Abstract
Ordered gene maps of mammalian species are becoming increasingly valued in assigning gene variants to function in human and animal models, as well as recapitulating the natural history of genome organization. To extend this power to the domestic cat, a radiation hybrid (RH) map of the cat was constructed integrating 424 Type I-coding genes with 176 microsatellite markers, providing coverage over all 20 feline chromosomes. Alignment of parallel RH maps of human and cat reveal 100 conserved segments ordered (CSOs) between the species, nearly three times the number observed with reciprocal chromosome painting analyses. The observed number is equivalent to theoretical predictions of the number of conserved segments to be found between cat and human, implying that 300–400 Type I gene markers is sufficient to reveal nearly all conserved segments for species that exhibit the most frequently observed “slow” rate of genome reorganization. The cat–human RH map comparisons provide a new genomic tool for comparative gene mapping in the cat and related Felidae, and provide confirmation that the cat genome organization is remarkably conserved compared with human. These data demonstrate that ordered RH-based gene maps provide the most precise assessment of comparing genomes, short of contig construction or full-sequence determination.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AZ081249–AZ081287 and AW646734–AW646956.]
The comparative method remains one of the most powerful approaches in the biological sciences (Harvey and Pagel 1991). The field of comparative genomics represents an invaluable extension of the Human Genome initiative, whereby genomic structure and function are interpreted in the evolutionary context of more manipulatable related animal species (O'Brien et al. 1997b,1999). Comparative gene maps serve two primary functions. First, they allow transfer of information from the gene-rich species maps of human and mouse to moderate-resolution gene maps of animal species that serve as models for biomedical, economical, and agricultural traits. Second, they provide a basis for understanding the rates and patterns of genome evolution (O'Brien et al. 1993; Andersson et al. 1996; VandeBerg and Graves 1998; O'Brien et al. 1999). Both of these endeavors require moderately dense comparative gene maps demarcating boundaries of conserved synteny among representative species.
At present, three species, human, mouse, and rat, have gene-dense, ordered Type I maps for every chromosome (Copeland et al. 1993; Dietrich et al. 1997; Deloukas et al. 1998; Watanabe et al. 1999), producing a strong framework for comparing ordered gene maps of other mammalian species. However, the rodent genome comparisons appear to be rearranged two- to threefold more extensively than genomes from most other compared mammalian species genomes that display a highly conserved pattern of genome-wide conservation among mammalian Orders (Graves 1996; Carver and Stubbs 1997; O'Brien et al. 1999). Other than rodents, most whole genome comparisons to humans have been limited to partial maps on the basis of unordered somatic cell hybrid or Zoo-FISH (chromosome painting)-based synteny, which do not reveal intrachromosomal inversions that have occurred during the evolution of these species (Chowdhary et al. 1998; Ferguson-Smith et al. 1998; Wienberg and Stanyon 1998; Finelli et al. 1999). It is becoming clear that a complete view of the extent and character of genome rearrangements that have occurred during species evolution must include precise orders of compared Type I-coding genes on each chromosome in different mammalian genomes (O'Brien 1991; Copeland et al. 1993; Womack and Kata 1995; Schibler et al. 1997; Yang and Womack 1998; Murphy et al. 1999a,b; Watanabe et al. 1999).
The domestic cat has long played a role in comparative medicine beginning with comparative anatomy, physiology, and biochemistry, extending to the present, in which the species provides numerous models for human heritable and infectious disease (http://www.angis.org.au/Databases/BIRX/omia/) including lysosomal storage diseases (Yogalingam et al. 1996; Berg et al. 1997; Fyfe et al. 1999), polycystic kidney disease (Biller 1996), and AIDS (FIV-feline immunodeficiency virus; Willett et al. 1997). The domestic cat also provides a valuable reference for comparative genome analysis, displaying remarkable syntenic conservation relative to the human genome (O'Brien et al. 1997a,b, 1999; Rettenberger et al. 1995; Wienberg et al. 1997). As for most published comparative mapping efforts in other species, these cat–human synteny observations are based on somatic cell-hybrid mapping, FISH, and Zoo-FISH results, but cannot reveal underlying subchromosomal conservation or rearrangement relative to other species.
Whole-genome radiation hybrid (RH) mapping has become a powerful and expedient method for gene ordering in vertebrate species by largely eliminating the need to identify within-species polymorphism (Cox et al. 1990; Walter et al. 1994). To date, this approach has been used to generate genome-spanning maps for the human, rat, and mouse genomes (Stewart et al. 1997; Deloukas et al. 1998; Steen et al. 1999; Van Etten et al. 1999; Watanabe et al. 1999), although maps are beginning to be developed for other mammalian species (Radiation Hybrid Database: www.linkage.rockefeller.edu/tara/rhmap). Using the feline RH panel (Murphy et al. 1999a), we present an RH map for the domestic cat that integrates 424 Type I (coding) loci with 176 selected Type II microsatellite loci ordered previously in an interspecies backcross-generated linkage map (Menotti-Raymond et al. 1999). This map represents the first ordered, full-genome parallel RH comparison to the human genome for a non-rodent species. The integrated 600 marker map has a mean density of 5 cM/26 cR5000 now useful for comparative candidate positional clone analysis of the >200 described feline hereditary phenotypes. A comparison of human to feline conserved segments revealed 100 ordered segments across both species genomes, more than three times the number revealed by previous comparative genomic methodologies (O'Brien et al. 1997a,b; Rettenberger et al. 1995; Wienberg et al. 1997). These data demonstrate the increased comparative precision required for accurate evolutionary inference and the achievement of this precision by moderately dense RH maps of Type I-coding gene markers.
RESULTS
Map Construction and Characterization
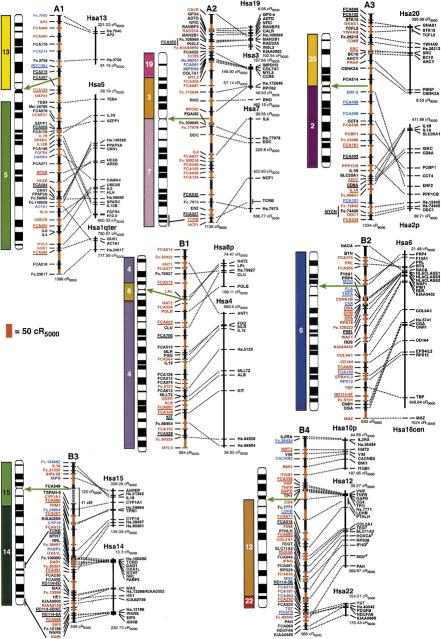
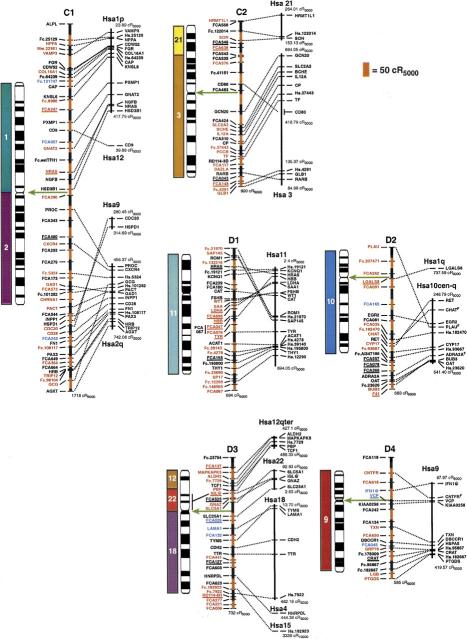
The cat RH map contains a total of 600 Type I-coding loci and Type II microsatellite markers (Table 1) typed on the 93-hybrid, 5000-rad feline RH panel (Murphy et al. 1999a). The 424 coding loci placed on the final map (Fig.1) were comprised of the following markers: 228 feline-specific ESTs, 70 feline genes deposited in GenBank, 70 comparative anchor locus-derived STSs (Venta et al. 1996; Lyons et al. 1997; Jiang et al. 1998), 40 newly developed or published (Murphy et al. 1999a,b) locus-specific STSs developed in our laboratory, seven human ESTs (Chen et al. 1999), and nine STSs for feline-specific endogenous retroelement insertions (RD-114). A detailed list of primers, sources, and locus information is available from the following website (http://lgd.nci.nih.gov). The 176 microsatellites were selected to represent an equivalently spaced subset of loci that would provide an initial cross reference to the feline linkage map (Menotti-Raymond et al. 1999). Together, these markers cover all 18 feline autosomes and X and Y chromosomes, with all but two loci occupying unique coordinates on the map. Two-hundred ninety-one loci have been ordered at odds ≥1000:1, with an additional 54 loci ordered at odds ≥100:1 (Fig. 1). The average intermarker spacing is estimated at 25.7 cR5000.
Table 1.
Feline RH Map Statistics
| Chr. | Total loci | Loci | Phys.length (Mb)a | cR | kb/cR | Avg. ret. freq. | |
|---|---|---|---|---|---|---|---|
| type I | type II | ||||||
| A1 | 44 | 27 | 17 | 288 | 1398 | 206 | 0.30 (0.11–0.71) |
| A2 | 40 | 28 | 12 | 210 | 1136 | 185 | 0.35 (0.17–0.74) |
| A3 | 36 | 26 | 10 | 168 | 1034 | 162 | 0.32 (0.12–0.46) |
| B1 | 34 | 18 | 16 | 231 | 994 | 232 | 0.39 (0.15–0.80) |
| B2 | 33 | 29 | 4 | 174 | 883 | 197 | 0.53 (0.25–0.83) |
| B3 | 41 | 33 | 8 | 168 | 725b | 231 | 0.40 (0.30–0.65) |
| B4 | 43 | 29 | 14 | 162 | 966 | 167 | 0.43 (0.24–0.76) |
| C1 | 53 | 39 | 14 | 258 | 1718 | 150 | 0.31 (0.14–0.59) |
| C2 | 30 | 18 | 12 | 174 | 800 | 218 | 0.37 (0.24–0.52) |
| D1 | 31 | 23 | 8 | 144 | 694 | 207 | 0.39 (0.17–0.57) |
| D2 | 23 | 14 | 9 | 120 | 683 | 176 | 0.37 (0.18–0.64) |
| D3 | 29 | 18 | 11 | 120 | 732 | 164 | 0.47 (0.18–0.75) |
| D4 | 19 | 13 | 6 | 111 | 585 | 190 | 0.26 (0.17–0.41) |
| E1 | 33 | 26 | 7 | 111 | 553b | 232 | 0.66 (0.15–1.00)c |
| E2 | 24 | 18 | 6 | 90 | 508 | 178 | 0.49 (0.31–0.73) |
| E3 | 20 | 15 | 5 | 69 | 471 | 146 | 0.46 (0.27–0.72) |
| F1 | 18 | 14 | 4 | 87 | 412 | 211 | 0.37 (0.29–0.46) |
| F2 | 11 | 4 | 7 | 87 | 380 | 229 | 0.41 (0.24–0.63) |
| X | 30 | 24 | 6 | 150 | 679 | 221 | 0.26 (0.13–0.48) |
| Y | 8 | 8 | 0 | 69 | 73b | —b | 0.39 (0.33–0.46) |
| Total | 600 | 424 | 176 | 2991 | 15,424 | ||
| Avg. | 30 | 21 | 9 | 150 | 771 | 195 | 0.39 (0.11–1.00) |
Estimated assuming a genome size of 3000 Mb and the percentage of the total cytogenetic length represented by each chromosome (W. Nash, unpubl.).
Values are underestimates due to gaps in map (e.g. B3, E1) or poor physical coverage (e.g. Y).
Bears the selectable locus, thymidine kinase (TK).
Figure 1.
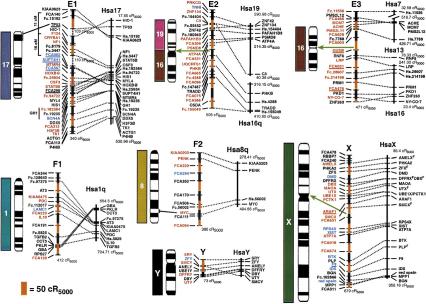
(See pages 693–695.) RH map of the domestic cat and human RH comparative map. Ideograms for each cat chromosome (Cho et al. 1997d) are shown to the left of each map. Hybridization results from human on cat chromosome painting (Rettenberger et al. 1995; Wienberg et al. 1997) are depicted to the left of each ideogram for comparison to the RH-derived syntenies. The extent of RH-based physical coverage of each chromosome is not implied, except where indicated by cytogenetic localization of Type I markers on the RH map (Cho et al. 1997a–c, 1998; Lee and Cho 1999). Orientation of RH maps relative to the chromosome is established except for the following chromosomes: D1, E3, F1, and Y. Green arrows indicate the inferred position of the centromere on the RH map, as determined by maximum retention frequency-based centromeric effects and syntenic boundaries. Loci labeled in red were ordered at odds ≥1000:1, those in blue ⩾100:1, those in black <100:1. The distances on the cat RH map are represented by alternating black and red vertical bars, with each bar corresponding to 50 cR5000. The length of each feline RH linkage group is listed below each map. The human RH maps are shown on the right. RH coordinates for the outermost markers of each human conserved homology block are listed. Human loci positioned on the Stanford G3-based RH map are depicted by a bar spanning the range of the same map interval in the Genebridge-4-based RH map (see Methods). Human ESTs that have not been associated with a named gene of known function are listed by their UniGene cluster identifier. Feline homologs of these loci are indicated by the prefix Fc. preceding the human cluster identifier number. The majority of feline microsatellites are denoted by the prefix FCA or an F, followed by a number (Menotti-Raymond et al. 1999). Broken lines connect homologous Type I loci in both cat and human RH maps. Gaps in the RH maps of B3 and E1 are connected by broken lines (no distance implied) and are bracketed by linked microsatellite loci in the genetic linkage map (Menotti-Raymond et al. 1999).
The average genome-wide retention frequency is 0.39, a value identical to that calculated previously from a much smaller number of loci (Murphy et al. 1999a). Marker retention frequencies varied along the chromosomes (Table 1), with prominent peaks near the centromeres of most bi-armed chromosomes. These regions of high-retention frequency are prominent and mark the centromeres of most chromosomes. Due to this effect, however, we were unable to detect linkage across the putative centromeric region on cat chromosome B3 (Fig. 1). Analyses of other chromosomes using a centromeric model (see Methods) allowed for reliable ordering across those showing marked centromeric effects. Feline chromosome E1, which bears the selectable locus thymidine kinase (TK), also contained two gaps in continuity in the RH map. These gaps result from the confounding effects of drastic changes in retention frequency, with quite low values (0.11) near the p-terminus increasing to high values (>0.70) covering much of the chromosome, likely due to the coupling of centromeric and selectable-locus effects. We tentatively connect and order these locus clusters on the basis of the orientation of included microsatellites from these same regions in the linkage map (Menotti-Raymond et al. 1999). Selectable locus models (the S2 model provided the greatest likelihood values; see Methods) facilitated ordering across the TK region, characterized by extremely high retention frequencies.
A total of 105 autosomal and 2 X-linked loci on the RH map have been mapped physically using the cat–rodent somatic cell hybrid panel (O'Brien et al. 1997a), allowing assignment to every feline chromosome except E2 and F1, which were poorly represented in the somatic cell hybrid panel (O'Brien et al. 1997a; Menotti-Raymond et al. 1999). RH linkage groups containing large tracks of Type I loci homologous to Hsa16/19 and Hsa1q were tentatively assigned to E2 and F1, respectively, on the basis of reciprocal full-genome Zoo-FISH data (Rettenberger et al. 1995; Wienberg et al. 1997). The assignment of the eight Y-linked loci has been reported previously (Murphy et al. 1999b). The orientation of linkage groups relative to each chromosome was determined for all but four chromosomes (D1, E3, F1, and Y) on the basis of reciprocal chromosome painting, FISH, or retention frequency-inferred centromere position on submetacentric chromosomes.
Comparison with the Microsatellite-Based Interspecies Genetic Map
The derived feline RH map (Fig. 1) revealed the resolution and orientation of marker order for previously unconnected linkage groups assigned to the same chromosome constructed from the interspecies backcross (Menotti-Raymond et al. 1999), as well as allowing placement of physically assigned but unlinked singleton loci to specific chromosomal positions. Discrepancies in marker order between the linkage and RH maps in most cases involved closely linked adjacent loci, loci ordered with lower confidence (LOD <2.0) than other markers in the recombination map (e.g., A2, B4), or chromosome translocation differences between the parent species of the interspecies backcross (e.g., D2). Four microsatellite linkage groups physically assigned on a tentative basis were reassigned in the present analysis to chromosomes D4, E2, E3, and F1 with high confidence, and further supported by comparative agreement of included Type I gene homologs with the predictions of Zoo-FISH (Rettenberger et al. 1995; Wienberg et al. 1997). A detailed comparison of the linkage and RH maps will be presented elsewhere, including an analysis of RH panel-retention patterns of both maternal and paternal chromosome segments revealed by microsatellite haplotype analysis (S. Sun et al., in prep).
Cat–Human Comparative Map
A comparison of genome reorganization revealed by previous reciprocal chromosome painting, which specify 30–32 homology segments between human and feline genomes (depending on the direction of comparison) is summarized by colored homology blocks in Figures 1 and 2. However, when the segments of homology between the two species are assessed by examining tracks of conserved Type I marker synteny (irrespective of gene order), the number of conserved segments unordered (CSUs) increases to 63–64 homology segments (Fig.1, Table 2). Further, when inversions or rearrangements that alter gene order are considered, we count 100–102 CSOs between cat and human genome organization. Thus, the actual number of differences in genome ordering of Type I genes is two to three times more abundant than has been observed by chromosome painting analyses (Rettenberger et al. 1995; Wienberg et al. 1997).
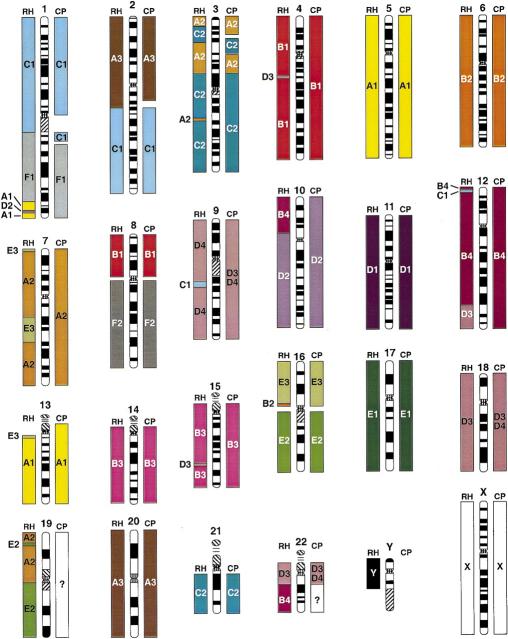
Figure 2.
Comparison of feline gene mapping (RH) and chromosome painting (CP) identified conserved segments relative to human chromosomes. The width and placement of the bars is approximate on the basis of cytogenetic assignment of loci in the blocks or proximity of ESTs to assigned human loci in the human RH maps. The smaller blocks do not imply coverage on the chromosome, but rather the number of genes per block.
Table 2.
Comparative Mapping Statistics for Observed Cat–Human Homologies
| Cat | Human | ||||||
|---|---|---|---|---|---|---|---|
| paint | RH | paint | RH | ||||
| CSU | CSU | CSO | CSU | CSU | CSO | ||
| A1 | 2 | 5 (4) | 10 (6) | Hsa1 | 2 | 5 (3) | 7 (6) |
| A2 | 3 | 7 (4) | 9 (4) | Hsa2 | 4 | 2 | 9 (6) |
| A3 | 2 | 2 | 7 (5) | Hsa3 | 4 | 6 (4) | 8 (3) |
| B1 | 3 | 3 (2) | 5 (4) | Hsa4 | 1 | 4 (2) | 4 (2) |
| B2 | 1 | 2 (1) | 4 (3) | Hsa5 | 1 | 1 | 6 (5) |
| B3 | 2 | 6 (4) | 7 (5) | Hsa6 | 1 | 1 | 4 |
| B4 | 2 | 5 (4) | 7 (5) | Hsa7 | 1 | 4 (3) | 5 (3) |
| C1 | 2 | 6 (5) | 9 (6) | Hsa8 | 2 | 2 | 3 |
| C2 | 2 | 3 | 4 (3) | Hsa9 | 1 | 3 | 4 |
| D1 | 1 | 1 | 4 | Hsa10 | 1 | 4 (2) | 6 (2) |
| D2 | 1 | 3 (1) | 6 (2) | Hsa11 | 1 | 1 | 4 |
| D3 | 3 | 6 (3) | 7 (3) | Hsa12 | 1 | 4 (2) | 6 (3) |
| D4 | 1 | 1 | 2 | Hsa13 | 1 | 2 (1) | 3 (2) |
| E1 | 1 | 1 | 4 | Hsa14 | 1 | 3 | 4 (3) |
| E2 | 2 | 4 (2) | 4 (3) | Hsa15 | 1 | 5 (2) | 6 (2) |
| E3 | 1 | 4 (2) | 4 (2) | Hsa16 | 2 | 3 (2) | 3 (2) |
| F1 | 1 | 1 | 2 | Hsa17 | 1 | 1 | 4 |
| F2 | 1 | 1 | 1 | Hsa18 | 1 | 2 (1) | 2 (1) |
| X | 1 | 1 | 1 | Hsa19 | 0 | 4 (2) | 5 (3) |
| Y | 0 | 1 | 3 (2) | Hsa20 | 1 | 1 | 1 |
| Hsa21 | 1 | 1 | 1 | ||||
| Total | 32 | 63 (44) | 100 (68) | Hsa22 | 2 | 3 | 3 |
| HsaX | 1 | 1 | 1 | ||||
| HsaY | 0 | 1 | 3 (2) | ||||
| Total | 30 | 64 (45) | 102 (70) | ||||
Numbers indicate direct counts of chromosome comparisons of feline chromosomes to human chromosomes considering homologous Type I gene locations and order in each species (see Methods). (CSU) Conserved segments unordered; (CSO) conserved segments ordered. In parentheses, CSUs and CSOs containing ≥2 mapped homologous loci.
The increased number and precision of conserved segments can be explained by two observations. The first involves the resolution of homology segments not revealed by chromosome painting. For example, we identified two sizable homology segments not observed with reciprocal Zoo-FISH, Hsa 10 on B4p and Hsa 7 on E3 (Fig. 1). The feline chromosome B4 association of segments from human chromosomes 10, 12, and 22 is observed in other carnivore species, and the chromosome E3 association of Hsa7 and Hsa16 is found in other mammalian Orders (Bielec et al. 1998; Chowdhary et al. 1998; Volleth et al. 1999). In addition, seven smaller segments of one to three loci were also discovered on feline chromosomes A1, B2, C1, D2, D3, and E3 (Fig. 1).
The second observation is the high frequency of intrachromosomal inversions (or translocations) that have reordered genes in the two species' maps. Every cat chromosome except for the X and F2 displays evidence of within-chromosome reordering compared with the homologous human gene segment. A chromosome by chromosome summation of the number of CSUs and CSOs observed by comparing the RH gene order in both cat and human is presented in Table 2.
A comparison of the conserved segments (CSUs) revealed by chromosome painting and by RH mapping of cat and human gene homologs is illustrated in Figure 2. It is clear from this analysis that RH mapping results in greater resolution of conserved segments than chromosome painting, in addition to revealing putative intrachromosomal rearrangements that cause uninterrupted conserved homology segments to become interrupted with homology segments from a different cat chromosome. For example, human chromosomes 1, 7 and 9 display interruptions of homology segments with at least two genes affirming the interruptions, whereas human chromosomes 1, 3, 4, 12, 15, 16, and 19 have an interstitial segment defined by a single marker.
If we consider gene order as the basis for segment homology (CSOs), we detected 100 conserved segments between cat and human genomes, with 68 of these involving two or more genes that span intervals greater than ∼20 cR. We did not consider ordering within smaller intervals because of decreased ordering power below this level. Using the model of Nadeau and Taylor (1984), we estimated the mean conserved segment length to be 153 (±12) cR5000, producing an expected estimate of 100 conserved segments between cat and human. This estimate is similar to the observed number of CSOs (Table 2), suggesting that most conserved segments found in the cat–human common ancestor is identified here. A comparison to mouse, rat, and other mammalian species with ordered maps would be useful here, but is not yet possible because of the low number of Type I markers mapped to date in rodent species that are homologous to feline Type I markers listed in Figure 1.
DISCUSSION
The domestic cat represents an ideal species for addressing a critical question concerning comparative maps: To what degree does conservation of synteny translate into conservation of gene order? Of all non-primate species examined by Zoo-FISH to date, the cat displays the fewest number of chromosomal changes relative to human (O'Brien et al. 1999), thus providing a base line for establishing whether such a correlation exists between conserved synteny and conserved order. Previous comparisons (of rat to human and goat to human) involving entire genomes have used orders inferred from FISH mapping, which can be ambiguous for ordering markers that are assigned to the same or overlapping chromosome bands (Schibler et al. 1997; Watanabe et al. 1999). Until now, the more precise parallel RH mapping has been applied only to single chromosomes (Yang and Womack 1998; Murphy et al. 1999b; Rexroad and Womack 1999). We present here the first ordered genome-wide comparison to humans for a non-rodent model species allowing parallel comparisons across the majority of both cat and human genomes. The results (Figs. 1 and 2, Table 2) provide a detailed, whole-genome view of the extent of genomic conservation and change that has occurred in ∼90 million years since primates and carnivores diverged from a common ancestor (Kumar and Hedges 1998).
Of the 425 mapped feline Type I loci, 353 of these had counterparts positioned on the human RH map, allowing for quite precise alignment of the two genomes. The observed number of conserved segments agrees with theoretical predictions (Nadeau and Taylor 1984), suggesting that a gene map of 300–400 Type I markers may offer sufficient density to identify the majority of conserved segments between any two species with genomes that are moderately conserved. The observed number of conserved segments relative to human were remarkably similar in comparisons of human to cat (100) and human to goat (107) maps (Schibler et al. 1997). However, 69% of conserved human segments represent intrachromosomal changes in the cat, compared with 53% in the goat (assuming 32 and 50 conserved human syntenies, respectively). In comparison, the rat and mouse genomes appear to have a far greater proportion of interchromosomal changes (76% and 75%–90%, respectively; Nadeau and Sankoff 1998; Wantanabe et al. 1999) relative to the human genome, probably because the shorter human-rodent conserved segments are less likely to contain intrachromosomal rearrangements. The rat comparison, however, is probably an overestimate because gene order was not taken into account (Wantanabe et al. 1999). Further, the actual numbers of conserved segments for both ruminants and rodents are probably somewhat higher than currently observed, due to the lower number of genes mapped in the goat and the use of lower resolution human cytogenetic positions as a reference in those comparisons. Nonetheless, the Nadeau-Taylor approximation for the estimated number of linkage disruptions since divergence from a common ancestor with human are 79 for the cat, >100 for the goat (Schibler et al. 1997), 132 for the rat (Watanabe et al. 1999), and 180 for the mouse (Nadeau and Taylor 1984; Nadeau and Sankoff, 1998). These results affirm and extend the impressions that the cat genome displays among the fewest rearrangements relative to human, implying that the human and cat genomes are rather close to the ancestral mammalian genome organization (O'Brien et al. 1997a,b, 1999; Rettenberger et al. 1997; Wienberg et al. 1997).
Although Zoo-FISH has been instrumental in our understanding of cross-genome homologies, gene mapping data such as ours further provide an important assessment of the detection sensitivity for heterologous chromosome painting. We discovered several small segments of one to three genes that were not observed by the Zoo-FISH results. Whereas it remains a possibility that some of the single gene assignments represent mapping errors or erroneous homology assessments, it is more likely that such small segments fall below the range of detection of Zoo-FISH (Wienberg and Stanyon 1998). Mapping of additional loci from these same small regions will be necessary to confirm the presence of these small conserved segments. Thirty to thirty-two conserved segments (conserved linkage with undetermined order) had been identified previously between cat and human on the basis of Zoo-FISH. The present analysis identified 63–64 CSUs and 100–102 CSOs between the two species. Clearly, Zoo-FISH results have underestimated the number of conserved segments at least twofold, a result that emphasizes the necessity for moderate resolution, ordered Type I-coding gene maps for comparative positional cloning (Womack and Kata 1995) and for evolutionary comparative inference (O'Brien et al. 1999). A similar comparative analysis of goat and human genome organization (using FISH-assigned goat Type I markers) also uncovered nearly twice the number of conserved segments as were imputed by Zoo-FISH of the same species (Schibler et al. 1997).
The parallel RH mapping data presented strongly emphasizes the need for constructing well-ordered comparative maps to extend Zoo-FISH analyses by delimiting boundaries of homology disrupted by intrachromosomal rearrangements, which can then be applied to phylogenetic or genome evolution studies (O'Brien et al. 1999). For example, the Hsa7–Hsa16 association observed on cat chromosome E3 was not detected by Zoo-FISH (O'Brien et al. 1997a,b; Rettenberger et al. 1995; Wienberg et al. 1997), although this association has been observed in another carnivore, the mink, in several Cetartiodactyls examined (e.g., cow, pig, Indian muntjac, and dolphin) and in bats (Bielec et al. 1998; Chowdhary et al. 1998; Volleth et al. 1999), providing a potentially valuable phylogenetic character for resolving the hierarchy of mammalian evolution. Failure of Zoo-FISH to consistently reveal this association in cat (and probably mink) may be a result of either sporadic fragmentation of the original association in independent carnivore lineages, or more likely, a reduction in heterologous probe sensitivity over increasing evolutionary divergence time (e.g., note that Hsa19 did not paint with any feline chromosome, Fig. 2). Another example of the increased resolution of the RH map is the detection of a Hsa10 segment on cat chromosome B4p associated with human chromosome 12 and 22 segments (Fig. 1). This same association was detected previously by Zoo-FISH in both mink and seal homologous chromosomes (Chowdhary et al. 1998), but in neither cat-human Zoo-FISH study (Rettenberger et al. 1995; Wienberg et al. 1997). In the absence of gene-mapping data, this character would have been misinterpreted as a derived character uniting pinnipeds and mustelids, when it actually represents a carnivore-specific synapomorphy. Hence, without reliable gene mapping data, it appears that a certain number of larger conserved chromosomal segments may go undetected, particularly as more ancient mammalian divergences are examined.
RH mapping foreseeably provides the most efficient tool for feasibly constructing genome-wide maps of a large number of representative mammalian species in the future. Recent development of high-throughput RH-based genotyping technologies for microsatellites (Van Etten et al. 1999) coupled with sets of conserved comparative markers (Venta et al. 1996; Lyons et al. 1997; Jiang et al. 1998) should facilitate the creation of integrated Type I and Type II gene maps in a diversity of species. Development of these maps integrating cross-genome comparisons with microsatellite and SNP diversity in a sufficient breadth of mammalian taxa will allow a multidimensional approach correlating genomic diversity with disease, morphological adaptations, and speciation in both domestic and exotic species.
METHODS
cDNA Library Construction and EST Development
The EST mapping strategy roughly follows the COMPASS approach described in Ma et al. (1998). Feline ESTs were generated by random sequencing of clones from a prepared domestic cat testis cDNA library, and a commercially available domestic cat heart cDNA library (Stratagene). The testis library was constructed by synthesizing cDNA from mRNA and unidirectionally cloning these products into the Uni-ZAP XR vector (Stratagene). The packaged library was amplified prior to mass excision of phagemids. Minipreps were performed on positive clones (on the basis of blue-white selection) using a 96-well format alkaline lysis protocol, and subsequently sequenced using ABI Big-Dye terminator chemistry on an ABI-377 sequencer. The sequences generated from both 5′ and 3′ ends were aligned to both nr and dbEST databases of GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Of the 479 high-quality sequences produced, 36 were of either mitochondrial DNA or repetitive element origin, and another 90 sequences produced no significant match (<80% sequence identity >200 bp in length) to the nr or EST database. Of the remaining 353 sequences, 244 matched known human genes or ESTs that had a known chromosomal assignment or location, and the remainder had not been assigned. Primers were designed for the majority of ESTs with known human chromosome locations using the Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primers were most frequently designed in 3′ untranslated regions (UTRs), although occasionally primers were designed from 5′-end sequences due to poor sequence generated through long poly-A tails. The majority of STSs designed were between 100 and 200 bp in length.
Comparative Loci, GenBank Loci, and Human EST Development
Primer pairs for new comparative loci were created using alignments of human and, when available, rodent sequences from GenBank. In cases in which intron size information was not available, we designed primers within large exons or UTRs that showed internal variation with available rodent sequence. The conserved segments were amplified and sequenced, and a second internal primer or primer pair was designed that was mismatched with the rodent sequence near the 3′ end to specifically amplify cat target sequence. Isolation and development of Y-specific loci were described previously (Murphy et al. 1999b). For feline sequences acquired from GenBank, primers were usually designed in 3′ UTRs, 5′-UTRs, or larger coding regions, depending on length and quality of information available. Primers amplifying a small number of human ESTs were tested in the cat for positive results. Loci for which a feline product was produced and out-competed hamster in pretests (see below) were sequenced and verified using BLAST. Loci giving positive matches were then typed on the feline RH panel.
Feline RH Panel Typing and Map Construction
PCR-based typing was performed on the 93-hybrid 5000-rad feline whole genome RH panel (Murphy et al. 1999a). All primer pairs were screened for performance prediction on the RH panel by pretesting on cat, hamster, and a 1:10 mix of cat and hamster DNA. Only those primer pairs producing robust cat amplification products accompanied by a negative or faint hamster product of different mobility were genotyped on the RH panel. Amplification and electrophoresis conditions were performed as described (Murphy et al. 1999a,b). All newly designed Type I amplification products were end sequenced for verification by BLAST search and deposited in GenBank. Microsatellite loci selected from the feline interspecies linkage map were amplified with fluorescently labeled primer pairs, genotyped on an Applied Biosystems-377 apparatus, and analyzed as described previously (Menotti-Raymond et al. 1999; Murphy et al. 1999a). All feline markers were genotyped and scored in duplicate, with discordant genotypes coded as 2. Locus vectors containing more than four discordancies between duplicate genotypes were discarded. Map construction was performed with the RHMAP software (Boehnke et al. 1991; Lunetta et al. 1995). Two-point linkage analysis was performed with RH2PT on a chromosome by chromosome basis, using information from the microsatellite-based linkage map, the somatic cell hybrid-based gene map, and reciprocal chromosome painting-based syntenies (O'Brien et al. 1997a,b; Rettenberger et al. 1995; Wienberg et al. 1997; Menotti-Raymond et al. 1999). A lower LOD threshold of 4.0 was used initially, due to low density of markers in some regions of the map. Those loci that did not display a 2-point LOD score ≥4.0 against their predicted chromosome were then screened against all previously identified linkage groups on other chromosomes at an increased threshold LOD≥6.0 to alleviate issues of spurious cross-chromosome linkage. To bridge a few gaps, a small number of loci were linked at LOD≥3.0, although these loci always resided on the chromosome predicted by Zoo-FISH and served to link multiple linkage groups observed at LOD≥4.0. Loci exhibiting significant linkage to multiple chromosomes were dropped. In all, > 97 % of loci exhibited pair-wise LOD scores ≥6.0, and 92% ≥8.0 to another marker. Linkage groups were ordered in RHMINBRK and RHMAXLIK, using the RHMAP 3.0 software mounted on a DEC-alpha high-speed computer. SAVMAX was initially set to 9 in RHMAXLIK, and increased in successive runs to increase the likelihood of finding the best orders. RH groups from larger chromosomes were successively divided into overlapping subsets for secondary ordering on the basis of the preliminary orders. Centromeric and selectable locus models were used for linkage groups spanning centromeres and the TK region of chromosome E1, respectively. Retention frequencies and other statistics were generated in RH2PT. Mapping information for some loci on cat chromosomes B4, D3, X, and all of Y were reported previously (Murphy et al. 1999a,b). New physical assignments to feline chromosomes (B4: Fc.26434, NMT2; D2: LGALS8; D4: CRAT, VCP) were performed on the rodent–cat somatic cell hybrid panel as described (O'Brien et al. 1997a; Menotti-Raymond et al. 1999). Investigators interested in mapping with the feline panel are encouraged to contact W.J. Murphy regarding panel distribution.
Human RH Comparative Mapping
The majority of the human RH data are from the UniGene (www.ncbi.nlm.nih.gov/UniGene/Hs.Home.html) and GeneMap '99 (www.ncbi.nlm.nih.gov/genemap) databases. The primary reference for human loci was the Genebridge-4-based map. When data were only available for the Stanford G3-based map, these were converted to a Genebridge-4-estimated range on the basis of the corresponding map interval (www.ncbi.nlm.nih.gov/genemap). Fourteen human loci had been assayed previously in our laboratory (Chen et al. 1999; Murphy et al. 1999b) using the human Genebridge-4 panel (Research Genetics) and positioned onto the current Sanger Center framework (www.sanger.ac.uk/RHserver). In most cases, primers were designed within 3' UTRs of sequences acquired from GenBank.
Estimating Comparative Mapping Statistics
Estimates of the number of conserved segments were divided into two categories, CSUs and CSOs. We chose to develop these terms to replace the more conventional, although frequently misused term, conserved linkage, which can imply either conserved or unconserved order. CSUs were defined as uninterrupted tracks of homologous genes residing on a single chromosome in two species, irrespective of internal order (conserved linkage, unconserved order). CSOs are analogous to CSUs, although they take into account tracks of genes for which the order is preserved between two species (conserved linkage, conserved order). Estimates for the actual number of conserved segments (ordered and unordered) between human and cat are based on the methods of Nadeau and Taylor (1984), which are based on conserved segments of two or more genes, assuming a random model of chromosomal breakage. We conservatively chose not to examine rearrangements within intervals < 20 centirays, due to the few number of chromosome breaks within segments of such length, and the saturation of these breaks with a small number of markers. Empirical observation of small map intervals generally showed very little difference in likelihood scores for alternative orders within such small intervals, justifying our conservative approach. Estimates for the number of rearrangements are calculated by subtracting the hypothesized number of chromosomes in the common ancestor of the two species being compared from the number of conserved segments. In the case of cat and human we used the midpoint, 21, between 19 and 23 haploid chromosomes, respectively.
Acknowledgments
We thank T. Beck, S. Cevario, V. David, J. Martenson, J. Menninger, K. Menotti, W. Nash, A. Snyder, and M. Thompson for assistance at various stages of this project. We acknowledge B. Stephens at the FCRDC supercomputer center for allotting usage time and space. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL murphywi@mail.ncifcrf.gov; FAX 301-846-6327.
REFERENCES
- Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Comparative genome organization of vertebrates. The first international workshop on comparative genome organization. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- Berg T, Tollersrud OK, Walkley SU, Siegel D, Nilssen O. Purification of feline lysosomal alpha-mannosidase, determination of its cDNA sequence and identification of a mutation causing alpha- mannosidosis in Persian cats. Biochem J. 1997;328:863–870. doi: 10.1042/bj3280863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielec PE, Gallagher DS, Womack JE, Busbee DL. Homologies between human and dolphin chromosomes detected by heterologous chromosome painting. Cytogenet Cell Genet. 1998;81:18–25. doi: 10.1159/000015002. [DOI] [PubMed] [Google Scholar]
- Biller DS, DiBartola SP, Eaton KA, Pflueger S, Wellman ML, Radin MJ. Inheritance of polycystic kidney disease in Persian cats. J Hered. 1996;87:1–5. doi: 10.1093/oxfordjournals.jhered.a022945. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Lange K, Cox DR. Statistical methods for multipoint radiation hybrid mapping. Am J Hum Genet. 1991;49:1174–1188. [PMC free article] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Chen ZQ, Lautenberger JA, Lyons LA, McKenzie L, O'Brien SJ. A human genome map of comparative anchor tagged sequences. J Hered. 1999;90:477–484. doi: 10.1093/jhered/90.4.477. [DOI] [PubMed] [Google Scholar]
- Cho KW, Okuda M, Endo Y, Satoh H, Kang CB, Watari T, Tsujimoto H, Hasegawa A. Assignment of the cat p53 tumor suppressor gene (TP53) to cat chromosome E1p14→p13 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1997a;79:145–146. doi: 10.1159/000134704. [DOI] [PubMed] [Google Scholar]
- Cho KW, Satoh H, Youn HY, Watari T, Tsujimoto H, Hasegawa A. Assignment of the feline c-myc gene (MYC) to cat chromosome F2q21.2 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1997b;78:135–136. doi: 10.1159/000134647. [DOI] [PubMed] [Google Scholar]
- Cho KW, Satoh H, Youn HY, Watari T, Tsujimoto H, O'Brien SJ, Hasegawa A. Assignment of the cat immunoglobulin heavy chain genes IGHM and IGHG to chromosome B3q26 and T cell receptor chain gene TCRG to A2q12→q13 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1997c;79:118–120. doi: 10.1159/000134696. [DOI] [PubMed] [Google Scholar]
- Cho KW, Youn HY, Watari T, Tsujimoto H, Hasegawa A, Satoh H. A proposed nomenclature of the domestic cat karyotype. Cytogenet Cell Genet. 1997d;79:71–78. doi: 10.1159/000134686. [DOI] [PubMed] [Google Scholar]
- Cho KW, Tsujimoto H, Hasegawa A, Satoh H. The cat immunoglobulin lambda light chain gene maps to chromosome D3p12– p11. Mamm Genome. 1998;9:178–179. doi: 10.1007/s003359900719. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Fronicke L, Scherthan H. Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo-FISH. Genome Res. 1998;8:577–589. doi: 10.1101/gr.8.6.577. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, et al. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: A somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith M, Yang F, O'Brien PCM. Comparative mapping using chromosome sorting and painting. ILAR J. 1998;39:68–76. doi: 10.1093/ilar.39.2-3.68. [DOI] [PubMed] [Google Scholar]
- Finelli P, Stanyon R, Plesker R, Ferguson-Smith MA, O'Brien PC, Wienberg J. Reciprocal chromosome painting shows that the great difference in diploid number between human and African green monkey is mostly due to non-Robertsonian fissions. Mamm Genome. 1999;10:713–718. doi: 10.1007/s003359901077. [DOI] [PubMed] [Google Scholar]
- Fyfe JC, Kurzhals RL, Lassaline ME, Henthorn PS, Alur PR, Wang P, Wolfe JH, Giger U, Haskins ME, Patterson DF, et al. Molecular basis of feline beta-glucuronidase deficiency: An animal model of mucopolysaccharidosis VII. Genomics. 1999;58:121–128. doi: 10.1006/geno.1999.5825. [DOI] [PubMed] [Google Scholar]
- Graves JA. Mammals that break the rules: Genetics of marsupials and monotremes. Annu Rev Genet. 1996;30:233–260. doi: 10.1146/annurev.genet.30.1.233. [DOI] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press; 1991. [Google Scholar]
- Jiang Z, Priat C, Galibert F. Traced orthologous amplified sequence tags (TOASTs) and mammalian comparative maps. Mamm Genome. 1998;9:577–587. doi: 10.1007/s003359900821. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cho KW. Assignment of TCRB encoding the T-cell receptor beta chain gene to cat chromosome A2q25-q26 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1999;84:109–110. doi: 10.1159/000015232. [DOI] [PubMed] [Google Scholar]
- Lunetta KL, Boehnke M, Lange K, Cox DR. Experimental design and error detection for polyploid radiation hybrid mapping. Genome Res. 1995;5:151–163. doi: 10.1101/gr.5.2.151. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O'Brien SJ. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- Ma RZ, van Eijk MJT, Beaver JE, Guérin G, Mummery CL, Lewin HA. Comparative analysis of 82 expressed sequence tags from a cattle ovary cDNA library. Mamm Genome. 1998;9:545–549. doi: 10.1007/s003359900816. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK, O'Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O'Brien SJ. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999a;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Shan S, Chen ZQ, Pecon-Slattery J, O'Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999b;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. The lengths of undiscovered conserved segments in comparative maps. Mamm Genome. 1998;9:491–495. doi: 10.1007/s003359900806. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ. Mammalian genome mapping: lessons and prospects. Curr Opin Genet Dev. 1991;1:105–111. doi: 10.1016/0959-437x(91)80050-v. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Womack JE, Lyons LA, Moore KJ, Jenkins NA, Copeland NG. Anchored reference loci for comparative genome mapping in mammals. Nat Genet. 1993;3:103–112. doi: 10.1038/ng0293-103. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Cevario SJ, Martenson JS, Thompson MA, Nash WG, Chang E, Graves JA, Spencer JA, Cho KW, Tsujimoto H, Lyons LA. Comparative gene mapping in the domestic cat (Felis catus) J Hered. 1997a;88:408–414. doi: 10.1093/oxfordjournals.jhered.a023127. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Wienberg J, Lyons LA. Comparative genomics: Lessons from cats. Trends Genet. 1997b;13:393–399. doi: 10.1016/s0168-9525(97)01297-3. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Marshall Graves JA. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- Rettenberger G, Klett C, Zechner U, Bruch J, Just W, Vogel W, Hameister H. ZOO-FISH analysis: Cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chromosome Res. 1995;3:479–486. doi: 10.1007/BF00713962. [DOI] [PubMed] [Google Scholar]
- Rexroad CE, III, Womack JE. Parallel RH mapping of BTA1 with HSA3 and HSA21. Mamm Genome. 1999;10:1095–1097. doi: 10.1007/s003359901168. [DOI] [PubMed] [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: A fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. doi: 10.1101/gr.8.9.901. [DOI] [PubMed] [Google Scholar]
- Steen RG, Kwitek-Black AE, Glenn C, Gullings-Handley J, Van Etten W, Atkinson OS, Appel D, Twigge S, Muir M, Mull T, et al. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 1999;9:AP1–AP8. [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Van Etten WJ, Steen RG, Nguyen H, Castle AB, Slonim DK, Ge B, Nusbaum C, Schuler GD, Lander ES, Hudson T J. Radiation hybrid map of the mouse genome. Nat Genet. 1999;22:384–387. doi: 10.1038/11962. [DOI] [PubMed] [Google Scholar]
- VandeBerg, J.L., and J.A.M. Graves. 1998. Comparative Gene Mapping. ILAR J 39: entire issue.
- Venta PJ, Brouillette JA, Yuzbasiyan-Gurkan V, Brewer GJ. Gene-specific universal mammalian sequence-tagged sites: Application to the canine genome. Biochem Genet. 1996;34:321–341. doi: 10.1007/BF02399951. [DOI] [PubMed] [Google Scholar]
- Volleth M, Klett C, Kollak A, Dixkens C, Winter Y, Just W, Vogel W, Hameister H. Zoo-FISH analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae) Chromosome Res. 1999;7:57–64. doi: 10.1023/a:1009227428727. [DOI] [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R. Comparative chromosome painting of primate genomes. ILAR J. 1998;39:77–91. doi: 10.1093/ilar.39.2-3.77. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R, Nash WG, O'Brien PC, Yang F, O'Brien SJ, Ferguson-Smith MA. Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet Cell Genet. 1997;77:211–217. doi: 10.1159/000134579. [DOI] [PubMed] [Google Scholar]
- Willett BJ, Flynn JN, Hosie MJ. FIV infection of the domestic cat: An animal model for AIDS. Immunol Today. 1997;18:182–189. doi: 10.1016/s0167-5699(97)84665-8. [DOI] [PubMed] [Google Scholar]
- Womack JE, Kata SR. Bovine genome mapping: Evolutionary inference and the power of comparative genomics. Curr Opin Genet Dev. 1995;5:725–733. doi: 10.1016/0959-437x(95)80004-o. [DOI] [PubMed] [Google Scholar]
- Yang YP, Womack JE. Parallel radiation hybrid mapping: A powerful tool for high-resolution genomic comparison. Genome Res. 1998;8:731–736. doi: 10.1101/gr.8.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogalingam G, Litjens T, Bielicki J, Crawley AC, Muller V, Anson DS, Hopwood JJ. Feline mucopolysaccharidosis type VI. Characterization of recombinant N-acetylgalactosamine 4-sulfatase and identification of a mutation causing the disease. J Biol Chem. 1996;271:27259–27265. doi: 10.1074/jbc.271.44.27259. [DOI] [PubMed] [Google Scholar]