Abstract
The success of the ongoing Human Genome Project has resulted in accelerated plans for completing the human genome sequence and the earlier-than-anticipated initiation of efforts to sequence the mouse genome. As a complement to these efforts, we are utilizing the available human sequence to refine human-mouse comparative maps and to assemble sequence-ready mouse physical maps. Here we describe how the first glimpses of genomic sequence from human chromosome 7 are directly facilitating these activities. Specifically, we are actively enhancing the available human-mouse comparative map by analyzing human chromosome 7 sequence for the presence of orthologs of mapped mouse genes. Such orthologs can then be precisely positioned relative to mapped human STSs and other genes. The chromosome 7 sequence generated to date has allowed us to more than double the number of genes that can be placed on the comparative map. The latter effort reveals that human chromosome 7 is represented by at least 20 orthologous segments of DNA in the mouse genome. A second component of our program involves systematically analyzing the evolving human chromosome 7 sequence for the presence of matching mouse genes and expressed-sequence tags (ESTs). Mouse-specific hybridization probes are designed from such sequences and used to screen a mouse bacterial artificial chromosome (BAC) library, with the resulting data used to assemble BAC contigs based on probe-content data. Nascent contigs are then expanded using probes derived from newly generated BAC-end sequences. This approach produces BAC-based sequence-ready maps that are known to contain a gene(s) and are homologous to segments of the human genome for which sequence is already available. Our ongoing efforts have thus far resulted in the isolation and mapping of >3,800 mouse BACs, which have been assembled into >100 contigs. These contigs include >250 genes and represent ∼40% of the mouse genome that is homologous to human chromosome 7. Together, these approaches illustrate how the availability of genomic sequence directly facilitates studies in comparative genomics and genome evolution.
The rapid advances and monumental achievements of the Human Genome Project are igniting a new era in biomedical research. The availability of the complete genomic sequence for the model organism Caenorhabditis elegans (The C. elegans Sequencing Consortium 1998), in conjunction with the impending completion of the Drosophila and human genomic sequences, provides an unprecedented view of the genetic essence of multicellular organisms. In the coming century, interpretation and utilization of complete genome sequences will provide new insights into old problems and inspire the development and testing of novel hypotheses. Adapting to a scientific environment with such rapidly changing and powerful new resources presents a significant but rewarding challenge to those studying biology.
Genome mapping is historically dependent on species-specific resources and reagents as well as species-independent technologies. For instance, genome mapping in mouse has been performed principally with genetic mapping methods using inbred strains (Reeves & D'Eustachio 1999). This valuable mouse-specific resource combined with technological advances (e.g., PCR) has increased the number of available genetic markers (Dietrich et al. 1992; Dietrich et al. 1994; Dietrich et al. 1996) and the rate at which they can be assembled into maps. The recent development of radiation hybrid mapping (Cox et al. 1990; Matise et al. 1999) and markers that are conserved between species (Lyons et al. 1997) reduces the dependence on species-specific resources, thereby increasing the number of genomes amenable to systematic mapping. The complete sequence of the human genome will provide a reference vertebrate ‘genetic blueprint’ and, as such, should accelerate the mapping of other genomes.
Here we provide a general overview of the approaches we are developing and implementing to exploit available human genomic sequence for the purpose of mapping a related mammal, the mouse. Importantly, our preliminary results demonstrate how genomic sequence greatly enhances comparative genome analyses.
Overview of Experimental Strategy
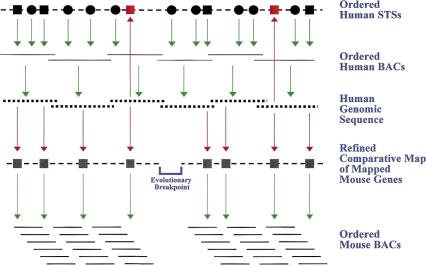
As a complement to the ongoing and accelerated efforts to sequence the human genome (Collins et al. 1998; Waterston & Sulston 1998; The Sanger Centre & The Washington University Genome Sequencing Center 1998) , we are developing a generalized strategy for using human sequence data to facilitate human-mouse comparative mapping. A schematic overview of our evolving approach is depicted in Figure 1. In the Human Genome Project, individual human bacterial-based clones [mostly bacterial artificial chromosomes (BACs) (Shizuya et al. 1992)] are being sequenced (Waterston & Sulston 1998; The Sanger Centre & The Washington University Genome Sequencing Center 1998). Most often, these have been mapped relative to existing sets of sequence-tagged sites (STSs) (Olson et al. 1989), ordered by yeast artificial chromosome (YAC)-based STS-content mapping (Green & Olson 1990; Green & Green 1991) and/or radiation hybrid mapping (Cox et al. 1990). By simple electronic detection of STSs within the genomic sequence, the BACs and their corresponding sequence can be reliably organized relative to the starting STS map. In addition to providing base-pair-level resolution, the genomic sequence enhances the STS map by identifying and positioning additional genes, expressed-sequence tags (ESTs), and other sequence elements. Such efforts, which require electronic analyses only, produce a more detailed and accurate sequence-based map.
Figure 1.
Overview of strategy for using human genomic sequence to facilitate human-mouse comparative mapping. Mapped and ordered human STSs (circles and squares correspond to random sequences and genes/ESTs, respectively) are used to isolate overlapping sets of human BACs, which in turn are sequenced. The resulting human genomic sequence can be readily aligned with the STS map by the electronic detection of mapped STSs. Also detected in the human sequence are previously unmapped sequences (e.g., genes/ESTs; depicted in red), thereby yielding an even more detailed STS map. Traditional comparative mapping can be enhanced with the human genomic sequence, specifically by the electronic detection of previously mapped mouse sequences (most often genes/ESTs). This allows refined comparative maps to be constructed that are more detailed than the starting human STS maps. The resulting linear order of markers on the comparative map allows more precise localization of evolutionary breakpoints at the ends of conserved segments. Finally, orthologous mouse sequences can be used to isolate corresponding mouse BACs and to assemble clone contigs. Red arrows reflect steps involving electronic analyses only, while green arrows reflect steps involving laboratory-based experimental analyses.
A variant of the above strategy can also be used to assemble refined comparative maps. Comparative mapping involves establishing the relative genomic positions of orthologous gene pairs in two species, such as human and mouse. Traditionally, this information comes from genetic, physical, radiation hybrid, and/or cytogenetic mapping data and can include genes/ESTs present on the human STS map that have also been mapped in mouse. A comparative map built from a single human STS map has the virtue of providing a high confidence marker order but is limited by the number of included genes (Hudson et al. 1995; Nagaraja et al. 1997; Bouffard et al. 1997). In contrast, an integrated STS-sequence map represents a robust framework for including a much larger set of genes (theoretically all genes) in the comparative map of a human genomic segment. Specifically, this involves the identification of genes in the human sequence that have been mapped either in human and mouse or only in mouse (Fig. 1). In the case of an ortholog pair already mapped in both species, only the identification of the gene in the human genomic sequence is required. In the case of a mapped mouse gene whose ortholog has not yet been mapped in human, the gene pair can be included on the comparative map provided the sequences are determined to be orthologous.
For the comparative mapping studies described below, we have established a hierarchical, stepwise approach to determine if a mouse gene sequence is orthologous to a human sequence. First, a given human genomic sequence is compared by BLAST with all available mouse mRNA and EST sequences, with any human-mouse DNA matches at or above the reported average for human-mouse orthologs (Makalowski et al. 1996; Makalowski & Boguski 1998) selected for further consideration. Second, candidate orthologs are evaluated with respect to their chromosomal location (if known) in mouse. As a DNA sequence-independent resource, such mapping data helps to remove nonorthologous mouse sequences that are similar to related gene family members or are pseudogenes as well as those detected because of some artifact associated with the human sequence (e.g., a chimeric clone). Third, candidate mouse orthologs with the highest similarity to a specific human sequence are then compared to all other available human genomic sequence; if no greater similarity is found, then the matching mouse sequence is operationally considered to be orthologous to the initial human sequence (used in the first step). This last criterion will become increasingly relevant and powerful as the human genome sequence reaches completion in the coming months. It is worth noting that even a mapped mouse EST with no known human ortholog can be readily placed on the comparative map based on these criteria.
A virtue of a refined human-mouse comparative map is that it provides accurate and detailed insights into the patterns of conservation between orthologous regions of the human and mouse genomes. Such patterns provide the basis for defining conserved segments containing genes linked in both human and mouse. By aligning groups of mouse genes with the human map, the boundaries between the conserved segments can be localized. These boundaries define genomic segments that have undergone rearrangement (i.e., evolutionary breakpoints; see Fig. 1) in either the mouse or human lineage since their last common ancestor.
In a similar fashion, human genomic sequence can be used for organizing and constructing higher resolution physical maps of mouse DNA. Specifically, the human sequence can be analyzed for the presence of orthologous mouse genes and ESTs (Marra et al. 1999) (also see http://www.ncbi.nlm.nih.gov/dbEST). Mouse-specific hybridization probes can then be designed from these sequences and used to construct mouse BAC contigs (Fig. 1). Importantly, the refined comparative map (described above) can be used as an organizational framework for mapping all regions of the mouse genome orthologous to a particular region of the human genome.
The general approaches described above and in Figure 1 thus produce two immediate products: refined human-mouse comparative maps and nascent mouse BAC physical maps. Our experience implementing these strategies has focused on human chromosome 7, which contains ∼5% of the human genome and is associated with a detailed STS map [containing 2150 mapped STSs (Bouffard et al. 1997) (see http://genome.nhgri.nih.gov/chr7)] and considerable amounts of sequence data. In fact, as of this writing, >50% of the ∼170-Mb chromosome is available as high-accuracy, finished sequence, while an additional 15–20% of the chromosome is available as ‘working draft’ sequence (see http://www.ncbi.nlm.nih.gov/genome/seq). Below we summarize our comparative mapping of the mouse genomic regions corresponding to human chromosome 7.
Assembly of a Refined Human-Mouse Comparative Map
Human–mouse comparative maps provide an important resource for the analysis of mammalian genomes (Copeland et al. 1993; Carver & Stubbs 1997; Nadeau & Sankoff 1998) (see http://www.informatics.jax.org). As such, the human gene database OMIM (http://ncbi.nlm.nih.gov/omim) and the Mouse Genome Informatics database (MGI; http://www.informatics.jax.org) include comparative mapping information. In particular, MGI provides an updated and reliable database of human-mouse comparative mapping data. Unfortunately, the predicted human gene locations catalogued in MGI are cytogenetically based, precluding the high-confidence ordering of genes along a human chromosome. Another, older source of human-mouse (and mouse-human) comparative mapping data is the Davis Human/Mouse Homology Map compiled by DeBry and Seldin (DeBry & Seldin 1996) (see http://www.ncbi.nlm.nih.gov/Homology); for example, this map depicts 14 segments of the mouse genome known to be orthologous to human chromosome 7. However, the rapid generation of human chromosome 7 sequence coupled with ongoing mouse mapping efforts makes these various sources of comparative mapping data incomplete. Thus, we are routinely using the available STS map and evolving human chromosome 7 sequence to assemble a more detailed and up-to-date human-mouse comparative map. This refined comparative map, which at present contains 154 genes, is available as a Web supplement to this paper (see http://genome.nhgri.nih.gov/chr7/comparative).
The genes on our current refined comparative map were included based on publicly available mouse mapping data and the human chromosome 7 STS (Bouffard et al. 1997) and sequence (see http://genome.nhgri.nih.gov/chr7) maps. Specifically, genes included both in previous comparative maps (see http://www.informatics.jax.org and http://www.ncbi.nlm.nih.gov/Homology) and in either the human chromosome 7 STS map (n=58) or mapped chromosome 7 genomic sequence (n=58) were ordered first. As part of a comprehensive effort to compare all mouse mRNA sequences against human chromosome 7 genomic sequence, additional mapped mouse genes orthologous to human chromosome 7 sequence were then added to the map (n=25). To better define regions of interest and enhance the comparative map, a small number of mouse orthologs of genes detected in human genomic sequence were genetically (n=8) or cytogenetically (n=5) mapped in mouse.
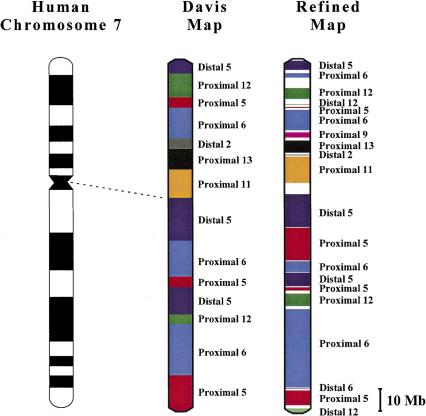
A small region of the resulting comparative map is depicted in Figure 2 to illustrate the types of differences seen between the MGI cytogenetic-based map, the Davis Human/Mouse Homology Map, and our refined map. Numerous new genes are present on the MGI-derived and refined maps but absent on the older Davis map, with very few genes uniquely present on either the MGI or Davis map compared to the refined map. Within the individual conserved genomic segments, a comparison of gene order is either not possible (e.g., between the MGI and refined maps) or reveals multiple differences [e.g., as seen between the Davis and refined maps (Fig. 2)]. These differences were expected because many of the human genes had not been ordered previously relative to one another, and thus in the Davis map, their positions were based on the inferred mouse order (rather than the human order), which could differ between species (DeBry & Seldin 1996). In addition, the order of conserved segments is occasionally different between the Davis and refined maps. For instance, the positions of the conserved segments from proximal MMU6 and proximal MMU5 are inverted between the Davis and refined maps (see below). Finally, in some cases, the Davis map incorrectly assigns a gene to a conserved segment. For example, GNB2 is placed in the distal MMU5 conserved segment near the top of Figure 2 (corresponding to human 7q11), but in fact resides within a more telomeric segment with homology to distal MMU5 (corresponding to 7q22).
Figure 2.
Small region of the refined human-mouse comparative map corresponding to a segment of human chromosome 7. On the left is a cytogenetic human-mouse comparative map of human chromosome 7q11-q22, based on information derived from MGI (ftp://ftp.informatics.jax.org/pub/informatics/reports/HMD_Human1.sq1.rpt).The predicted order of many of the same genes within this region of chromosome 7, as depicted on the Davis Human/Mouse Homology Map (DeBry & Seldin 1996) (see http://www.ncbi.nlm.nih.gov/Homology), is shown in the middle. On the right is our refined comparative map of the identical region, assembled with the aid of an available STS map (Bouffard et al. 1997) and genomic sequence from human chromosome 7 (see text for details). The complete refined comparative map is available at http://genome.nhgri.nih.gov/chr7/comparative. Note that the MGI-derived map includes a broader region than that shown for the Davis and refined maps. No definitive human gene order can be readily deduced from the crude cytogenetic positions available from the MGI-derived map. Using older information, the Davis map combined human cytogenetic mapping data with mouse genetic mapping data to deduce an order for human genes. All three maps indicate that this region of human chromosome 7 is orthologous to at least three different regions of the mouse genome. However, comparison of the Davis and refined maps reveals that the order of the proximal MMU6 and proximal MMU5 segments are inverted, the gene order within these segments is significantly different, and two genes included in the Davis map (ICA1 and RELN) are not present in this region. In general, human gene symbols are shown, except in cases where the human gene has not been named, in which case the mouse symbol or accession number is used. Lines connect identical genes present on both the Davis and refined maps. Genes depicted in bold are unique to one map, while those depicted in green on the MGI-derived map are located on MMU12.
Taken together, our efforts to date have provided a more refined human-mouse comparative map for human chromosome 7. A global view of this is provided in Figure 3, which for illustrative purposes shows the sizes and relative order of the orthologous segments on the Davis map and our refined map. The former contains 14 conserved segments from seven distinct regions of the mouse genome, while the latter (to date) contains 20 conserved segments from ten distinct regions of the mouse genome. The six unique conserved segments on the refined map reflect four instances where new mapping data led to the addition of a conserved segment and two instances where simple misordering of genes on the Davis map resulted in a failure to detect conserved segments.
Figure 3.
Overview of human-mouse comparative maps for human chromosome 7. The refined comparative map of human chromosome 7 (see text for details) is shown on the right, along with the corresponding Davis Human/Mouse Homology Map (DeBry & Seldin 1996) (see http://www.ncbi.nlm.nih.gov/Homology) in the middle, and the chromosome 7 cytogenetic map on the left. The Davis map contains 14 orthologous segments from seven regions of the mouse genome. The refined map contains 20 orthologous segments from ten regions of the mouse genome. Evidence for all but one segment, MMU9 (which was defined by new genetic mapping data; J.W. Thomas and E.D. Green, unpubl.), is present in the MGI database. The refined map is drawn to scale, with the size of each established orthologous segment (in color) or unassigned region (white) estimated based on the number of mapped human STSs within the interval [assuming an average inter-STS spacing of 79 kb (Bouffard et al. 1997)]. The sizes of the conserved segments in the Davis map are not drawn to scale.
Comparison of the previous comparative maps with our refined map reveals two instances where the relative order of adjacent conserved segments is inverted: one on 7p between proximal MMU13 and distal MMU2 and the other shown in Figure 2 involving proximal MMU5 and proximal MMU6 (discussed above). Interestingly, we have encountered only a single case of potential conflict in the assignment of a gene to a conserved segment. Calumenin (CALU), whose cytogenetic-based assignment to 7q32 (Yabe et al. 1998) can be confirmed by its identification in genomic sequence from that region, has been mapped to MMU7 by genetic mapping (Yabe et al. 1997) but falls in the middle of a large conserved segment homologous to proximal MMU6. Preliminary physical mapping in the mouse is consistent with the linkage of calumenin (Calu) to proximal MMU6; as such, this gene is currently not included on the refined comparative map. It is also worth noting that three conserved segments (the proximal MMU5 segment on 7p, the distal MMU2 segment on 7p, and the distal MMU6 segment on 7q) are based on a single gene, making them somewhat tentative. However, indirect support for the presence of the proximal MMU5 segment comes from other conserved segments homologous to this region on human chromosome 7.
Although there are 20 conserved segments on the refined comparative map, these correspond to just ten nonadjacent regions of the mouse genome. This reflects the fact that five conserved segments in mouse (proximal MMU5, proximal MMU6, distal MMU5, distal MMU12, and proximal MMU12) are homologous to 15 distinct segments on human chromosome 7. The latter provides strong evidence for the occurrence of intrachromosomal rearrangements, particularly inversions, during the evolution of human chromosome 7. For example, at least three inversions likely occurred to produce the four human chromosome 7 segments homologous to proximal MMU5. Thus, intrachromosomal rearrangements can be hypothesized to be the mechanism producing roughly half of the conserved segments on human chromosome 7.
Because the refined comparative map is based on the linear STS and sequence maps, the size of each conserved segment can be estimated, as can the sizes of the gaps (corresponding to regions for which the orthologous mouse region is unknown) (Fig. 3). Based on STS content [and assuming an average inter-STS spacing of 79 kb (Bouffard et al. 1997)], the average size of the conserved segments is ∼7.1 Mb, with the largest and smallest being ∼37 and <0.1 Mb, respectively. It is not possible to compare directly the sizes of the chromosome 7 conserved segments between the Davis and refined comparative maps since the former reflects a compilation of data from multiple maps. However, the average size of conserved segments across the mouse genome has been estimated at 8.1 (±1.6) cM (Nadeau & Taylor 1984). Given their assumption of a ∼1600-cM mouse genome (Nadeau & Taylor 1984) and the additional assumption that the mouse genome spans ∼3000 Mb, one might postulate that the average size of a conserved segment in the mouse genome is 15.2 ( ± 3.0) Mb. This genome-wide estimate is over twice that observed with the current refined comparative map of human chromosome 7, even after inflating our estimated average conserved segment size by ∼15% to account for the portion of the chromosome that cannot currently be assigned to a position in mouse (see below). Whether this difference reflects refined mapping data, specific features of chromosome 7, or some combination of these should become clearer with additional mapping and sequencing of both genomes.
Our efforts have resulted in the predicted assignment of ∼85% of human chromosome 7 to a defined position in the mouse genome. The remaining portions of the chromosome (‘gaps’) correspond to regions residing between conserved segments; these average ∼1.4 Mb, with the largest and smallest estimated to be ∼5 and <0.1 Mb, respectively. To reduce the amount of chromosome 7 without an established corresponding position in mouse, orthologous genes and ESTs within these gaps are being identified in human genomic sequence and then genetically mapped in mouse. In five of the first seven cases, such an effort simply extended one of the adjacent conserved segments; in the remaining two cases, a new conserved segment (homologous to proximal MMU9 on 7p) was defined. The amount of chromosome 7 not represented on the refined comparative map should be reduced further by the additional mapping of orthologous genes and ESTs in the mouse (e.g., by the clone-based physical mapping detailed below) and eventually by the sequencing of the corresponding regions of the mouse genome.
Construction of Mouse BAC-based Sequence-ready Maps
A major program to sequence the mouse genome as part of the ongoing Human Genome Project has recently been launched (Battey et al. 1999) (see http://www.nih.gov/science/models/mouse). To facilitate the sequencing of regions of the mouse genome corresponding to human chromosome 7, we are extending our comparative mapping efforts to include the construction of mouse BAC-based physical maps (Fig. 1).
Similar to constructing refined comparative maps, the key computational step for this endeavor involves analyzing all new human genomic sequence for the presence of orthologous mouse sequences. In this case, however, the orthologous sequences (occasionally known genes but most often ESTs) need not be mapped in the mouse genome for use in the construction of corresponding mouse physical maps. Although by no means assured, the central working assumptions are that gene content and order within a conserved segment are identical between human and mouse and that a single mouse BAC contig can be assembled for that region. Such assumptions provide a framework for evaluating probe order and content in the assembled mouse contigs, directing contig mergers, estimating the sizes of gaps between adjacent mouse contigs, and detecting potential differences in gene order between human and mouse. This general approach also allows blocks of human genomic sequence as well as orthologous, unmapped mouse genes/ESTs to be assigned predicted positions in the mouse genome based on their relationship with already mapped, flanking genes. In addition, the relative confidence for such predicted assignments can be made based on proximity to the informative flanking markers.
The analysis of human genomic sequence for detecting the presence of orthologous mouse sequences involves the following steps. Available human sequence, either in a preliminary working draft or high-accuracy finished form, is compared to a series of GenBank databases (nr, month, htgs, and dbEST; see http://www.ncbi.nlm.nih.gov) using the program PowerBLAST (Zhang & Madden 1997) (Fig. 4) and/or to a local, customized database consisting of all publicly available mouse and rat mRNA sequences using WebBLAST (Ferlanti et al. 1999). The resulting sequence alignments provide valuable overviews of the gene content and structure of each region (see Fig. 4).
Figure 4.
Accelerated construction of a mouse sequence-ready BAC contig using human genomic sequence. Roughly 180 kb of genomic sequence (top line) from human chromosome 7q22 (GenBank AC004022 and AC005021) was analyzed by RepeatMasker (A.F.A. Smit and P. Green, unpubl.; see http://www.genome.washington.edu/UWGC/analysistools/repeatmask.htm) to mask repetitive elements and then compared to GenBank using PowerBLAST (Zhang & Madden 1997). The small red dots below the human sequence represent a simplified view of matching mouse and rat mRNA/EST sequences. Three complete genes (PON1, PON2, PON3) and the 3′ end of a gene similar to rat neurabin (GenBank U72994) were detected in the region, with arrows indicating the direction of transcription. Based on this analysis, four overgo-type hybridization probes were designed from the matching mouse sequences (indicated by dashed arrows, with their names reflecting the corresponding GenBank accession numbers), optimizing for gene content and spacing (see text for details). These probes along with others designed from flanking human sequence were used to screen the mouse RPCI-23 BAC library, with the resulting probe-content data allowing assembly of three nascent contigs. The subsequent development and mapping of BAC insert end-specific overgo probes (indicated by squares) allowed the merger of the three contigs into the depicted >1-Mb contig from mouse chromosome 6.
While PowerBLAST output provides a highly detailed and comprehensive analysis of a region, the actual computer time and file storage can be limiting when applied on a large scale. Thus, by reviewing carefully the results from PowerBLAST analysis of ∼200 BAC sequences, we have been able to establish stringent criteria for establishing human-mouse orthology. These criteria can be applied to the more streamlined and time-efficient WebBLAST program to compare our local rodent mRNA database with new human genomic sequence.
Based mainly on our PowerBLAST analyses and using a cutoff value of E=1e-25, we have found that ∼50% of human BAC sequences contain at least one matching orthologous rodent sequence. This figure is highly variable from region to region, reflecting significant differences in gene density and an incomplete collection of rodent mRNA sequences. However, these findings suggest that our general approach should yield nascent physical maps for mouse genomic segments corresponding to at least 50% of sequenced human regions. Our analyses can also be used to estimate the percentage of all human genes with at least one matching orthologous rodent mRNA sequence. We have found this number to be >80%, consistent with the findings of Marra et al. (1999).
For mouse BAC library screening, we have emphasized the development and use of ‘overgo’-type hybridization probes. Such probes are derived by subjecting two ∼22-base oligonucleotides with an 8-base complementary overlap to a radioactive primer extension reaction, thereby producing a double-stranded ∼36-bp probe (Vollrath 1999) (see http://genome.wustl.edu/gsc). Specifically, the aligned rodent gene/EST and human genomic sequence is examined, and overgo probes that are fully contained within individual exons are designed (so as to insure contiguity of the probe and mouse genomic sequence). Probes are generated for all independent genes within a region and, when possible, an attempt is made to develop probes at somewhat regular spacing (Fig. 4), albeit with the caveat that the mouse gene structure, order, and size are inferred. For the RPCI-23 mouse BAC library (see http://bacpac.med.buffalo.edu), with its average BAC size of ∼200 kb (J.W. Thomas, V.V. Braden Maduro, and E.D. Green, unpubl.), an inter-probe spacing of ∼65 kb would be desirable (since clones would then contain an average of at least two probe sequences). In cases where more than one probe are developed for a gene, a fixed probe order can be imposed, given that exon order between orthologs should not vary. This data becomes useful in evaluating the integrity of the nascent BAC contig maps. Similarly, achieving high probe densities helps to verify that positive clones contain the true orthologous sequence as opposed to a closely related sequence (e.g., a pseudogene). For example, clones containing the authentic gene should often be positive for another nearby probe(s), unlike clones containing only a related sequence. Finally, in our experience >95% of the designed mouse overgo probes successfully identify at least one positive mouse clone within the RPCI-23 library.
To screen the mouse RPCI-23 BAC library on a large scale, we have implemented a two-stage hybridization-based approach. The first round involves hybridizing filters containing arrays of all the clones in the library with pools of 30–50 overgo probes. Single-colony isolates of all candidate positive clones are then rearrayed onto new (secondary) filters, which in turn are analyzed with combinatorial pools derived from the starting set of overgo probes. These second-round combinatorial pools are constructed by arraying the overgo probes and pooling individual rows and columns (i.e., two-dimensional pooling). Each row/column pool is then hybridized to an individual secondary filter. The data from the second round of hybridizations can then be disambiguated to produce the probe-clone relationships. This entire two-stage screening procedure is greatly facilitated by use of the program ComboScreen (Jamison et al. 2000) (see http://genome.nhgri.nih.gov/comboscreen), which has been developed in conjunction with our ongoing mouse mapping efforts. This program is tailored specifically for assimilating the data from both rounds of hybridization, including the design of secondary filters and combinatorial pools, as well as analyzing the results to deduce probe-clone relationships.
The resulting probe-content data can be analyzed with the program SEGMAP (Green & Green 1991) (see http://www.genome.washington.edu/UWGC/analysistools/segmap.htm) to assemble nascent contig maps that reflect both the predicted probe order and the relative overlaps among BACs. Such contigs can then be readily expanded by developing new overgo probes from insert-end sequences of BACs residing at contig ends [e.g., generated by bubble-PCR (Riley et al. 1990)] and rescreening the BAC library. Figure 4 depicts a representative example of a SEGMAP-constructed BAC contig spanning >1 Mb of proximal MMU6 containing Pon1, Pon2, Pon3, and Dncic1. Newly mapped and ordered genes in this contig include the orthologs of the rat neurabin gene and human PDK4 and SLC25A13. Note that the deduced order for the six mapped genes is the same in human and mouse.
In addition to being a tool for contig expansion, BAC insert-end sequences can often provide mapping information based on their similarity to human genomic sequence. For example, ∼7% of our nonredundant BAC-end sequences (n= >1000) have significant similarity to human genomic sequence. Such matching sequence can be utilized to solidify the evolving human-mouse comparative map. Another cross-species resource involves designing overgo probes from rat mRNA sequences. In our preliminary experience, >60% of such rat probes can be used to isolate corresponding mouse BACs (with a similar percentage of mouse probes successfully isolating corresponding rat BACs).
The above strategy for constructing BAC contigs has been implemented for mapping mouse genomic regions homologous to human chromosome 7. To date, we have analyzed the sequence for greater than half of human chromosome 7 (∼100 Mb), resulting in the design of >400 gene/EST-based overgo probes. In addition, we have developed >400 probes from generated BAC-end sequences. Together, these probes have been used to assemble >3800 BACs from the RPCI-23 library into 110 contigs that are estimated to span a total of ∼61 Mb of the mouse genome. Thus, our firstpass mapping efforts appear to have yielded mouse BAC contigs corresponding to more than half of the human sequence analyzed. Additional rounds of contig expansion coupled with the increasing availability of genome-wide mapping resources (http://www.nih.gov/science/models/mouse), human genomic sequence (http://www.ncbi.nlm.nih.gov/genome/seq), and mouse ESTs (http://www.ncbi.nlm.nih.gov/dbEST) should facilitate expansion of these mouse physical maps. Finally, groups of BACs already assigned to individual contigs (such as those depicted in Fig. 4) can be readily subjected to restriction enzyme digest-based fingerprint analysis (Marra et al. 1997), allowing the assembly of higher-resolution contig maps (i.e., ‘sequence-ready maps’) and the rational selection of minimally overlapping clones suitable for systematic sequencing.
In summary, the approaches we describe here represent effective strategies for utilizing newly generated reference genomic sequence from one species, in this case human, to refine comparative maps and to facilitate the rapid construction of ‘sequence-ready’ physical maps. These methods are efficient and can be implemented in a relatively small-scale fashion, with all the results mentioned above generated with ∼3 person-years of effort. Together, these mapping studies yield detailed information about the relationships among homologous mouse and human genomic segments, provide well-characterized clone sets for use in genomic sequencing, and, in the long run, should markedly enhance our understanding of mammalian genome structure and evolution. Finally, while human-mouse comparative mapping has been emphasized here, our approaches can be readily adapted for use in the comparative analyses of other vertebrate species.
Acknowledgments
We thank the Washington University Genome Sequencing Center and University of Washington Genome Center for generating human chromosome 7 sequence data. We also thank Bill Pavan for critical review of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL egreen@nhgri.nih.gov; FAX (301) 402–4735.
REFERENCES
- Battey J, Jordan E, Cox D, Dove W. An action plan for mouse genomics. Nat Genet. 1999;21:73–75. doi: 10.1038/5012. [DOI] [PubMed] [Google Scholar]
- Bouffard GG, Idol J R, Braden VV, Iyer LM, Cunningham AF, Weintraub LA, Touchman JW, Mohr-Tidwell RM, Peluso DC, Fulton RS, et al. A physical map of human chromosome 7: An integrated YAC contig map with average STS spacing of 79 kb. Genome Res. 1997;7:673–692. doi: 10.1101/gr.7.7.673. [DOI] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L Members of the DOE and NIH Planning Groups. New goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, et al. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- Dietrich W, Katz H, Lincoln SE, Shin H-S, Friedman J, Dracopoli NC, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Miller JC, Steen RG, Merchant M, Damron D, Nahf R, Gross A, Joyce DC, Wessel M, Dredge RD, et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat Genet. 1994;7:220–225. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Dietrich W F, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls K A, O'Connor T J, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Ferlanti ES, Ryan JF, Makalowska I, Baxevanis AD. WebBLAST 2.0: An integrated solution for organizing and analyzing sequence data. Bioinformatics. 1999;15:422–423. doi: 10.1093/bioinformatics/15.5.422. [DOI] [PubMed] [Google Scholar]
- Green ED, Olson MV. Chromosomal region of the cystic fibrosis gene in yeast artificial chromosomes: a model for human genome mapping. Science. 1990;250:94–98. doi: 10.1126/science.2218515. [DOI] [PubMed] [Google Scholar]
- Green ED, Green P. Sequence-tagged site (STS) content mapping of human chromosomes: theoretical considerations and early experiences. PCR Methods Appl. 1991;1:77–90. doi: 10.1101/gr.1.2.77. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu S-H, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Jamison, D.C., J.W. Thomas, and E.D. Green. 2000. ComboScreen facilitates the multiplex hybridization-based screening of high-density clone arrays. Bioinformatics (in press). [DOI] [PubMed]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O'Brien SJ. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Zhang J, Boguski MS. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M, Hillier L, Kucaba T, Allen M, Barstead R, Beck C, Blistain A, Bonaldo M, Bowers Y, Bowles L, et al. An encyclopedia of mouse genes. Nat Genet. 1999;21:191–194. doi: 10.1038/5976. [DOI] [PubMed] [Google Scholar]
- Matise TC, Wasmuth JJ, Myers RM, McPherson JD. Somatic cell genetics and radiation hybrid mapping. In: Birren B, et al., editors. Genome analysis: A laboratory manual. Vol. 4 Mapping genomes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 259–302. [Google Scholar]
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. Counting on comparative maps. Trends Genet. 1998;14:495–501. doi: 10.1016/s0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- Nagaraja R, MacMillan S, Kere J, Jones C, Griffin S, Schmatz M, Terrell J, Shomaker M, Jermak C, Hott C, et al. X chromosome map at 75-kb STS resolution, revealing extremes of recombination and GC content. Genome Res. 1997;7:210–222. doi: 10.1101/gr.7.3.210. [DOI] [PubMed] [Google Scholar]
- Olson M, Hood L, Cantor C, Botstein D. A common language for physical mapping of the human genome. Science. 1989;245:1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Reeves RH, D'Eustachio P. Genetic and comparative mapping in mice. In: Birren B, et al., editors. Genome analysis: A laboratory manual. Vol. 4 Mapping genomes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 71–133. [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- The Sanger Centre and The Washington University Genome Sequencing Center. Toward a complete human genome sequence. Genome Res. 1998;8:1097–1108. doi: 10.1101/gr.8.11.1097. [DOI] [PubMed] [Google Scholar]
- Vollrath D. DNA markers for physical mapping. In: Birren B, et al., editors. Genome analysis: A laboratory manual. Vol. 4 mapping genomes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 187–215. [Google Scholar]
- Waterston R, Sulston JE. The Human Genome Project: Reaching the finish line. Science. 1998;282:53–54. doi: 10.1126/science.282.5386.53. [DOI] [PubMed] [Google Scholar]
- Yabe D, Nakamura T, Kanazawa H, Tashiro K, Honjo T. Calumenin, a Ca2 +-binding protein retained in the endoplasmic reticulum with a novel carboxyl-terminal sequence, HDEF. J Biol Chem. 1997;272:18232–18239. doi: 10.1074/jbc.272.29.18232. [DOI] [PubMed] [Google Scholar]
- Yabe D, Taniwaki M, Nakamura T, Kanazawa N, Tashiro K, Honjo T. Human calumenin gene (CALU): cDNA isolation and chromosomal mapping to 7q32. Genomics. 1998;49:331–333. doi: 10.1006/geno.1998.5245. [DOI] [PubMed] [Google Scholar]
- Zhang J, Madden TL. PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]