Abstract
The application of the gene finder HMMGene to the Adh region of the Drosophila melanogaster is described, and the prediction results are analyzed. HMMGene is based on a probabilistic model called a hidden Markov model, and the probabilistic framework facilitates the inclusion of database matches of varying degrees of certainty. It is shown that database matches clearly improve the performance of the gene finder. For instance, the sensitivity for coding exons predicted with both ends correct grows from 62% to 70% on a high-quality test set, when matches to proteins, cDNAs, repeats, and transposons are included. The specificity drops more than the sensitivity increases when ESTs are used. This is due to the high noise level in EST matches, and it is discussed in more detail why this is and how it might be improved.
It is difficult to accurately identify all the genes in the eukaryotic DNA sequences that are pouring out of the large-scale sequencing laboratories these days. At present, the most reliable way to find genes is by close similarity to proteins from the same or other organisms, by cDNAs from the same or a closely related organism, or by comparison between closely (but not too closely) related genomes, such as human and mouse. Homology information does, of course, not solve the annotation problem completely, because many genes have no significant similarity with other known sequences. In addition, the homology information is often incomplete, so that only part of the gene can be localized because ESTs are incomplete or a protein only shows similarity to part of the sequence.
Most work on automated gene finding has focussed on de novo prediction of genes with no homology. These efforts have not yet produced the perfect method, at least not for most eukaryotes. For a gene finder to be really useful for large-scale annotation, it has to be able to combine database matches with de novo prediction so as to give a reasonable prediction of gene structures compatible with the matches. This is more complicated than it sounds because many database matches are quite uncertain, in particular, those involving ESTs.
This paper describes the application of the HMMGene gene finder (Krogh 1997, 1998a) to the 3-Mb Adh region of Drosophila melanogaster (Ashburner et al. 1999). The expert annotation of this region was held back, and a blind prediction experiment called GASP was done. Several groups submitted predictions (Reese et al. 2000a), and this paper is a description of my submission to GASP.
HMMGene is based on a probabilistic model called a hidden Markov model (Durbin et al. 1998). The probabilistic framework allows for a very simple, and yet powerful, way of including database matches. The advantage of the hidden Markov model, as compared with many other models, is that it can model the grammatical structure of genes (Krogh 1998b), which means that the prediction will always splice correctly, always start with a start codon and end with a stop codon (if the gene is complete), always obey whatever consensus splice sites you specify, and so on. (HMMGene very rarely violate the grammar of a gene by predicting an intron in the middle of a stop codon, meaning that after splicing there will be an in-frame stop codon. This problem is difficult to fix in the standard HMM framework.) In addition, it has modules for recognition of both coding regions and the common signals associated with genes built into it. This combination of “everything you want for a gene finder” in one model is the reason for the recent popularity of HMMs for this problem (Krogh et al. 1994; Kulp et al. 1996; Burge and Karlin 1997; Henderson et al. 1997; Lukashin and Borodovsky 1998).
In this paper I will focus on the improvements obtained when database matches are included.
RESULTS
The Adh region was searched for matches to proteins, cDNAs, ESTs, repeats, and transposons. Each match was recorded with the type of match (coding, EST, repeat, etc.), beginning and end of the match (on the DNA), the strand on which it occurs, and a confidence. If the confidence is close to 1, the feature is considered certain, whereas if it is close to 0, it is considered very uncertain. For the ESTs matching the sequence, it was checked whether it was likely that the match was on the wrong strand (see Methods). Introns were annotated when a match was split in two segments in an intron-like manner (see Methods). After resolving overlapping matches, the number of different regions resulting from the searches are shown in Table 1.
Table 1.
Number of Various Types of Regions Annotated in the Adh Sequence after Database Searches
| Type of match | No. of matches | No. of bases covered | |
|---|---|---|---|
| Coding | 130 | 120647 | (4.13%) |
| cDNA | 48 | 8682 | (0.30%) |
| EST | 337 | 91010 | (3.12%) |
| Intron | 176 | 158868 | (5.44%) |
| Repeat | 50 | 65999 | (2.26%) |
| Total | 741 | 445206 | (15.25%) |
A hidden Markov model was estimated from a set of 514 sequences with a total of ∼4 Mb (see Methods). The model has states for intergenic regions, 5′ and 3′-untranslated regions (UTRs), coding regions, and introns in both UTRs and coding regions. It contains states designed to recognize translation start and stop, splice sites, branchpoints, and poly(A) sites.
The model was used to predict genes in the Adh sequence. For each strand, the most probable gene structures were found given the model and using the various types of database matches.
To assess the effect of using database matches, exactly the same procedure was repeated once without any database matches and with all database matches, except for ESTs. Here, I have given the standard statistics, that is, the sensitivity and specificity at the base level, the exon level, and gene level. Note that “exon” here means only the coding part of an exon—UTRs are not included in the statistics. For exons and whole genes, the number of missed ones and wrong ones are also given. The sensitivity calculated at the gene level is the percentage of annotated genes with a correctly predicted coding region. This means that the annotated and predicted coding regions agree exactly. The specificity is defined as the percent of the total number of predicted genes that are correct.
The sensitivity and specificity at the whole gene level was also calculated with a less stringent measure of correctness, 99% and 95% correct coding bases. This is for two reasons. First, a 95% correctly predicted gene is almost as good as a 100% correctly predicted one for most uses, and second, for a 99% correct gene it is quite likely that the prediction is the correct one and the annotation is a bit off, because coding regions of a few base pairs or the exact start codon can easily be wrongly annotated even from cDNA. These numbers, I believe, are of more practical relevance than those for exactly correct genes.
The performance was measured on the two annotations provided by the GASP organizers: std1 that contains 43 genes that are quite certain and std3 that contains 222 genes of which many are less certain. The numbers for the std1 and std3 annotations are shown in Tables 2 and 3. The annotations in std1 are nonexhaustive, so numbers on specificity are not given. For std3, which is probably close to being exhaustive, but of less certainty, I have given both sensitivity and specificity. Because of software limitations, the predictions were done on pieces of ∼300 kb of the Adh sequence, and while making the statistics for this paper, I discovered that the program had failed on the reverse strand of the piece between bases 1,000,001 and 1,300,000. This has been rectified, so I give the numbers from both the original submission and the corrected one, which contains eight additional genes in that region.
Table 2.
The Performance on the std1 Set
| No database matches | All but EST matches | Including all matches | Original submission | |
|---|---|---|---|---|
| Base level | ||||
| Sensitivity | 76.9 | 95.9 | 96.8 | 96.3 |
| Exon level | ||||
| Sensitivity | 61.8 | 69.9 | 69.9 | 68.3 |
| Missing exons | 19.5 | 7.3 | 4.1 | 5.7 |
| Gene level | ||||
| Sensitivity 100% | 37.2 | 39.5 | 37.2 | 34.9 |
| Sensitivity 99% | 41.9 | 44.2 | 44.2 | 41.9 |
| Sensitivity 95% | 46.5 | 48.8 | 55.8 | 53.5 |
| Missing genes | 18.6 | 4.7 | 4.7 | 7.0 |
The last column corresponds to the original submission to GASP.
Table 3.
The Performance on the std3 Set
| No database matches | All but EST matches | Including all matches | Predictions >200 | Original submission | |
|---|---|---|---|---|---|
| Base level | |||||
| Sensitivity | 74.0 | 80.6 | 81.1 | 81.1 | 77.9 |
| Specificity | 91.3 | 92.5 | 91.1 | 91.5 | 90.7 |
| Exon level | |||||
| Sensitivity | 48.3 | 50.4 | 51.4 | 51.4 | 50.1 |
| Specificity | 58.1 | 56.7 | 52.7 | 54.5 | 53.2 |
| Missing exons | 29.8 | 24.2 | 23.1 | 23.1 | 25.6 |
| Wrong exons | 15.6 | 15.1 | 21.3 | 18.7 | 20.9 |
| Gene level | |||||
| Sensitivity 100% | 26.1 | 28.8 | 30.2 | 30.2 | 29.3 |
| Sensitivity 99% | 28.8 | 32.0 | 33.3 | 33.3 | 32.4 |
| Sensitivity 95% | 33.3 | 39.2 | 42.3 | 42.3 | 41.4 |
| Specificity 100% | 34.3 | 33.9 | 30.2 | 32.4 | 30.4 |
| Specificity 99% | 37.9 | 37.6 | 33.3 | 35.7 | 33.6 |
| Specificity 95% | 43.8 | 46.0 | 42.3 | 45.4 | 43.0 |
| Missing genes | 16.7 | 9.9 | 9.9 | 9.9 | 13.2 |
| Wrong genes | 7.7 | 6.9 | 16.7 | 10.6 | 16.4 |
In the column Predictions >200, 15 predicted genes with a coding region <200 bp long were removed from the predictions using all database matches. Other columns correspond to Table 2.
DISCUSSION
The performance of the method compares well with the other submissions to GASP (Reese et al. 2000a). It is interesting that the sensitivity is significantly higher on the std1 set than on std3. This might be an indication of more errors in std3 than should be expected, but it could of course also be a random fluctuation, because std1 contains only 43 genes. Another possible explanation is that std1 may contain “easier” genes than std3 for some reason. More than half of the genes are >95% correct in std1 (when including all database matches), which is a quite encouraging number.
There is a general increase in sensitivity when more of the database matches are included, and there is an accompanying drop in the number of missed genes. The only exception is that sensitivity drops by one gene for 100% correct genes for std1 when EST matches are used. However, when EST matches are added, the specificity consistently drops significantly, and the number of wrong predictions increases for std3 (wrong predictions are those where the predictions do not overlap any annotated coding region).
The drop in specificity is probably an indication that the method is sensitive to ESTs matching in unexpected places or being on the wrong strand. This is confirmed when looking at the lengths of the coding regions of predicted genes. When not using ESTs, the two shortest predicted coding regions are 183 and 270 bases. With ESTs there are nine predictions of a coding region <100 and six between 100 and 200. To accommodate the strange EST matches, the program predicts silly little genes. When excluding the 15 predicted genes with a coding region <200, the specificity increases without affecting the sensitivity on either of the standard sets (see Table 3).
It is still unclear to what extent it pays to include EST matches in the way done here. The inclusion of ESTs is a complicated affair because of the high noise level. HMMGene assigns a probability to a base in a database match, and the total probability of the region is this probability raised to the power of the length of the match. This means that ignoring a long EST is very improbable, whereas it is quite probable to ignore a short one. The way database matches are included in the prediction seems quite sensible for “safe” database matches, such as proteins and cDNA, but for ESTs experimentation with other types of length dependences is necessary. Another possible improvement is to include information about the position of the EST, that is, whether it is 5′ or 3′, which was not done here.
The problem of ESTs being on the wrong strand is a serious one for a method like HMMGene. If a long EST is on the wrong strand, a gene is likely to be predicted on that strand. It does not exclude a correct prediction on the opposite strand, however. For each EST on one strand, I put a weak EST match on the other strand to permit the information to be used, but that of course makes it possible to get a gene on the wrong strand when the EST is correctly placed, so it has to have a very low confidence. Another possibility would be to have the matches with equal confidence on both strands and then in a post-processing step choose between overlapping genes.
It is a nontrivial task to postprocess database hits. For instance, How should overlapping hits be dealt with? and How should the confidence be assigned? The rules I have used are quite ad hoc and are essentially just a first shot. It is very likely that better procedures can be found.
There is perhaps also room for improvements in the underlying gene model. Comparing the results of the gene predictions without database matches with those obtained with, for example, Genie (Reese et al. 2000b) suggests that it could be better. A comparison of the length distribution of predicted exons with that of annotated exons (data not shown) suggests that this is one place where an improvement could be obtained, and this and other improvements will be tested in future work.
In conclusion, this study shows that gene predictions can certainly be improved significantly by including database matches, as expected. However, ESTs are not easy to deal with in an automated manner, and more work is needed to fully benefit from these.
METHODS
The Adh sequence was downloaded from http://whitefly.lbl.gov/GASP1/data/data.html. The std1 and std3 sets were obtained after the experiment from http://whitefly.lbl.gov/GASP1/data/standard.html.
Database Searches
All database searches were done with BLAST version 2.0.9 (Altschul et al. 1997) doing gapped alignments. All databases mentioned below, except for SWISS-PROT, were supplied by the GASP1 organizers at http://whitefly.lbl.gov/GASP1/data/data.html.
Below, matching a “bad word” means that the description matches the word transposon, transposable, repetitive, retrovirus, or transcriptase.
Proteins: The SWISS-PROT database release 35.0 (Bairoch and Apweiler 1998) was searched using BLASTX with default settings. All matches with a score >100 bits and not matching a bad word were kept and annotated as coding. The fraction of identities was used as the confidence of the match. If a protein match had a gap in the DNA sequence, an intron was annotated if the matching parts of the protein were <20 amino acids apart and overlapped with <20 amino acids and if the intron was between 40 bp and 20,000 bp long. The confidence of the intron was set to the average of the flanking protein matches defining it.
cDNA: The Adh region was searched against the cDNA database with BLASTN and a maximum E-value of 1.e − 10. All matches with a score >100 bits and not matching a bad word were kept and annotated as cDNA. The confidence was calculated in the same way as for proteins, and the introns were annotated as for the protein matches.
EST: The Adh region was searched against the EST database with BLASTN. All matches with a score >100 bits, not matching a bad word, and having an identity >93% were kept and annotated as EST. The confidence was set to the fraction of identical residues minus 0.9 times 10 (i.e., ranging from 0.3 to 1). Some ESTs were moved to the other strand (see below).
Repeats: The Adh region was searched against the repeat database with BLASTN and a maximum E-value of 1.e − 10. All matches with a score >100 bits were kept and annotated as repeat. The confidence was calculated in the same way as for proteins.
Transposons: The Adh region was searched against the Transposons database with BLASTN. All matches with a score >100 bits were kept and annotated as repeat. The confidence was calculated in the same way as for proteins.
It is a serious problem that some ESTs occur on the wrong strand, and an attempt was made to correct some of them. A copy of the gene model was made and modified to an EST model by removing states for intergenic regions and allowing frameshift errors and stop codons in the reading frame (both with very low probabilities). For each EST, the probability of it being one of these classes was calculated: 5′ → 5′, 5′ → coding, 5′ → 3′, coding → coding, coding → 3′, or 3′ → 3′. Here, for example, 5′ → coding means that the EST starts in the 5′ UTR of a gene and ends in the coding region. These probabilities were also calculated for the reverse strand. If the log probability of the most probable class on the reverse strand was 5 higher than the best on the direct strand, then the EST was moved to the other strand.
All of these database matches were combined in the following way (outline): every database match within cDNA introns was deleted. Repeats and transposons were combined into one group called repeats, and overlapping matches were combined. EST matches overlapping with repeats or introns (defined by cDNAs or proteins) were removed. All overlapping ESTs were combined. Parts of EST or cDNA that overlapped coding regions were discarded.
Finally, repeat matches were copied to the other strand, protein and cDNA matches were labeled intergenic on the other strand, and ESTs were copied to the other strand with a confidence scaled by 0.01.
Data Sets for Training
Two data sets were obtained with GenBank annotations of reasonable quality. The first was supplied by Staffan Bergh and Anneli Attersand who had carefully checked it. The second one was supplied by the GASP1 organizers and is available at the GASP1 Web site. These two sets were combined into one such that identical GenBank entries were avoided. The two sets may contain some homologous genes, but not very many. Sequences longer than 20 kb were split into two or more. The final set contains 4 Mb in 514 sequences. For the GASP1 experiment it was not required that only the data set provided by the organizers was used, so it was supplemented in this way simply to enlarge it. The larger the data set, the better the gene finder tends to perform.
HMMGene
HMMGene builds on a hidden Markov model (Durbin et al. 1998) with states for intergenic regions, 5′ and 3′ UTRs, coding regions, and introns in both UTRs and coding regions. It contains states designed to recognize translation start and stop, splice sites, branchpoints, and poly(A) sites. The model is estimated by conditional maximum likelihood from the training data (Krogh 1997).
Each state of the model is labeled as belonging to one of the nine classes: intergenic, 5′ UTR, 3′ UTR, coding, intron of phase 0, 1, or 2 in coding region, intron in 5′ UTR, or intron in 3′ UTR. Each path through the model gives a labeling of the DNA sequence. The total probability of a labeling is the sum over all paths giving that labeling. Genes are predicted as the most probable labeling given the model by the N-best algorithm (Krogh 1997). This is an approximative algorithm, because there is no efficient way to do it exactly, but the approximation is very good. A sequence of states (a path) π = π1,…,πL has the probability
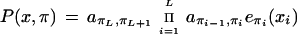 |
where x = x1,…,xL is the DNA sequence, akl is the probability of making a transition from state k to state l, and ek(a) is the probability of emitting base a in state k. State π0 is the begin state, and state πL+1 is the end state.
To include database matches, a probability distribution over labels is assigned to each base in the sequence. In regions with no database matches, the probabilities are uniform. In a region with a hit to a protein for instance, the probability for coding is set higher than the rest, and similarly for other types of matches. If the probability for label d at position i is called pi(d), the probability of a path and a labeling is
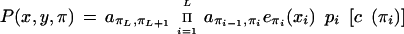 |
Here c(k) means the label of state k.
In Table 4 the probabilities of the various types of matches used in this study are given under the assumption that the confidence is 1. If the confidence is not 1, a probability P is scaled by (1 − confidence) / 9 + confidence × P, meaning that the distribution is uniform (1 / 9) at a confidence of 0.
Table 4.
Label Probabilities for Database Matches of the Six Types Considered
| Type of annotation | Coding | Intron in coding | Intron in UTR | Intergenic | 5′ UTR | 3′ UTR |
|---|---|---|---|---|---|---|
| Coding | 0.300 | 0.150 | 0.050 | 0.050 | 0.050 | 0.050 |
| cDNA | 0.320 | 0.007 | 0.007 | 0.007 | 0.320 | 0.320 |
| Intron | 0.088 | 0.130 | 0.130 | 0.088 | 0.088 | 0.088 |
| EST | 0.120 | 0.107 | 0.107 | 0.107 | 0.120 | 0.120 |
| Repeat | 0.000 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 |
| Intergenic | 0.013 | 0.013 | 0.013 | 0.990 | 0.013 | 0.013 |
There are three different labels for introns in coding regions (corresponding to the three possible phases) and two intron labels for UTRs, those in 5′ UTRs, and those in 3′ UTRs. Therefore, the rows sum to 1 (e.g., for coding 0.3 + 3 × 0.15 + 2 × 0.05 + 0.05 + 0.05 + 0.05 = 1).
These probabilities are multiplied along a path, so the probability of not using a path consistent with a database match drops exponentially with the length of the match. Therefore, some of the probabilities used are very close to uniform. This is most notable for ESTs.
Because of software limitations, the sequence of 3 Mb was split-up into pieces of up to 300 kb in size, and genes were predicted on these pieces. Afterwards, the predictions were transformed back to the original sequence coordinates. The sequence was split either at a repeat/transposon position or at positions where an initial scan had not predicted any genes to try to avoid splitting in the middle of genes.
Performance Measures
Performance was measured by several measures. Below, exon means the coding part of an exon. UTRs are ignored in the statistics. A correct prediction means one that agrees with one of the standard sets disregarding the possibility of errors in these:
Base level sensitivity: the percent of bases annotated as coding, which are predicted as coding.
Base level specificity: the percent of bases predicted as coding, which are correct.
Exon level sensitivity: the percent of annotated exons predicted correctly (with both ends correct).
Exon level specificity: the percent of predicted exons, which are correct.
Missed exons: the percent of annotated exons with which no predicted exons overlap.
Wrong exons: the percent of predicted exons not overlapping any annotated exons.
Gene level sensitivity: the percent of annotated genes predicted >X% correct. A gene is X% correct if at least X% of the annotated coding bases are predicted and X% of the bases predicted as coding are also annotated as coding (i.e., base level sensitivity and specificity >X% for that gene). Numbers are reported for X = 95, 99, and 100.
Gene level specificity: the percent of predicted genes, which are X% correct.
Missed genes: the percent of annotated genes with coding regions not overlapping any predicted coding regions.
Wrong genes: the percent of predicted genes with coding regions not overlapping any annotated coding regions.
Acknowledgments
I thank Staffan Bergh, Anneli Attersand, and Luis Parodi at Pharmacia and Upjohn in Stockholm for supplying training data. Thanks also to the organizers of GASP1 for this excellent initiative. This work was supported by a grant from the Danish National Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL krogh@cbs.dtu.dk; FAX 45 4593 1585.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1998. Nucleic Acids Res. 1998;26:38–42. doi: 10.1093/nar/26.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Durbin RM, Eddy SR, Krogh A, Mitchison G. Biological sequence analysis. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- Henderson J, Salzberg S, Fasman KH. Finding genes in DNA with a hidden Markov model. J Comput Biol. 1997;4:127–141. doi: 10.1089/cmb.1997.4.127. [DOI] [PubMed] [Google Scholar]
- Krogh A. Two methods for improving performance of a HMM and their application for gene finding. In: Gaasterland T, Karp P, Karplus K, Ouzounis C, Sander C, Valencia A, editors. Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1997. pp. 179–186. [PubMed] [Google Scholar]
- ————— . An introduction to hidden Markov models for biological sequences. In: Salzberg SL, Searls DB, Kasif S, editors. Computational methods in molecular biology. 1998a. , chapter 4, pp. 45–63. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- ————— . Gene finding: Putting the parts together. In: Bishop MJ, editor. Guide to human genome computing. 1998b. , chapter 11, pp. 261–274. Academic Press, San Diego, CA. [Google Scholar]
- Krogh A, Mian IS, Haussler D. A hidden Markov model that finds genes in E. coli DNA. Nucleic Acids Res. 1994;22:4768–4778. doi: 10.1093/nar/22.22.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. A generalized hidden Markov model for the recognition of human genes in DNA. In: States D, Agarwal P, Gaasterland T, Hunter L, Smith R, editors. Proceeding of the Conference on Intelligent Systems in Molecular Biology. Menlo Park, CA: AAAI Press; 1996. pp. 134–142. [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. Genemark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese, M.G., G. Hartzell, N.L. Harris, U. Ohler, and S.E. Lewis. 2000a. Genome annotation assessment in Drosophila melanogaster. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Reese, M.G., D. Kulp, H. Tammana, and D. Haussler. 2000b. Genie—gene finding in Drosophila melanogaster. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]


