Bartolomeo A, Fuccelli R, Pelli MA, Morozzi G. Genotoxic effect of bile acids on human normal and tumour colon cells and protection by dietary antioxidants and butyrate. Eur J Nutr. 2008;47:301–309. doi: 10.1007/s00394-008-0725-8. [DOI] [PubMed] [Google Scholar]
- 144.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 145.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 146.Theisen J, Peters JH, Fein M, et al. The mutagenic potential of duodenoesophageal reflux. Ann Surg. 2005;241:63–68. doi: 10.1097/01.sla.0000150072.55037.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fein M, Peters JH, DeMeester TR. Carcinogenesis in reflux disease – in search for bile-specific effects. Microsurgery. 2007;27:647–650. doi: 10.1002/micr.20424. 2007. [DOI] [PubMed] [Google Scholar]
- 148.Narahara H, Tatsuta M, Iishi H, et al. K-ras point mutation is associated with enhancement by deoxycholic acid of colon carcinogenesis induced by azoxymethane, but not with its attenuation by all-trans-retinoic acid. Int J Cancer. 2000;88:157–161. doi: 10.1002/1097-0215(20001015)88:2<157::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 149.Watabe J, Bernstein H. The mutagenicity of bile acids using a fluctuation test. Mutat Res. 1985;158:45–51. doi: 10.1016/0165-1218(85)90096-5. [DOI] [PubMed] [Google Scholar]
- 150.Assinder SJ, Upshall A. Mitotic aneuploidy induced by sodium deoxycholate in Aspergillus nidulans. Mutat Res. 1982;93:101–108. doi: 10.1016/0027-5107(82)90128-2. [DOI] [PubMed] [Google Scholar]
- 151.Ferguson LR, Parry JM. Mitotic aneuploidy as a possible mechanism for tumour promoting activity in bile acids. Carcinogenesis. 1984;5:447–451. doi: 10.1093/carcin/5.4.447. [DOI] [PubMed] [Google Scholar]
- 152.Albertini S, Friederich U, Wurgler FE. Induction of mitotic chromosome loss in the diploid yeast Saccharomyces cerevisiae D61.M by genotoxic carcinogens and tumor promoters. Environ Mol Mutagen. 1988;11:497–508. doi: 10.1002/em.2850110410. [DOI] [PubMed] [Google Scholar]
- 153.Fenech M, Crott J, Turner J, Brown S. Necrosis, apoptosis, cytostasis and DNA damage in human lymphocytes measured simultaneously within the cytokinesis-block micronucleus assay: description of the method and results for hydrogen peroxide. Mutagenesis. 1999;14:605–612. doi: 10.1093/mutage/14.6.605. [DOI] [PubMed] [Google Scholar]
- 154.Fenech M, Bonassi S, Turner J, et al. Intra- and inter-laboratory variation in the scoring of micronuclei and nucleoplasmic bridges in binu-cleated human lymphocytes. Results of an international slide-scoring exercise by the HUMN project. Mutat Res. 2003;534:45–64. doi: 10.1016/s1383-5718(02)00248-6. [DOI] [PubMed] [Google Scholar]
- 155.Hunt CR, Sim JE, Sullivan SJ, et al. Genomic instability and catalase gene amplification induced by chronic exposure to oxidative stress. Cancer Res. 1998;58:3986–3992. [PubMed] [Google Scholar]
- 156.Limoli CL, Giedzinski E. Induction of chromosomal instability by chronic oxidative stress. Neoplasia. 2003;5:339–346. doi: 10.1016/S1476-5586(03)80027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Samper E, Nicholis DG, Melov S. Mitochondrial oxidative stress causes chromosomal instability of mouse embryonic fibroblasts. Aging Cell. 2003;2:277–285. doi: 10.1046/j.1474-9728.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 158.D’Angiolella V, Santarpia C, Grieco D. Oxidative stress overrides the spindle checkpoint. Cell Cycle. 2007;6:576–579. doi: 10.4161/cc.6.5.3934. [DOI] [PubMed] [Google Scholar]
- 159.Fang L, Fang G. Centromere cohesion: regulating the guardian. Cell Res. 2007;17:664–665. doi: 10.1038/cr.2007.68. [DOI] [PubMed] [Google Scholar]
- 160.Barber TD, McManus K, Yuen KW, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Green RA, Kaplan KB. Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by a dominant mutation in APC. J Cell Biol. 2003;163:949–961. doi: 10.1083/jcb.200307070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tomonaga T, Nomura F. Chromosome instability and kinetochore dysfunction. Histol Histopathol. 2007;22:191–197. doi: 10.14670/HH-22.191. [DOI] [PubMed] [Google Scholar]
- 163.Dash BC, El-Deiry WS. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- 164.Kops GJPL, Weaver BAA, Cleveland DW. On the road to cancer:aneuploidy and the mitotic checkpoint. Nature Rev. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 165.Perez de Castro I, de Carcer G, Malumbres M. A census of mitotic cancer genes:new insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- 166.Wang X, Cheung HW, Chun AC, Jin DY, Wong YC. Mitotic checkpoint defects in human cancers and their implications to chemotherapy. Front Biosci. 2008;13:2103–2114. doi: 10.2741/2827. [DOI] [PubMed] [Google Scholar]
- 167.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 168.Plentz RR, Wiemann SU, Flemming P, et al. Telomere shortening of epithelial cells characterises the adenomacarcinoma transition of human colorectal cancer. Gut. 2003;52:1304–1307. doi: 10.1136/gut.52.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 170.Bourke E, Dodson H, Merdes A, et al. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Burum-Auensen E, DeAngelis PM, Schjolberg AR, Roislien J, Andersen SN, Clausen OPF. Spindle proteins Aurora A and BUB1B, but not Mad2, are aberrantly expressed in dysplastic mucosa of patients with longstanding ulcerative colitis. J Clin Pathol. 2007;60:1403–1408. doi: 10.1136/jcp.2006.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Burum-Auensen E, De Angelis PM, Schjolberg AR, Kravik KL, Aure M, Clausen OPF. Subcellular localization of the spindle proteins aurora A, Mad2, and BUBR1 assessed by immunohistochemistry. J Histochem Cytochem. 2007;55:477–486. doi: 10.1369/jhc.6A7077.2007. [DOI] [PubMed] [Google Scholar]
- 173.Burum-Auensen E, De Angelis PM, Schjolberg AR, Roislien J, Mjaland O, Clausen OPF. Reduced level of the spindle checkpoint protein BUB1B is associated with aneuploidy in colorectal cancers. Cell Prolif. 2008;41:645–659. doi: 10.1111/j.1365-2184.2008.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barber TD, McManus K, Yuen KWY, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kang HC, Kim I-J, Jang S-G, et al. Coding region polymorphisms in the CHFR mitotic stress checkpoint gene are associated with colorectal cancer risk. Cancer Lett. 2008;260:170–179. doi: 10.1016/j.canlet.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 176.Spitz DR, Mackey MA, Li GC, Elwell JH, McCormick ML, Oberley LW. Relationship between changes in ploidy and stable cellular resistance to hydrogen peroxide. J Cell Physiol. 1989;139:592–598. doi: 10.1002/jcp.1041390320. [DOI] [PubMed] [Google Scholar]
- 177.Tarin JJ, Vendrell FJ, Ten J, Blanes R, Van Blerkom J, Cano A. The oxidizing agent tertiary butyl hydroperoxide induces disturbances in spindle organization, c-meiosis, and aneuploidy in mouse oocytes. Mol Human Reprod. 1996;2:895–901. doi: 10.1093/molehr/2.12.895. [DOI] [PubMed] [Google Scholar]
- 178.Karlhuber GM, Bauer HC, Eckl PM. Cytotoxic and genotoxic effects of 4-hydroxynonenal in cerebral endothelial cells. Mutat Res. 1997;381:209–216. doi: 10.1016/s0027-5107(97)00170-x. [DOI] [PubMed] [Google Scholar]
- 179.Tarin JJ, Vendrell FJ, Ten J, Cano A. Antioxidant therapy counteracts the disturbing effects of diamide and maternal ageing on meiotic division and chromosomal segregation in mouse oocytes. Mol Human Reprod. 1998;4:281–288. doi: 10.1093/molehr/4.3.281. [DOI] [PubMed] [Google Scholar]
- 180.Hagstrom KA, Meyer BJ. Condensin and cohesin: More than chromosome compactor and glue. Nature Reviews Genetics. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 181.Gerton J. Chromosome cohesion: A cycle of holding together and falling apart. PloS Biol. 2005;3:371–374. doi: 10.1371/journal.pbio.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim DS, Franklyn JA, Smith VE, et al. Securin induces genetic instability in colrectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis. 2007;28:749–759. doi: 10.1093/carcin/bgl202. [DOI] [PubMed] [Google Scholar]
- 183.Eytan E, Braunstein I, Ganoth D, et al. Two different mitotic checkpoint inhibitors of the anaphase-promoting complex/cyclosome antagonize the action of the activator Cdc20. Proc Natl Acad Sci U S A. 2008;105:9181–9185. doi: 10.1073/pnas.0804069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang X, Cheung HW, Chun AC, Jin DY, Wong YC. Mitotic checkpoint defects in human cancers and their implications to chemotherapy. Front Biosci. 2008;13:2103–2114. doi: 10.2741/2827. [DOI] [PubMed] [Google Scholar]
- 185.Emanuele MJ, Lan W, Jwa M, et al. Aurora B kinase and protein phos-phatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 187.Chang TS, Jeong W, Lee DY, Cho CS, Rhee SG. The RING-H2-finger protein APC11 as a target of hydrogen peroxide. Free Radic Biol Med. 2004;37:521–530. doi: 10.1016/j.freeradbiomed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 188.Ariyoshi K, Suzuki K, Goto M, Watanabe M, Kodama S. Increased chromosome instability and accumulation of DNA double-strand breaks in Werner syndrome cells. J Radiat Res. 2007;48:219–231. doi: 10.1269/jrr.07017. [DOI] [PubMed] [Google Scholar]
- 189.Zheng ZY, Bernstein H, Bernstein C, Payne CM, Martinez JD, Gerner EW. Bile acid activation of the gadd153 promoter and of p53-independent apoptosis: relevance to colon cancer. Cell Death Differ. 1996;3:407–414. [PubMed] [Google Scholar]
- 190.Bernstein H, Payne C, Bernstein C, Beard S, Schneider J, Crowley C. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108:37–46. doi: 10.1016/s0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 191.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Li X, Ding X, Adrian TE. Arsenic trioxide causes redistribution of cell cycle, caspase activation, and GADD expression in human colonic, breast, and pancreatic cancer cells. Cancer Invest. 2004;22:389–400. doi: 10.1081/cnv-200029068. [DOI] [PubMed] [Google Scholar]
- 193.Li J, Holbrook NJ. Elevated gadd153/chop expression and enhanced c-Jun N-terminal protein kinase activation sensitizes aged cells to ER stress. Exp Gerontol. 2004;39:735–744. doi: 10.1016/j.exger.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 194.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 195.Gotoh T, Mori M. Nitric oxide and endoplasmic reticulum stress. Arterioscler Thromb Vasc Biol. 2006;26:1439–1446. doi: 10.1161/01.ATV.0000223900.67024.15. [DOI] [PubMed] [Google Scholar]
- 196.Tamaki N, Hatano E, Taura K, et al. CHOP deficiency attenuates cholestasis-induced liver fibrosis by reduction of hepatocyte injury. Am J Physiol Gastrointest Liver Physiol. 2008;294:G498–G505. doi: 10.1152/ajpgi.00482.2007. [DOI] [PubMed] [Google Scholar]
- 197.Tagawa Y, Hiramatsu N, Kasai A, Hayakawa K, Okamura M. Induction of apoptosis by cigarette smoke via ROS-dependent endoplasmic reticulum stress and CCAAT/enhancer-binding protein-homologous protein (CHOP) Free Radic Biol Med. 2008;45:50–59. doi: 10.1016/j.freeradbiomed.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 198.Wu Y, Zhang H, Dong Y, Park YM, Ip C. Endoplasmic reticulum stress signal mediators are targets of selenium action. Cancer Res. 2005;65:9073–9079. doi: 10.1158/0008-5472.CAN-05-2016. [DOI] [PubMed] [Google Scholar]
- 199.Payne CM, Crowley-Weber CL, Dvorakova K, et al. Mitochondrial perturbation attenuates bile acid-induced cytotoxicity. Cell Biol Toxicol. 2005;21:215–231. doi: 10.1007/s10565-005-0166-6. [DOI] [PubMed] [Google Scholar]
- 200.Gerasimenko JV, Flowerdew SE, Voronina SG, et al. Bile acids induce Ca2+ release from both the endoplasmic reticulum and acidic intracellular calcium stores through activation of inositol triphosphate receptors and ryanodine receptors. J Biol Chem. 2006;281:40154–40163. doi: 10.1074/jbc.M606402200. [DOI] [PubMed] [Google Scholar]
- 201.Kawahara K, Oyadomari S, Gotoh T, Kohsaka S, Nakayama H, Mori M. Induction of CHOP and apoptosis by nitric oxide in p53-deficient microglial cells. FEBS Lett. 2001;506:135–139. doi: 10.1016/s0014-5793(01)02898-8. [DOI] [PubMed] [Google Scholar]
- 202.Arrington DD, Schnellmann RG. Targeting of the molecular chaperone oxygen-regulated protein 150 (ORP150) to mitochondria and its induction by cellular stress. Am J Physiol Cell Physiol. 2008;294:C641–C650. doi: 10.1152/ajpcell.00400.2007. [DOI] [PubMed] [Google Scholar]
- 203.Kass GE, Price SC. Role of mitochondria in drug-induced cholestatic injury. Clin Liver Dis. 2008;12:27–51. doi: 10.1016/j.cld.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 204.Rolo AP, Oliveira PJ, Moreno AJ, Palmeira CM. Chenodeoxycholate induction of mitochondrial permeability transition pore is associated with increased membrane fluidity and cytochrome c release: protective role of carvedilol. Mitochondrion. 2003;2:305–311. doi: 10.1016/S1567-7249(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 205.Wachs FP, Krieg RC, Rodrigues CMP, et al. Bile salt-induced apoptosis in human colon cancer cell lines involves the mitochondrial transmembrane potential but not CD95 (Fas/Apo-1) receptor. Int J Colorectal Dis. 2005;20:103–113. doi: 10.1007/s00384-004-0616-2. [DOI] [PubMed] [Google Scholar]
- 206.Lemasters JJ. Modulation of mitochondrial membrane permeability in pathogenesis, autophagy and control of metabolism. J Gastroenterol Hepatol. 2007;(Suppl 1):S31–S37. doi: 10.1111/j.1440-1746.2006.04643.x. [DOI] [PubMed] [Google Scholar]
- 207.Schmucker DL, Ohta M, Kanai S, Sato Y, Kitani K. Hepatic injury induced by bile salts: correlation between biochemical and morphological events. Hepatology. 1990;12:1216–1221. doi: 10.1002/hep.1840120523. [DOI] [PubMed] [Google Scholar]
- 208.Gumpricht E, Devereaux MW, Dahl RH, Sokol RJ. Glutathione status of isolated rat hepatocytes affects bile acid-induced cellular necrosis but not apoptosis. Toxicol Appl Pharmacol. 2000;164:102–111. doi: 10.1006/taap.2000.8894. [DOI] [PubMed] [Google Scholar]
- 209.Rolo AP, Palmeira CM, Holy JM, Wallace KB. Role of mitochondrial dysfunction in combined bile acid-induced cytotoxicity: the switch between apoptosis and necrosis. Toxicol Sci. 2004;79:196–204. doi: 10.1093/toxsci/kfh078. [DOI] [PubMed] [Google Scholar]
- 210.Borgognone M, Perez LM, Basiglio CL, Ochoa JE, Roma MG. Signaling modulation of bile salt-induced necrosis in isolated rat hepatocytes. Toxicol Sci. 2005;83:114–125. doi: 10.1093/toxsci/kfi012. [DOI] [PubMed] [Google Scholar]
- 211.Kobak GE, Dahl R, Devereaux MW, et al. Increased susceptibility of fat-laden Zucker-rat hepatocytes to bile acid-induced oncotic necrosis: an in vitro model of steatocholestasis. J Lab Clin Med. 2005;145:247–262. doi: 10.1016/j.lab.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 212.Garewal H, Bernstein H, Bernstein C, Sampliner R, Payne C. Reduced bile acid-induced apoptosis in “normal” colorectal mucosa: a potential biological marker for cancer risk. Cancer Res. 1996;56:1480–1483. [PubMed] [Google Scholar]
- 213.Bernstein H, Holubec H, Warneke JA, et al. Patchy field defects of apoptosis resistance and dedifferentiation in flat mucosa of colon resections from colon cancer patients. Ann Surg Oncol. 2002;9:505–517. doi: 10.1007/BF02557276. [DOI] [PubMed] [Google Scholar]
- 214.Hague A, Elder DJE, Hicks DJ, Paraskeva C. Apoptosis in colorectal tumour cells: Induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int J Cancer. 1995;60:400–406. doi: 10.1002/ijc.2910600322. [DOI] [PubMed] [Google Scholar]
- 215.Marchetti MC, Migliorati G, Moraca R, et al. Possible mechanisms involved in apoptosis of colon tumor cell lines induced by deoxycholic acid, short-chain fatty acids and their mixtures. Nutr Cancer. 1997;28:74–80. doi: 10.1080/01635589709514555. [DOI] [PubMed] [Google Scholar]
- 216.Milovic V, Teller IC, Faust D, Caspary WF, Stein J. Effects of deoxycholate on human colon cancer cells: apoptosis or proliferation. Eur J Clin Invest. 2002;32:29–34. doi: 10.1046/j.0014-2972.2001.00938.x. [DOI] [PubMed] [Google Scholar]
- 217.Martinez JD, Stratagoules ED, LaRue JM, et al. Different bile acids exhibit distinct biological effects: The tumor promoter deoxycholic acid induces apoptosis and the chemopreventive agent ursodeoxycholic acid inhibits cell proliferation. Nutr Cancer. 1998;31:111–118. doi: 10.1080/01635589809514689. [DOI] [PubMed] [Google Scholar]
- 218.Yui S, Saeki T, Kanamoto R, Iwami K. Characteristics of apoptosis in HCT116 colon cancer cells induced by deoxycholic acid. J Biochem (Tokyo) 2005;138:151–157. doi: 10.1093/jb/mvi106. [DOI] [PubMed] [Google Scholar]
- 219.Crowley CL, Payne CM, Bernstein H, Bernstein C, Roe D. The NAD+ precursors, nicotinic acid and nicotinamide protect cells against apoptosis induced by a multiple stress inducer, deoxycholate. Cell Death Diff. 2000;7:314–326. doi: 10.1038/sj.cdd.4400658. [DOI] [PubMed] [Google Scholar]
- 220.Washo-Stultz D, Crowley C, Payne CM, et al. Increased susceptibility of cells to inducible apoptosis during growth from early to late log phase: an important caveat for in vitro apoptosis research. Toxicol Lett. 2000;116:199–207. doi: 10.1016/s0378-4274(00)00225-3. [DOI] [PubMed] [Google Scholar]
- 221.Haza AI, Glinghammar B, Grandien A, Rafter J. Effect of colonic luminal components on induction of apoptosis in human colonic cell lines. Nutr Cancer. 2000;36:79–89. doi: 10.1207/S15327914NC3601_12. [DOI] [PubMed] [Google Scholar]
- 222.Schlottmann K, Wachs FP, Krieg RC, Kullmann F, Scholmerich J, Rogier G. Characterization of bile salt-induced apoptosis in colon cancer cell lines. Cancer Res. 2000;60:4270–4276. 2000. [PubMed] [Google Scholar]
- 223.Powolny A, Xu J, Loo G. Deoxycholate induces DNA damage and apoptosis in human colon epithelial cells expressing either mutant or wild-type p53. Int J Biochem Cell Biol. 2001;33:193–203. doi: 10.1016/s1357-2725(00)00080-7. 2001. [DOI] [PubMed] [Google Scholar]
- 224.Payne CM, Waltmire CN, Crowley C, et al. Caspase-6 mediated cleavage of guanylate cyclase alpha 1 during deoxycholate-induced apoptosis: Protective role of nitric oxide signaling module. Cell Biol Toxicol. 2003;19:373–392. doi: 10.1023/b:cbto.0000013331.70391.0e. [DOI] [PubMed] [Google Scholar]
- 225.Krahenbuhl S, Talos C, Fischer S, Reichen J. Toxicity of bile acids on the electron transport chain of isolated rat liver mitochondria. Hepatology. 1994;19:471–479. doi: 10.1002/hep.1840190228. 1994. [DOI] [PubMed] [Google Scholar]
- 226.Rolo AP, Oliveira PJ, Moreno AJ, Palmeira CM. Bile acids affect liver mitochondrial bioenergetics: possible relevance for cholestasis therapy. Toxicol Sci. 2000;57:177–185. doi: 10.1093/toxsci/57.1.177. [DOI] [PubMed] [Google Scholar]
- 227.Palmeira CM, Rolo AP. Mitochondrially-mediated toxicity of bile acids. Toxicology. 2004;203:1–15. doi: 10.1016/j.tox.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 228.Sola S, Brito MA, Brites D, Moura JJ, Rodrigues CM. Membrane structural changes support the involvement of mitochondria in the bile salt-induced apoptosis of rat hepatocytes. Clin Sci (Lond) 2002;103:475–485. doi: 10.1042/cs1030475. [DOI] [PubMed] [Google Scholar]
- 229.Sokol RJ, Dahl R, Devereaux MW, Yerushalmi B, Kobak GE, Gumpricht E. Human hepatic mitochondria generate reactive oxygen species and undergo the permeability transition in response to hydrophobic bile acids. J Pediatr Gastroenterol Nutr. 2005;41:235–243. doi: 10.1097/01.mpg.0000170600.80640.88. [DOI] [PubMed] [Google Scholar]
- 230.Tan KP, Yang M, Ito S. Activation of nuclear factor (erythroid-2 like) factor 2, by toxic bile acids provokes adaptive defense responses to enhance cell survival at the emergence of oxidative stress. Mol Pharmacol. 2007;72:1380–1390. doi: 10.1124/mol.107.039370. [DOI] [PubMed] [Google Scholar]
- 231.Payne CM, Bernstein C, Bernstein H. Apoptosis overview emphasizing the role of oxidative stress, DNA damage and signal-transduction pathways. Leuk Lymphoma. 1995;19:43–93. doi: 10.3109/10428199509059662. [DOI] [PubMed] [Google Scholar]
- 232.Longpre JM, Loo G. Paradoxical effect of diphenyleneiodonium in inducing DNA damage and apoptosis. Free Radic Res. 2008;42:533–543. doi: 10.1080/10715760802126692. [DOI] [PubMed] [Google Scholar]
- 233.Rokutan K, Kawahara T, Kuwano Y, Tominaga K, Sekiyama A, Teshima-Kondo S. NADPH oxidases in the gastrointestinal tract: a potential role of Nox1 in innate immune response and carcinogenesis. Antioxid Redox Signal. 2006;8:1573–82. doi: 10.1089/ars.2006.8.1573. [DOI] [PubMed] [Google Scholar]
- 234.Komatsu D, Kato M, Nakayama J, Miyagawa S, Kamata T. NADPH oxidase 1 plays a critical mediating role in oncogenic Ras-induced vascular endothelial growth factor expression. Oncogene. 2008;27:4724–4732. doi: 10.1038/onc.2008.102. [DOI] [PubMed] [Google Scholar]
- 235.Laurent E, McCoy JW, 3rd, Macina RA, et al. Nox1 is over-expressed in human colon cancers and correlates with activating mutations in K-Ras. Int J Cancer. 2008;123:100–107. doi: 10.1002/ijc.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nisimoto Y, Tsubouchi R, Diebold BA, et al. Activation of NADPH oxidase 1 in tumor colon epithelial cell. Biochem J. 2008;415:57–65. doi: 10.1042/BJ20080300. [DOI] [PubMed] [Google Scholar]
- 237.Cocco T, Di Paola M, Papa S, Lorusso M. Arachidonic acid interaction with the mitochondrial electron transport chain promotes reactive oxygen species generation. Free Radic Biol Med. 1999;27:51–59. doi: 10.1016/s0891-5849(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 238.Becker S, Reinehr R, Grether-Beck S, Eberle A, Haussinger D. Hydrophobic bile salts trigger ceramide formation through endosomal acidification. Biol Chem. 2007;388:185–196. doi: 10.1515/BC.2007.021. [DOI] [PubMed] [Google Scholar]
- 239.Zhang XF, Li BX, Dong CY, Ren R. Apoptosis of human colon carcinoma HT-29 cells induced by ceramide. World J Gastroenterol. 2006;12:3581–3584. doi: 10.3748/wjg.v12.i22.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Magnuson BA, Shirtliff N, Bird RP. Resistance of aberrant crypt foci to apoptosis induced by azoxymethane in rats chronically fed cholic acid. Carcinogenesis. 1994;15:1459–1462. doi: 10.1093/carcin/15.7.1459. [DOI] [PubMed] [Google Scholar]
- 241.Bernstein H, Prasad A, Holubec H, et al. Reduced Pms2 expression in non-neoplastic flat mucosa from patients with colon cancer correlates with reduced apoptosis competence. Appl Immunohistochem Mol Morphol. 2006;14:166–172. doi: 10.1097/01.pai.0000170533.61579.b9. [DOI] [PubMed] [Google Scholar]
- 242.Hill MJ, Drasar BS, Aries VC, Crowther JS, Hawksworth GS, Williams REO. Bacteria and etiology of cancer of large bowel. Lancet. 1971;1:95–100. doi: 10.1016/s0140-6736(71)90837-3. [DOI] [PubMed] [Google Scholar]
- 243.Hill MJ, Taylor AJ, Thompson MH, Wait R. Faecal steroids and urinary volatile phenols in four Scandinavian populations. Nutr Cancer. 1982;4:67–73. doi: 10.1080/01635588209513740. [DOI] [PubMed] [Google Scholar]
- 244.Hill MJ. Bile flow and colon cancer. Mutat Res. 1990;238:313–320. doi: 10.1016/0165-1110(90)90023-5. [DOI] [PubMed] [Google Scholar]
- 245.Reddy BS, Wynder EL. Large bowel carcinogenesis: faecal constituents of populations with diverse incidence ratio of colon cancer. J Natl Cancer Inst. 1973;50:1437–1442. doi: 10.1093/jnci/50.6.1437. [DOI] [PubMed] [Google Scholar]
- 246.Reddy BS, Wynder EL. Metabolic epidemiology of colon cancer: fecal bile acids and neutral sterols in colon cancer patients and patients with adenomatous polyps. Cancer. 1977;39:2533–2539. doi: 10.1002/1097-0142(197706)39:6<2533::aid-cncr2820390634>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 247.Reddy BS, Hedges AR, Laakso K, Wyndeer EL. Metabolic epidemiology of large bowel cancer: fecal bulk and constituents of high-risk North American and low-risk Finnish populations. Cancer. 1978;42:2832–2838. doi: 10.1002/1097-0142(197812)42:6<2832::aid-cncr2820420644>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 248.Crowther JS, Drasar BS, Hill MJ, et al. Faecal steroids and bacteria and large bowel cancer in Hong Kong by socioeconomic groups. Br J Cancer. 1976;34:191–198. doi: 10.1038/bjc.1976.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jensen OM, MacLennan R, Wahrendorf J. Diet, bowel function, fecal characteristics, and large bowel cancer in Denmark and Finland. Nutr Cancer. 1982;4:5–19. doi: 10.1080/01635588209513733. [DOI] [PubMed] [Google Scholar]
- 250.Domellof L, Darby L, Hanson D, Mathews L, Simi B, Reddy BS. Fecal sterols and bacterial β-glucuronidase activity: A preliminary metabolic epidemiology study of healthy volunteers from Umea, Sweden, and metropolitan New York. Nutr Cancer. 1982;4:120–127. doi: 10.1080/01635588209513747. [DOI] [PubMed] [Google Scholar]
- 251.Cheah PY. Hypotheses for the etiology of colorectal cancer – An overview. Nutr Cancer. 1990;14:5–13. doi: 10.1080/01635589009514073. [DOI] [PubMed] [Google Scholar]
- 252.Munch A, Strom M, Soderholm JD. Dihydroxy bile acids increase mucosal permeability and bacterial uptake in human colon biopsies. Scand J Gastroenterol. 2007;42:1167–1174. doi: 10.1080/00365520701320463. [DOI] [PubMed] [Google Scholar]
- 253.Bernstein C, Bernstein H, Payne CM, Dvorak K, Garewal H. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260:1–10. doi: 10.1016/j.canlet.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction. Nature Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 255.Polley ACJ, Mulholland F, Pin C, et al. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal neoplasia. Cancer Res. 2006;66:6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- 256.Chen LC, Hao CY, Chiu YSY, et al. Alteration of gene expression in normal-appearing colon mucosa of APCmin mice and human cancer patients. Cancer Res. 2004;64:3694–3700. doi: 10.1158/0008-5472.CAN-03-3264. [DOI] [PubMed] [Google Scholar]
- 257.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 268.Hutchins JT, Reading CL, Giavazzi R, Hoaglund J, Jessup JM. Distribution of mono-, di, and tri-O-acetylated sialic acids in normal and neoplastic colon. Cancer Res. 1988;48:483–489. [PubMed] [Google Scholar]
- 259.Shen Y, Tiralongo J, Kohla G, Schauer R. Regulation of sialic acid O-acetylation in human colon mucosa. Biol Chem. 2004;385:145–152. doi: 10.1515/BC.2004.033. [DOI] [PubMed] [Google Scholar]
- 260.Shen Y, Kohla G, Lrhorfi AL, et al. O-acetylation and de-O-acetylation of sialic acids in human colorectal carcinoma. Eur J Biochem. 2004;271:281–290. doi: 10.1046/j.1432-1033.2003.03927.x. [DOI] [PubMed] [Google Scholar]
- 261.Anti M, Armuzzi A, Morini S, et al. Severe imbalance of cell proliferation and apoptosis in the left colon and in the rectosigmoid tract in subjects with a history of large adenomas. Gut. 2001;48:238–246. doi: 10.1136/gut.48.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Badvie S, Hanna-Morris A, Andreyev HJN, Cohen P, Saini S, Allen-Mersh TG. A “field change” of inhibited apoptosis occurs in colorectal mucosa adjacent to colorectal adenocarcinoma. J Clin Pathol. 2006;59:942–946. doi: 10.1136/jcp.2005.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bronner MP, Culin C, Reed JC, Furth EE. The bcl-2 proto-oncogene and the gastrointestinal epithelial tumor progression model. Am J Pathol. 1995;146:20–26. [PMC free article] [PubMed] [Google Scholar]
- 264.Sarela AI, Macadam RCA, Farmery SM, Markham AF, Guillou PJ. Expression of the antiapoptosis gene survivin predicts death from recurrent colorectal carcinoma. Gut. 2000;46:645–650. doi: 10.1136/gut.46.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Greaves LC, Preston SL, Tadrous PJ, et al. Mitochondrial DNA mutations are established in human clonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Payne CM, Holubec H, Bernstein C, et al. Crypt-restricted loss and decreased protein expression of cytochrome C oxidase subunit I as potential hypothesis-driven biomarkers of colon cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2066–2075. doi: 10.1158/1055-9965.EPI-05-0180. [DOI] [PubMed] [Google Scholar]
- 267.Scalmati A, Lipkin M. Proliferation and differentiation biomarkers in colorectal mucosa and their application to chemoprevention studies. Environ Health Perspect. 1993;99:169–173. doi: 10.1289/ehp.9399169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Meyerhardt JA, Niedzwiecki D, Hollis D, et al. Association of dietary patterns with cancer recurrence and survival in patients with stage III colon cancer. JAMA. 2007;298:754–764. doi: 10.1001/jama.298.7.754. [DOI] [PubMed] [Google Scholar]
- 269.Bartram HP, Scheppach W, Schmid H, et al. Proliferation of human colonic mucosa as an intermediate biomarker of carcinogenesis: effects of butyrate, deoxycholate, calcium, ammonia, and pH. Cancer Res. 1993;53:3283–3288. [PubMed] [Google Scholar]
- 270.Bartram HP, Englert S, Scheppach W, et al. Antagonistic effects of deoxycholic acid and butyrate on epithelial cell proliferation in the proximal and distal human colon. Z Gastroenterol. 1994;32:389–392. [PubMed] [Google Scholar]
- 271.Velazquez OC, Zhou D, Seto RW, et al. In vivo crypt surface hyper-proliferation is decreased by butyrate and increased by deoxycholate in normal rat colon: associated in vivo effects on c-Fos and c-Jun expression. JPEN J Parenter Enteral Nutr. 1996;20:243–250. doi: 10.1177/0148607196020004243. [DOI] [PubMed] [Google Scholar]
- 272.Velazquez OC, Seto RW, Choi J, et al. Butyrate inhibits deoxycho-late-induced increase in colonic mucosal DNA and protein synthesis in vivo. Dis Colon Rectum. 1997;40:1368–1375. doi: 10.1007/BF02050825. [DOI] [PubMed] [Google Scholar]
- 273.Rosignoli P, Fabiani R, De Bartolomeo A, et al. Protective ativity of butyrate on hydrogen peroxide-induced DNA damage in isolated human colonocytes and HT29 tumour cells. Carcinogenesis. 2001;22:1675–1680. doi: 10.1093/carcin/22.10.1675. [DOI] [PubMed] [Google Scholar]
- 274.Abrahamse SL, Pool-Zobel BL, Rechkemmer G. Potential of short chain fatty acids to modulate the induction of DNA damage and changes in the intracellular calcium concentration by oxidative stress in isolated rat distal colon cells. Carcinogenesis. 1999;20:629–634. doi: 10.1093/carcin/20.4.629. [DOI] [PubMed] [Google Scholar]
- 275.Heerdt BG, Halsey HK, Lipkin M, Augenlicht LH. Expression of mitochondrial cytochrome c oxidase in human colonic cell differentiation, transformation, and risk for colonic cancer. Cancer Res. 1990;50:1596–1600. [PubMed] [Google Scholar]
- 276.Heerdt BG, Augenlicht LH. Effects of fatty acids on expression of genes encoding subunits of cytochrome c oxidase and cytochrome c oxidase activity in HT29 human colonic adenocarcinoma cells. J Biol Chem. 1991;266:19120–19126. [PubMed] [Google Scholar]
- 277.Scheppach W, Muller JG, Boxberger F, et al. Histological changes in the colonic mucosa following irrigation with short-chain fatty acids. Eur J Gastroenterol Hepatol. 1997;9:163–168. doi: 10.1097/00042737-199702000-00010. [DOI] [PubMed] [Google Scholar]
- 278.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 279.Bordonaro M, Lazarova DL, Augenlicht LH, Sartorelli AC. Cell type- and promoter-dependent modulation of the Wnt signaling pathway by sodium butyrate. Int J Cancer. 2002;97:42–51. doi: 10.1002/ijc.1577. [DOI] [PubMed] [Google Scholar]
- 280.Williams EA, Coxhead JM, Mathers JC. Anti-cancer effects of butyrate: use of micro-array technology to investigate mechanisms. Proc Nutr Soc. 2003;62:107–115. doi: 10.1079/PNS2002230. [DOI] [PubMed] [Google Scholar]
- 281.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 282.Pool-Zobel BL, Sauer J. Overview of experimental data on reduction of colorectal cancer risk by inulin-type fructans. J Nutr. 2007;137(11 Suppl):2580S–2584S. doi: 10.1093/jn/137.11.2580S. [DOI] [PubMed] [Google Scholar]
- 283.Kim YS, Milner JA. Dietary modulation of colon cancer risk. J Nutr. 2007;137(11 Suppl):2576S–2579S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 284.Stempelj M, Kedinger M, Augenlicht L, Klampfer L. Essential role of the JAK/STAT1 signaling pathway in the expression of inducible nitric-oxide synthase in intestinal epithelial cells and its regulation by butyrate. J Biol Chem. 2007;282:9797–9804. doi: 10.1074/jbc.M609426200. [DOI] [PubMed] [Google Scholar]
- 285.Bordonaro M, Lazarova DL, Sartorelli AC. Butyrate and Wnt signaling: a possible solution to the puzzle of dietary fiber and colon cancer risk? Cell Cycle. 2008;7:1178–1183. doi: 10.4161/cc.7.9.5818. [DOI] [PubMed] [Google Scholar]
- 286.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 287.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 289.Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 290.Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 291.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterol. 2007;132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 292.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 293.Attene-Ramos MA, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 294.Attene-Ramos MA, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 295.Baskar R, Li L, Moore PK. Hydrogen sulfide-induces DNA damage and changes in apoptotic gene expression in human lung fibroblast cells. FASEB J. 2007;21:247–255. doi: 10.1096/fj.06-6255com. [DOI] [PubMed] [Google Scholar]
- 296.Christl SU, Eisner HD, Dusel G, Kasper H, Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci. 1996;41:2477–2481. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- 297.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 298.Christl SU, Bartram HP, Paul A, Kelber E, Scheppach W, Kasper H. Bile acid metabolism by colonic bacteria in continuous culture: effects of starch and pH. Ann Nutr Metab. 1997;41:45–51. doi: 10.1159/000177977. [DOI] [PubMed] [Google Scholar]
- 299.Blottiere HM, Buecher B, Galmiche JP, Cherbut C. Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proc Nutr Soc. 2003;62:101–106. doi: 10.1079/PNS2002215. [DOI] [PubMed] [Google Scholar]
- 300.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Biol Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 301.Wollowski I, Rechkemmer G, Pool-Zobel BL. Protective role of probiotics and prebiotics in colon cancer. Amer J Clin Nutr. 2001;73(2 Suppl):451S–455S. doi: 10.1093/ajcn/73.2.451s. [DOI] [PubMed] [Google Scholar]
- 302.Pool-Zobel B, Veeriah S, Bohmer FD. Modulation of xenobiotic metabolizing enzymes by anticarcinogens – focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mutat Res. 2005;591:74–92. doi: 10.1016/j.mrfmmm.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 303.Young GP, Hu Y, Le Leu RK, Nyskohus L. Review. Dietary fibre and colorectal cancer: A model for environment – gene interactions. Mol Nutr Food Res. 2005;49:571–584. doi: 10.1002/mnfr.200500026. [DOI] [PubMed] [Google Scholar]
- 304.Coxhead JM, Williams ES, Mathers JC. DNA mismatch repair status may influence anti-neoplastic effects of butyrate. Biochem Soc Trans. 2005;33:728–729. doi: 10.1042/BST0330728. [DOI] [PubMed] [Google Scholar]