Abstract
Inherited chronic renal diseases are associated with failures in glomerular filtration and tubular resorption. Such failures invariably result from defects in selective filtration and absorption in surface renal epithelium. Recently, we described an autosomal recessive chronic interstitial nephritis with diffuse zonal fibrosis (CINF) in cattle. Bovine CINF, characterized by increased blood urea nitrogen, creatinine, and urinary proteins, leads to lethality before puberty, usually within the first 6 months or year of life. Here, we demonstrate that the first four exons of PCLN-1/Claudin-16 (CL-16), which encodes a member of the claudin family of tight junction proteins, were deleted in CINF-affected cattle. CL-16 was expressed preferentially in kidneys of normal cattle, but transcripts were totally absent in affected offspring. This observation suggests that the lack of CL-16 protein contributes to the dysfunction of paracellular renal transport systems.
[The CL-16 cDNA sequence has been deposited at GenBank under accession no. AB030082.]
Recently, we described a novel autosomal recessive renal disorder, chronic interstitial nephritis with diffuse zonal fibrosis (CINF) in Japanese Black (Wagyu) cattle (Kobayashi et al. 2000). CINF was diagnosed preliminarily by increased blood urea nitrogen, creatinine, and urinary protein levels. The symptoms suggest that CINF results from defects in selective filtration and absorption in the surface renal epithelium. After examining pedigree structures, a sire was selected as a possible progenitor for the disease. Affected cattle were produced by selective matings between the sire and his daughter or between his son and daughter. The CINF locus was assigned to the central region of bovine chromosome 1 (BTA1; P < 3.4× 10−10) close to a microsatellite marker BM9019 (Kobayashi et al. 2000). In this paper, we describe the identification of a Claudin-16 (CL-16) mutation that is strongly associated with bovine CINF.
Claudin proteins are components of tight junction strands and have four transmembrane domains (Furuse et al. 1998; Morita et al. 1999; Tsukita et al. 1999). They show specific tissue distribution patterns depending on the claudin species. Recently Paracellin-1 (PCLN-1), a new member of the claudin gene family, was reported to be a causative gene for renal hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHN) and was localized to HSA3q (Simon et al. 1999). The human–cattle comparative map shows that HSA3q corresponds to BTA1 where CL-16 is located. The nucleotide sequence of CL-16 has ∼90% homology to that of PCLN-1. Thus, we conclude that CL-16 is a cattle ortholog of PCLN-1. Because FHHN has been characterized by renal Mg2+ and Ca2+ wastings (Praga et al. 1995), the product of PCLN-1/CL-16 is likely to be required for selective paracellular conductance and to form an intercellular pore permitting paracellular passage of Mg2+ and Ca2+ on tight junction of renal tubule. Although PCLN-1/CL-16 mutations cause renal disorders FHHN and CINF, the clinical features of both diseases are quite different. Therefore, these two clinically dissimilar diseases may be related through specific mutations/deletions in the same gene or through species specificity.
RESULTS
The CINF critical region was mapped to an ∼10-cM region between microsatellite markers BMS4030 and INRA119 (Fig. 1a). To locate the CINF region physically and to isolate additional informative microsatellite markers, bovine YAC clones harboring BM9019, a marker segregating with CINF, were isolated. Of the isolated YAC clones, clone 249E9, harboring both BM9019 and BMS4009, was chosen to prepare a cosmid library. Four cosmid clones (cos68, cos54, cos23, and cos48) were also mapped by FISH to the same region of BTA1. BM9019 was detected in cos68 and cos54, and BMS4009 was contained in cos54 and cos23. Digestion with restriction enzymes followed by Southern blot analysis confirmed that cos23 overlapped with cos48, and cos54 overlapped with both cos68 and cos23. To determine the physical relationship between cos68 and cos23, PCR primers 68-2F and 23-3R, which amplify an ∼250-bp PCR product, were designed for end sequences of cos68 and cos23, respectively. The sequence of the PCR product revealed that cos68 and cos23 stand end to end. The cosmid contig composed of cos68, cos23 and cos48 spans ∼100 kb.
Figure 1.
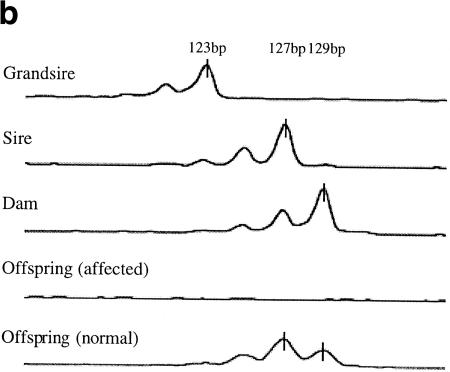
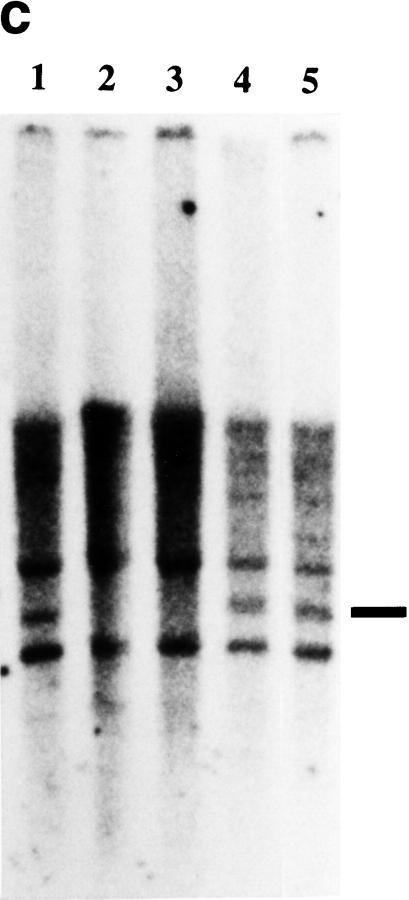
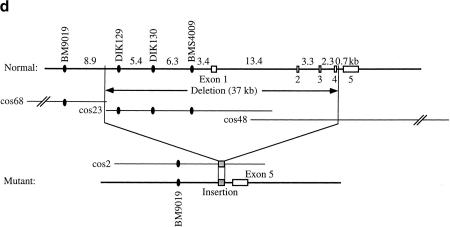
(a) Genetic (top) and physical (bottom) maps of the CINF critical region. The cosmid contig spans ∼100 kb. (E) EcoRI; (S) SacI; (X) XhoI. (b) Genotyping of DIK130. Sire and dam are offspring of grandsire. Both offspring are full siblings from sire and dam. The 123-bp allele of grandsire was not transmitted to sire and dam. (c) Southern blot of CINF-affected cattle. Genomic DNA from normal (lane 5), carrier (lanes 1,4) , and CINF-affected cattle (lanes 2,3) was digested with EcoRI. The blot was hybridized to a probe of 0.8 kb from the 5′ half of cDNA 55. Bar, 3.0-kb deleted DNA fragment. (d) Genomic structure of CL-16 and the deleted region. CL-16 is composed of five exons. The deleted region spans 37 kb and contains exons 1–4 of CL-16. In the mutant allele, there is an insertion at the deletion point. (e) Sequence of the deleted region. The G-rich inserted sequence is 31 bp.
Two microsatellite loci, DIK129 and DIK130, were isolated from cos23. DIK130 exhibited a single allele in the disease founder grandsire. However, the DIK130 allele of the founder grandsire was not transmitted to either his son or daughter, indicating transmission from the founder of a null allele (Fig. 1b). A null allele was also observed with BMS4009 and DIK129, respectively, strongly suggesting the presence of deletion in the cos23 region.
This finding suggested that the deletion was associated with a loss-of-function mutation of CINF. To isolate a CINF candidate gene, a bovine kidney cDNA library was screened with cos23, and cos48 as probes. A single 2.9-kb cDNA clone, number 55, was isolated with cos23 and cos48. To determine whether the coding region of cDNA 55 was deleted in CINF-affected cattle, Southern blot analysis was performed with 0.8 kb of the 5′ half of cDNA 55 as a probe. Figure 1c shows the 3.0-kb fragment deleted in CINF-affected cattle. It is likely that the CINF deletion is in the 100-kb cosmid contig. To compare the normal 100-kb cosmid contig sequence with the mutant sequence, a mutant cosmid library was prepared from CINF-affected cattle DNA. A mutant cosmid clone designated mutant-cos2 was isolated with BM9019 and cDNA 55 as probes. To identify the deleted region, cos68, cos23, cos48, and mutant-cos2 were shotgun sequenced. On the basis of the sequence of mutant-cos2, the deleted region was determined to contain all of cos23 and parts of cos68 and cos48 and to be 37 kb (Fig. 1d). The deleted region, however, contained a G-rich fragment at the deletion point (Fig. 1e).
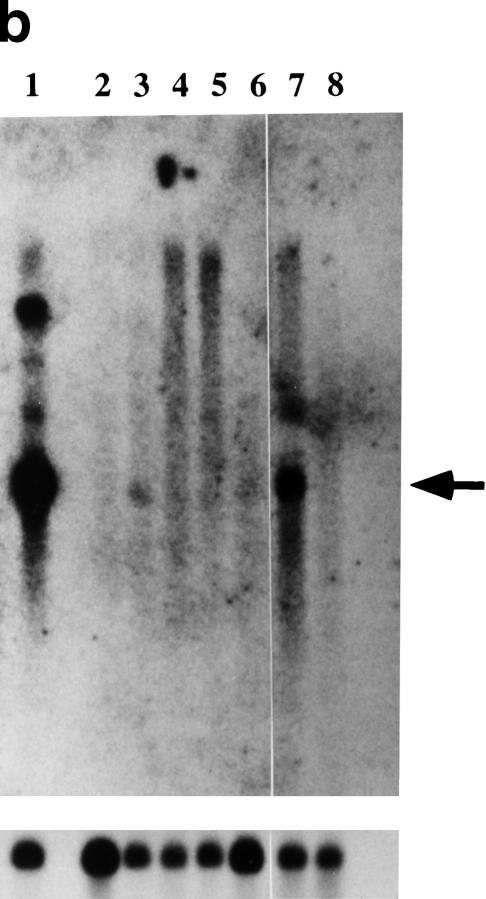
Because cDNA 55 was the only gene present in the 100-kb cosmid contig, the complete open reading frame was determined and shown to encode a 254-amino-acid protein with four transmembrane domains (Fig. 2a). The protein sequence of cDNA 55 showed sequence and structural similarity to the claudin family of tight-junction proteins (Furuse et al. 1998; Morita et al. 1999; Tsukita et al. 1999; Simon et al. 1999). Therefore, we named the gene responsible for CINF Claudin-16 (CL-16). From the genomic structure of CL-16, it was determined that exons 1–4 are deleted in CINF (Fig. 1d). This deletion was confirmed by Southern blot analysis with exon-specific probes (data not shown). CL-16 shows ∼90% homology to human PCLN-1, which was recently reported to be a causative gene for FHHN (Simon et al. 1999; Fig. 2a). The four transmembrane domains of CL-16 are 94% identical at the amino acid level to those of PCLN-1. In bovine CL-16, there is an in-frame termination codon (TAA) preceding two possible ATG initiation codons corresponding to methionine-1 and methionine-20. The amino-terminal CINF protein sequence from methionine-20 shows a low similarity to PCLN-1, as compared with the downstream sequence. It is likely that CL-16 encodes a 235-amino-acid protein from methionine-20 with 19%–23% identity to Claudin-1 through Claudin-8 and 92% identity to PCLN-1. CL-16 was expressed differentially in normal unaffected kidney, but not in CINF-affected kidney (Fig. 2b), suggesting that the CL-16 protein may have a renal function. Although CL-16 was also expressed minimally in the lungs, we did not observe any pathophysiological change in the lungs of CINF-affected cattle. Therefore, CL-16 may not be necessary for respiratory function, or, alternatively, other members of the claudin family may complement CL-16 function in the lungs.
Figure 2.
(a) Alignment of CL-16 and PCLN-1. (Asterisks) Amino acid similarity; (shaded amino acids) highly conserved residues among claudin family members; (TM1–TM4), putative transmembrane domains. (b) Kidney-specific expression of CL-16 in cattle. (Lane 1) Kidney; (lane 2) heart; (lane 3) lung; (lane 4) liver; (lane 5) brain; (lane 6) muscle; (lane 7) kidney; (lane 8) CINF-affected kidney. (Arrow) Position of the 2.9-kb CL-16 transcript. (Bottom) Control glycerol-3-phosphate dehydrogenase expression.
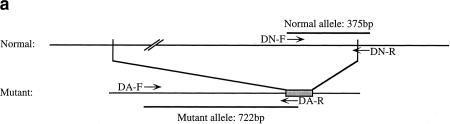
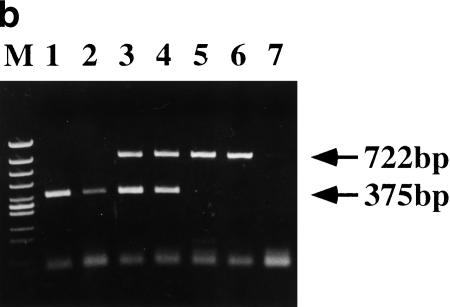
To establish a DNA test for CINF-carrier cattle, we designed PCR primers DN-F and DN-R for the 375-bp normal allele as shown in Figure 3a. The 722-bp CINF mutant allele was PCR amplified with primers DA-F and DA-R (for primer sequences, see Methods). Because the G-rich region was problematic for PCR amplification, the reverse primers DN-R and DA-R were designed for the deletion points. As shown in Figure 3b, the CINF mutant allele was present in CINF-affected and CINF-carrier cattle. In contrast, the mutant allele was not detected in control noncarrier DNA. DNA samples from 80 normal, 96 CINF-affected, and 26 CINF-carrier cattle were collected and subjected to DNA screening. All results correlated with observed phenotypes. Thus, the DNA test was 100% accurate in detecting the mutant allele.
Figure 3.
(a) DNA-based PCR test for CINF-carrier cattle. Reverse primers were designed for the deletion point. PCR primers DN-F and DN-R are for the normal allele (375 bp), and PCR primers DA-F and DA-R are for the mutant allele (722 bp). (b) Detection of the CINF mutant allele. (Lanes 1,2) Normal; (lanes 3,4) CINF carrier; (lanes 5,6) CINF-affected cattle; (lane 7) no DNA template as negative control. (M) size standard.
DISCUSSION
Renal glomerular filtration and tubular resorption are essential steps for maintaining homeostasis. We have identified CL-16 as a causative gene for bovine CINF, which is characterized by defective renal filtration and resorption. Therefore, the CL-16 protein most likely plays an important role in filtration and resorption. Claudin proteins are localized at the tight-junctions of epithelial cell layers (Furuse et al. 1998; Morita et al. 1999; Tsukita et al. 1999) and may regulate selective paracellular permeation of metabolites through self interaction or interaction with other protein components. FHHN in humans is characterized as a profound renal Mg2+ wasting disease (Praga et al. 1995). Simon et al. (1999) reported that point mutations of PCLN-1, the human ortholog of CL-16, in FHHN patients results in defective paracellular Mg2+ resorption. Their work provides the first evidence of claudin function in renal physiology. In contrast, our mutation had a total loss of function, resulting in more severe phenotypes. Impairment in mineral and/or protein resorption are correlated with renal epithelial intercellular spaces. CINF-affected cattle have increased urinary proteins as well as decreased serum calcium, the latter of which may be related to changes in bow posture of the affected phenotypes. It is possible that the mutated PCLN-1 protein in FHHN partially disorganizes the integrity of the renal epithelial structure, whereas the loss of CL-16 protein in CINF causes its complete disruption. Alternatively, different clinical phenotypes between FHHN and CINF may be accounted for by species specificity. Nevertheless, our data provide additional evidence that the tight-junction protein CL-16 plays an important role in maintaining homeostasis in the kidneys.
METHODS
Cosmid Preparation, Characterization by FISH, and Shotgun Sequencing
A bovine YAC library was screened by use of a PCR-based method as described (Takeda et al. 1998). An isolated YAC (249E9) and total genomic DNA from peripheral blood leukocytes of CINF-affected cattle were used for construction of cosmid and mutant cosmid libraries, respectively. DNA (100 μg) was partially digested with Sau3AI, and the resulting 20- to 30-kb fragments were collected by agarose gel electrophoresis followed by ligation into the pWe15 cosmid vector. Physical assignment of cosmid clones was done by FISH. Briefly, bovine metaphase chromosome spreads were hybridized with cosmid clones essentially as described (Wada et al. 1994) by use of reagents supplied in the Nick Translation Kit with Spectrum Green-dUTP (Visis, IL, USA). The physical relationship between cos68 and cos23 was determined by PCR with forward primer 68-2F, AATCATAGAACTCAGACACAG and reverse primer 23-3R, CACTAAGGATCTGATTCTGCC, located near the ends of cos68 and cos23, respectively. The size of the PCR product was ∼250 bp.
For shotgun sequencing, cosmid DNA was sheared by nebulization to an average size of ∼1.5 kb. The random fragments were cloned into pBluescript II SK(+) (Stratagene). The clones were PCR amplified with T3 and T7 primers and PCR products sequenced directly with T3 and T7 primers by use of ABI 377 DNA sequencers (Perkin-Elmer Applied Biosystems). The sequence reads were assembled and the contig sequences edited using AutoAssembler (Perkin-Elmer Applied Biosystems) and the GENETYX package.
Microsatellite development and genotyping
Microsatellite loci from cos23 were isolated by use of a poly[d(A-C)]poly[d(G-T)] Pharmacia) probe essentially as described (Hirano et al. 1996). The following primer pairs were synthesized: for DIK129, forward, 5′-TATTGCACTGATTACATTCTAC-3′ and reverse, 5′-GATTCTCAAACTTTATGAGGAC-3′; for DIK-130, forward, 5′-TTGGAATTTGACTTGCTCACC-3′ and reverse, 5′-CTCACTTGACTTCACATTAGG-3′. Both loci were PCR amplified at annealing temperatures of 55°C and 60°C. For other loci, the PCR conditions were optimized as recommended (Kappes et al. 1997), and other reaction conditions were set as recommended by the manufacturer. Total bovine genomic DNA was prepared from peripheral blood leukocytes. Microsatellite polymorphisms were analyzed by PCR amplification and gel electrophoresis with an ABI 377 automated DNA sequencer as described (Hirano et al. 1996). Genotype data were captured by means of GENESCAN and Genotyper software (Perkin-Elmer Applied Biosystems).
Mutation Detection
On the basis of DNA sequences that include the deletion region, the following primer pairs were synthesized: for the normal allele (annealing temperature, 57°C; PCR product, 375 bp), DN-F, 5′-TATGCTGTTGATGTTTATGTAG-3′ and DN-R, 5′CCCCCCCCCGCCTTTTTC-3′; and for the mutant allele (annealing temperature, 57°C; PCR product, 722 bp), DA-F, 5′-ATTGTATTTTTAGGAGTGACTC-3′ and DA-R, 5′-CCCCCCCCCACTCTATAC-3′. PCR mixtures (total volume of 15 μl) contained 40 ng of genomic DNA, 12.5 pmoles of each primer, 1.5 mm MgCl2 , 10 m Tris-HCl (pH 8.3), 50 m KCl, 200 μm of each dNTP, and 0.75 units of Taq DNA polymerase (TAKARA, Tokyo, Japan).
RNA Isolation, cDNA Library Preparation, and Northern Blot Analysis
Tissues from calves were frozen in liquid nitrogen. Total RNA was isolated by homogenization of each sample in Trizol reagent (Life Technologies). Poly(A)+ RNA was isolated from total RNA by use of Oligotex-dt30 (Roche). For preparation of a cDNA library, calf kidney poly(A)+ RNA was reverse-transcribed by use of the Superscript Lambda System for cDNA Synthesis, and cDNAs were cloned by use of the Lambda ZipLox (NotI-SalI arms) Cloning Kit (GIBCO BRL). Blots with poly(A)+ RNA were hybridized to the 3′-untranslated region of CL-16 cDNA, which was PCR amplified with the following primers: sense, 5′- TTCAAAGGAGGCCAGAGATT-3′ and antisense, 5′-GAATATACAAAATATGACAGAC-3′.
Acknowledgments
The authors wish to thank Laurence B. Schook for critical reading, and Ikuko Fujita, Michiyo Ishida, Chiyoko Itoh, Junko Mizuno, Ri-e Nihei, Harumi Suzuki, and Aki Takada for technical assistance. This study was supported by grants from the Japan Racing and Livestock Promotion Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kazusugi@cocoa.ocn.ne.jp; FAX 81-248-25-5725.
REFERENCES
- Furuse M, Fjita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Nakane S, Mizoshita K, Yamakuchi H, Inoue-Murayama M, Watanabe T, Barendse W, Sugimoto Y. Characterization of 42 highly polymorphic bovine microsatellite markers. Anim Genet. 1996;27:365–368. doi: 10.1111/j.1365-2052.1996.tb00980.x. [DOI] [PubMed] [Google Scholar]
- Kappes SM, Keel JW, Stone RT, McGraw RA, Sonstegard TS, Smith TPL, Lopez-Corrales NL, Beattie CW. A second-generation linkage map of the bovine genome. Genome Res. 1997;7:235–249. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- Kobayashi, N., T. Hirano, S. Maruyama, H. Matsuno, K. Mukoujima, H. Morimoto, H. Noike, H. Tomimatsu, K. Hara, T. Itoh et al. 2000. Genetic mapping of a locus associated with bovine chronic interstitial nephritis to chromosome 1. Anim. Genet. (in press). [DOI] [PubMed]
- Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praga M, Vara J, Gonzalez-Parra E, Andres A, Alamo C, Araque A, Ortiz A, Rodicio JL. Familial hypomagnesemia with hypercalciuria and nephrocalcinosis. Kidney Int. 1995;47:1419–1425. doi: 10.1038/ki.1995.199. [DOI] [PubMed] [Google Scholar]
- Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Paraga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- Takeda H, Yamakuchi H, Ihara N, Hara K, Watanabe T, Sugimoto Y, Oshiro T, Kishine H, Kano Y, Kohno K. Construction of a bovine yeast artificial chromosome (YAC) library. Animal Genet. 1998;29:216–219. doi: 10.1046/j.1365-2052.1998.00325.x. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M. Occludin and claudins in tight-junction strands: Leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- Wada M, Abe K, Okumura K, Taguchi H, Kohno K, Imamoto F, Schlessinger D, Kuwano M. Chimeric YACs were generated at unreduced rates in conditions that suppress coligation. Nucleic Acids Res. 1994;22:1651–1654. doi: 10.1093/nar/22.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]