Abstract
Persistent hepatitis C virus (HCV) infection is a leading cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma and the major indication for liver transplantation in adults. Current standard of care treatment (SOC) with pegylated-interferon-α 2 and ribavirin (RBV) has a limited efficacy and is associated with significant side effects frequently associated with poor compliance or treatment discontinuation, requiring specialized and frequent monitoring. To overcome the limited efficacy of SOC, more than 50 direct-acting antiviral agents (DAA) designed to target viral-encoded proteins essential in the HCV life cycle are currently under development. The rapid selection of resistant mutants associated with the quasispecies nature of HCV with high mutation and replication rates is one of the main challenges for the new HCV therapies. Predictive host and viral factors together with combination of DAAs with or without IFN and/or RBV need to be accurately evaluated to design the most effective individualized treatment strategy within the shortest time interval and with minimum side effects.
Keywords: HCV, treatment, quasispecies, resistance
Introduction
An estimated 200 million people are infected with the hepatitis C virus (HCV) worldwide, a prevalence fivefold higher than that of human immunodeficiency virus (HIV).1,2 Chronic HCV infection (CHC) is one of the leading causes of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). HCV-associated end-stage liver disease and/or HCC account for more than 50% of adult liver transplantation cases in Western countries,3 and HCV-associated disease burden is expected to exponentially increase in the next decades. Epidemiological studies draw a future gradual decline in the infected population; however, the number of patients with HCV-associated cirrhosis and its complications is expected to increase exponentially during the next decades.4 A mathematical model has been used to project, over the next 30 years, the HCV-related complications and costs in a cohort of 419,895 infected patients, representing the HCV-infected population in Spain. The model predicts a gradual decline in the infected population, but the proportion of patients with cirrhosis will increase by up to 14% and morbidity associated with HCV infection by up to 10% by the year 2030. The same model predicted that treating from 10% to 50% of the HCV population using standard therapy will result in a reduction of 6% and 26% in morbidity and 4% and 20% in mortality in the next decades.5
In contrast to HBV and HIV, HCV does not integrate into the host genome and treatment is aimed at viral eradication rather than suppression of viral replication.6 Indeed, sustained viral response (SVR), defined as undetectable serum HCV RNA 6 months after treatment completion, has been shown to halt progression to cirrhosis7 in the vast majority of patients. Furthermore, viral eradication among patients treated when cirrhosis has already established significantly reduces the risk of hepatic decompensation and HCC,8 increasing survival and reducing the need for liver transplantation.
The current standard of care (SOC) treatment for CHC is the combination of pegylated-interferon-α 2b (peg-IFN-α 2b, PegIntron®; Merck (MSD), Whitehouse Station, NJ) or 2a (peg-IFN-α 2a, Pegasys®; Roche, Berlin, Germany) with weight-adjusted ribavirin (RBV). The rates of SVR achieved with peg-IFN-α and RBV are genotype dependent, with ∼80% of genotypes 2- and 3-infected patients achieving SVR after 6 months of treatment and ∼50% of genotypes 1- and 4-infected patients treated for 48 weeks.9 Unfortunately, HCV genotype 1, the most prevalent worldwide,10–13 is also the less responsive to current treatment, with no significant differences in SVR rates between the two available peg-IFN + RBV formulations.12 In addition to its limited efficacy, SOC treatment is associated with significant side effects,14 frequently requiring dose reduction or treatment discontinuation (10%–15% of patients), with a significant decrease in SVR rates, or even precluding their use in patients with certain comorbidities (ie, major psychiatric conditions, renal insufficiency, and/or cardiac disease).4 Therefore, the limited efficacy of peg-IFN + RBV therapy, especially against HCV genotype 1, together with side effects and treatment contraindications in many patients and a lack of an alternative to present therapy have led to the use of many adjunctive pharmacotherapeutic agents.
This review will highlight the most promising new drug treatments under advanced clinical trials, the need to evaluate predictive host and viral factors, and new combination of agents with or without IFN and/or RBV to improve antiviral efficacy.
Emerging treatments
Emerging treatments have been focused on improving the existing armamentarium in the form of modified or alternative IFN and RBV preparations, identifying new inhibitors of HCV, defining new regimens of antiviral combinations, and looking for predicting factors of SVR.
New IFN and RBV preparations
The basic requirements for newly developed agents are to increase efficacy, to reduce duration of treatment, to reduce toxicity and facilitate accomplishment of treatment, and to allow treatment of patients with IFN contraindications (Table 1).
Table 1.
Novel interferon (IFN) and ribavirin (RBV) preparations
| Class | Agent | Phase of development | ||
|---|---|---|---|---|
| Type III IFN | IFN-λ1 (IL-29) | Phase IIb | ||
| IFN | Slow-release type I IFN | Albuferon | Zalbin (USA) | Under review |
| Joulferon (EU) | Halted | |||
| Locteron (BLX-883) | Phase II | |||
| Oral IFN-α | Belerofon | Phase II1 | ||
| RBV | RBV prodrug | TBV | Phase III |
Note:
Phase II 2007.
IFN-λs
IFN-λs are type III interferons (IFNs) that have been shown to upregulate major histocompatibility complex (MHC) class I antigen expression and induce antiviral protection, antiproliferative, antitumor, and immune responses, as well as activation of IFN-stimulated genes.15,16 The correlation between SVR rates and a single nucleotide polymorphism (SNP) situated in the IL28B genomic region (see below) in genotype 1 HCV-infected patients treated with SOC highlighted the IFN-λ signaling axis as a potential target for novel antiviral drug development. Initial studies showed that IFN-λ1 (IL29) and IFN-λ2 (IL28A) blocked HCV replication in human hepatocytic cell lines.17–19 In a Phase Ib trial of responders/relapsers chronic HCV patients, the drug showed a robust activity against HCV and a low toxicity, which may be explained by a much more restricted tissue distribution of the IFN-λ receptor compared to that of the IFN-α receptor. Phase IIb studies with different IFN-λ doses in combination with RBV (daily) have started including patients chronically infected with genotypes 1 to 4.
Interestingly, there is in vitro evidence that anti-HCV activity of IFN-λs and IFN-α is enhanced by a low dose of the other, which suggests that both IFNs may interact playing a complementary role in the suppression of HCV.17,20
Albumin-IFN-α
Recombinant human slow-release type 1 albumin-IFN-α is a novel 85.7-kD recombinant protein consisting of IFN-α-2b genetically fused to human serum albumin (Albuferon®; Human Genome Sciences Inc., Rockville, MD, USA/Novartis AG, Basel, Switzerland) named Zalbin®in the United States and Joulferon® in Europe. The advantage of Albuferon® is a longer mean life that allows regimens of dosage every 2–4 weeks. Phase III studies reported similar results in SVR rates21 than standard peg-IFN-α 2 in SOC treatment. However, observation of respiratory adverse events has prompted the US Food and Drug Administration (FDA) to raise some questions about the benefit–risk assessment. Recently (in June 2010), the manufacturer formally withdrew Albuferon® from European license procedure, and also in the United States following the 61st Meeting of AASLD in Boston.79
Other IFN preparations
Locteron® (Biolex, Pittsboro, NC/OctoPlus, Leiden, The Netherlands) is a microsphere-based controlled-release of unpegylated IFN-α 2b recombinant formulation that allows regimens of dosage every 2 weeks. Clinical trials combining Locteron® + RBV have reported similar results than peg-IFN + RBV over 12 weeks of treatment and with less fewer flu-like side effects.22 Some other promising preparations such as Belerofon® (Nautilus Biotech, Evry, France), a slow-release IFN preparation that can be orally administrated, omega IFN (Intarcia Therapeutics, Hayward, CA),23 a type1 IFN that has been tested for anti-HCV effect using an implantable infusion pump (Duros device) for the continuous delivery and consistent dose of drug for 3–12 months,24 or Consensus IFN (CIFN),25 an artificially engineered IFN, despite having promising reports, have not reached a final approval or been extensively commercialized for different reasons.
RBV
RBV is still an essential drug in all present anti-HCV regimens. RBV has antiviral efficacy, measured by clearance of HCV RNA, when used in combination with IFN.
The impact of RBV on HCV infection remains poorly understood. It has been suggested that RBV acts clinically by promoting nonviable HCV RNA mutation rates and is capable of driving replicating virus into error catastrophe.26–29 The combination therapy of RBV with IFN was superior than RBV monotherapy in inducing nucleic acid substitutions.29 The principal specific side effect caused by RBV is anemia, and it is frequently detected during combination peg-IFN + RBV treatment. Anemia is one of the reasons for discontinuation of treatment and makes many patients with contraindications, such as patients with renal insufficiency and/or cardiac diseases, ineligible.4 To improve efficacy, tolerability, and reduce RBV-associated anemia,30 liver-targeting RBV prodrugs are under development. Among them, taribavirin (TBV) (Viramidine®; Valeant Pharmaceuticals, Aliso Viejo, CA) is a prodrug that is metabolized by the liver to the active metabolite, thereby limiting the red blood cell effect and reducing anemia caused by standard RBV. A comparative clinical Phase IIb study of RBV versus TBV both combined with peg-IFN-α 2b revealed that when doses were adjusted to body weight, the combination peg-IFN-α 2b + TBV achieved comparable virological response rates than standard peg-IFN-α 2b + RBV treatment, but with the advantage that TBV caused less anemia (7%–15% of peg-IFN + TBV-treated patients compared with 24% treated with peg-IFN + RBV).31 Secondary effects such as diarrhea were more common with TBV but were generally mild and not dose limiting. Clinical Phase III trials are in process.
New inhibitors of HCV
Manipulation of immune response either innate or adaptive
Preliminary data exist for direct immune stimulants, both innate and adaptive, as well as therapeutic vaccines. Specifically enhancing adaptive immunity against HCV-specific epitopes with concomitant alteration of the cytokine milieu represents multiple opportunities for drug design, but manipulation of immune response which represents a tightly regulated system represents a delicate balance and can result in severe secondary effects (reviewed by Schinazi and coworkers32). A more specific therapeutical approach needs deeper knowledge of HCV biology since several HCV proteins (core, envelope, NS3, and NS5A) may interfere/inhibit IFN-α-induced activation of the JAK–STAT pathway, which is a critical transducer of IFN-α-mediated signaling and serves as a global director of the IFN-α-driven innate immune response.33–35
Many classes of immunomodulators are under various stages of clinical investigation, including polyclonal antibodies, interleukin therapy, broad-spectrum anti-inflammatory agents, and nonspecific immune enhancers such as toll-like receptor targets, thymosin α1, 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors, the monoclonal antiphosphatidylserine bavituximab (Peregrine Pharmaceuticals, Inc., Tustin, CA), the dipeptide l-Glu-l-Trp oglufanide (Implicit BioScience, Toowong, Australia), SCV-07 (SciClone Pharmaceuticals, Inc., Causeway Bay, Hong Kong), and the antiprotozoal nitazoxanide.32,36,37
Several therapeutic vaccines are under development. GI-5005 (GlobeImmune, Inc., Louisville, CO) has recently finished Phase IIb trial and results show that in triple therapy, peg-IFN + RBV + GI-5005 increased the SVR in genotype 1 IFN-naive patients (58%) compared to patients receiving peg-IFN + RBV alone (48%).
Inhibition of host-encoded targets essential for viral replication
A number of host-encoded targets essential for HCV replication have been identified, of which enzymatically active cyclophilin A is the most advanced. It is required for HCV replication and processing kinetics.38 One promising drug showing potent anti-HCV function is cyclosporine A (CsA) because of its high affinity to cyclophilins (CyPs), but it is a powerful immunosuppressive drug39 due to its ability to block the phosphatase calcineurin. Because of the nonimmunosuppressive properties combined with profound antiviral activity, CsA derivatives such as Debio-025 (alisporivir; Debiopharm SA, Research Triangle Park, NC),40,41 NIM811 (Novartis, Basel, Switzerland),42 and SCY-635 (Scynexis, Durham, NC) are more likely to be used as anti-HCV agents. Unlike CsA, these molecules bind to CyPs but do not display calcineurin inhibition and have demonstrated clinical antiviral activity against HCV.43 Clinical Phase IIb trials are ongoing.
Recent in vitro assays reveal that the combination of Debio-025 with either RBV or another inhibitor (see below) in the absence of IFN resulted in additive antiviral activity in short-term antiviral assays, delaying or preventing the development of resistance to HCV protease inhibitors as well as to nucleoside and nonnucleoside polymerase inhibitors. This result will improve efficacy in the treatment of patients with severe contraindication to IFN treatment.
Debio-025 is an attractive drug candidate for the treatment of HCV infections in combination either with standard IFN-based treatment and/or treatments that directly target the HCV polymerase and/or protease.44
Use of microRNAs to regulate HCV expression (microRNA-122)
MicroRNAs (miRNAs) are key regulators of gene expression at a post-transcriptional level. The liver-expressed microRNA-122 (miR-122) seems essential for HCV RNA accumulation in cultured liver cells since it stimulates its replication by binding to the 5′ noncoding RNA region (5®NCR), which is highly conserved in all six HCV genotypes. Treatment of chronically infected chimpanzees with a locked nucleic acid-modified oligonucleotide (SPC3649; Santaris Pharma, Hoersholm, Denmark) complementary to miR-122 leads to long-lasting suppression of HCV viremia, with no evidence of viral resistance or side effects in the treated animals. Conservation of both miR-122 seed sites in all HCV genotypes and subtypes suggests that such therapy will be genotype independent.45 However, miR-122 has been regarded as the negative regulator of gene expression in, for example, cell differentiation, proliferation, migration, and invasion. Recent studies have revealed that miR-122 plays an important role in the regulation of intrahepatic metastasis of HCC46 and that it is downregulated in both primary and metastatic HCC.47,48
Therefore, miR-122-based therapeutics might be a double-edged sword for the treatment of HCC in HCV-infected patients and inadvisable for the basic treatment of HCV-infected patients.
Inhibitors of HCV life cycle steps
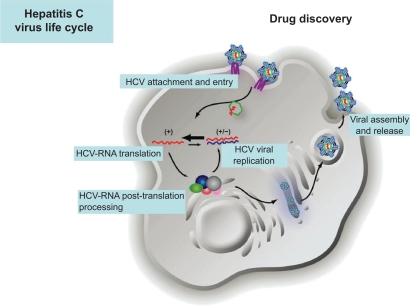
All proteins encoded in the small HCV genome are essential for viral propagation and very attractive for therapy since inhibition of these viral proteins does not mainly affect cellular functions. Putative targets to design specific drugs include inhibitors of all steps in the HCV life cycle: viral entry, HCV RNA translation and post-translational processing, HCV replication, and viral assembly and release (Figure 1 and Table 2).
Figure 1.
Hepatitis C virus life cycle and putative targets to design specific drugs.
Provided by courtesy of Dr Charles Rice and Dr Shihyun You.
Table 2.
Some of the most promising direct-acting antiviral agents
| Viral life cycle step | Target | Agent | Phase of development |
|---|---|---|---|
| Post-translational processing | NS3/4A protease | Telaprevir (VX-950) | Phase III1 |
| Boceprevir (SCH503034) | Phase III1 | ||
| GS-9256 | Phase II | ||
| BI 201335 | Phase II | ||
| RG7227 (Danoprevir) | Phase II | ||
| Vaniprevir (MK-7009) | Phase II | ||
| Narlaprevir (SCH900518) | Phase II | ||
| TMC435 | Phase IIa | ||
| BMS-650032 | Phase II | ||
| BMS-791325 | Phase II | ||
| Danoprevir | Phase II | ||
| ABT-450 | Phase II | ||
| NS3 helicase | BTN10/BTN11 | Preclinical | |
| Replication | NS5A | AZD-7295 | Phase II |
| BMS-790052 | Phase II | ||
| A-832 | Phase II | ||
| NS5B (NI) | RG-7128 | Phase II | |
| PSI-7977 | Phase II | ||
| Tegobuvir | Phase II | ||
| IDX184 | Phase II | ||
| NS5B (NNI) | GS-9190 | Phase II | |
| ABT-333 | Phase II | ||
| ANA598 | Phase II | ||
| BI-207127 | Phase II | ||
| ABT-072 | Phase II | ||
| Filibuvir (PF-00868554) | Phase II | ||
| VCH-916 | Phase II | ||
| VX-222 | Phase II | ||
| VX-759 | Phase II |
Note:
Putative final approval in 2011.
Inhibitors of viral attachment and entry
Several receptors have been associated with HCV-specific attachment and entry to the hepatocyte. It has been proposed that the first step includes the cell surface contact through the heterodimeric E1 and E2 proteins. Both glycoproteins are expressed on the virus envelope that is associated with low- and very-low-density lipoprotein (vLDL).49 Initial host cell contact is mediated by interaction with the low-density lipoprotein receptor (rLDL) and/or glycosaminoglycans. E2 also binds to dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin and liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin; the asialoglycoprotein receptor may also interact with HCV structural proteins.50–56 The virus subsequently binds to high-affinity receptors, including scavenger receptor class B type I and the tetraspanin protein CD81, through the ecto-domain of E2. It then binds to claudin 1 and occludin, both of which are expressed at tight junctions of polarized hepatocytes.57,58 Internalization of the virus proceeds through clathrin-dependent endocytosis, and viral membrane fusion takes place in the acidified endosome, a pH-dependent mechanism.59 This releases the viral core into the host cytosol, where uncoating and disassembly of the virus capsid releases the RNA genome. To date, numerous entry inhibitors are in various stages of preclinical development and are reviewed by Schinazi and coworkers.32 Among them d,l-a-peptides have shown activity in genotypes 1a, 1b, and 2a in vitro,60 and CD81mAb in liver-uPA-SCID mice model, but administration of anti-CD81 antibodies after viral challenge had no effect.61 Inhibition of viral entry into HCV-permissive cells could provide an efficacious mechanism for reduction or elimination of viral infection, but blocking cellular receptors with important biochemical functions for the hepatocyte without differentiating between infected and noninfected hepatocytes may cause important negative effects on the normal functionality of the liver.
Inhibitors of viral assembly
Core (191aa) acts as the viral nucleocapsid and RNA packaging. It has good sequence conservation across isolates, and the protein is poorly structured prior to assembly. However, core–core interaction avidity can reduce potency of any inhibitors in vivo. HCV assembly starts with recruitment of the core protein and NS5A to the surface of lipid droplets, followed by delivery of HCV RNA from the HCV replication complex to the nascent viral particle.62,63 However, nonspecific inhibitors targeting this region have not yet proved the proof of principle in clinical trials.
Alteration of proper envelope protein folding of many viruses has been obtained using α-1 glucosidase I inhibitors.64 Phase IIa clinical investigation using one of these inhibitors named celgosivir (Megenix Inc., Vancouver, Canada) has demonstrated potency against both HCV and HBV,64 but studies have been cancelled.
Inhibitors of RNA translation, post-translation, and replication
Internal ribosome entry site, p7, and NS2 inhibitors
The internal ribosome entry site (IRES), the initiation of cap-independent translation of HCV, serves as a direct regulator for assembly of initiation of translation complexes on viral mRNA. It is highly conserved and structured. As this mechanism is distinct from those observed in prototypical eukaryotic translation machineries, the inhibition of viral IRES could provide a virus-specific target for antiviral compounds. Specific inhibitors of IRES HCV translation, either RNA molecules (chimeric HH363 that target essential domain IV stem-loop position 363) that interfere with formation of a translationally active complex or DNAzymes (specific molecules with cleavage activity) or small molecules that bind p7 (63aa, ion channel or porin with low sequence conservation) and NS2 (217aa, cysteine autoprotease), have reported activity in cell culture,65 but none has yet reached development. Antisense RNAs specific for the IRES have not yet led to a proof of principle in clinical studies.32
NS3-NS4A-NS4B-NS5A-NS5B inhibitors
More than 50 new drugs specific against HCV named direct-acting antiviral agents (DAAs) (also known as specifically targeted antiviral therapy for hepatitis C or STAT-C) are under advanced clinical trials to be used either as an adjunct to current SOC therapy in triple, quadruple, or multiple combination or in place of it in patients with SOC severe contraindications. In the next 2 years, it will likely become standard for treatment either in naive or in pretreated patients.66 Among them, DAAs targeting NS3, NS5A, and NS5B have demonstrated proof of principle for viable targets.
NS3
NS3 is essential for viral replication. It is a multifunctional protein that harbors a serine protease located in the N-terminal one-third (189aa) involved in cis-cleavage of NS3–NS4A site, and the NS3 forming an heterodimer with NS4A is responsible for the downstream cleavage of the NS4A/4B, NS4B/5A, and NS5A/B junctions in the non-structural region. The C-terminal two-thirds of NS3 have an NTPase/RNA helicase domain (442aa).
NS3 protease
NS3 serine protease activity is essential for cleavage of the large HCV polyprotein into active peptides that in turn are critical in forming the replicative complex from which viral RNA synthesis occurs.67–70 The NS3/NS4A serine protease also blocks activity of IFN regulatory factor 3 (IRF3), the critical transcription factor required for virus-induced IFN production.71 Therefore, the NS3 represents a dual therapeutic target, the inhibition of which may both block viral replication and restore hepatocyte IRF3 control of HCV infection.72
The serine protease inhibitors telaprevir (VX-950; Vertex, San Diego, CA) and boceprevir (SCH503034; Schering-Plough, Whitehouse Station, NJ) are the most advanced DAAs in clinical studies.
Phase II clinical trials using triple therapy with peg-IFN-α + RBV + telaprevir (TPR) (termed PROVE 1 for the US study73 and PROVE 2 for the European one74) or boceprevir (BPR) (SPRINT) have shown an important increase in SVR rates and a decrease in breakthrough and relapse rates, being highest in patients who did not achieve a rapid virological response at week 4 with SOC therapy.75 The addition of a potent protease inhibitor to the SOC treatment considerably accelerates the first slope of viral decline during the first few days of therapy.76–78
The Phase IIb studies (PROVE 1 and PROVE 2) in HCV genotype 1 patients showed that double combination TPR and peg-IFN-α had low efficacy (SVR rates of 41%–46% PROVE 1 and 2, respectively) and higher relapse (22%–23%) than triple therapy adding RBV (SVR rates 61%–69% and relapse 2%–14%). Therefore, the first conclusion of this trial was that RBV is still an essential drug for these anti-HCV regimens. Another important conclusion was that administration of TPR for 12 weeks followed by 12 weeks of SOC yields superior SVR rates with shorter duration of therapy compared to SOC treatment.73,74
In the case of BPR, triple combination was compared with SOC treatment in the SPRINT-1 study. A key aspect of this trial was the assessment of a 4-week SOC lead-in phase before the introduction of BPR in triple therapy. The rationale for this approach was to establish antiviral activity before adding BPR with the hope of minimizing the risk of emergent drug resistance, followed by 44 weeks of triple therapy with BPR (for a total of 48 weeks of treatment). The most important finding from this analysis was that 44 weeks of BPR following the 4-week lead-in phase was associated with the highest SVR rate of 75% compared to 38% with SOC (P < 0.0001). The next highest SVR rate was 67%, which was achieved by patients receiving the 48-week triple-combination regimen that did not include lead-in treatment (P < 0.0001 versus SOC). The proportion of patients who achieved SVR with 24 weeks of triple therapy following a 4-week lead-in phase was 56% (P = 0.0005 versus SOC) versus 54% with 28 weeks of triple therapy and no lead-in treatment (P = 0.013 versus SOC). BPR seems to be quite effective in this context, and the triple-combination regimen with the 4-week lead-in phase has been selected for further evaluation in Phase III clinical trials.
In both TPR and BPR treatment clinical trials, the improved efficacy comes with additional side effects, particularly rash and increased anemia, and a risk of selecting drug-resistant viral variants that may limit subsequent therapeutic options.75,79 As a result, response-guided therapy has been adopted in the design of the Phase III trials either using TPR or BPR.
The recently reported Phase III study named ADVANCE (Vertex) using TPR is an example of a response-guided trial. Seventy-five percent of patients infected with HCV1 who were not previously treated achieved an SVR after receiving 12 weeks of TPR in triple therapy followed by a course of SOC therapy for at least another 12 weeks. In the ADVANCE trial, in the TPR group, if the virus was sufficiently suppressed after 4 weeks, patients received only 24 weeks of total treatment (half the standard treatment time). Notably, about 70% of those who achieved SVR only received 24 weeks of therapy. Patients in the control group underwent standard therapy for 48 weeks and 44% achieved an SVR.
In the case of BPR, Phase III trials have been focused on treating genotype 1 patients who were nonresponders or relapsers to prior therapy comparing SOC treatment (arm1) with response-guided therapy receiving BPR in triple therapy for 32 weeks, and those HCV RNA positive at week 8 received an additional 12 weeks of SOC (arm2), and BPR of 44 weeks (arm3). There was a significant absolute increase in SVR compared with SOC of 37.4% in arm2 and 45.2% in arm3.
Phase III results from either TPR or BPR in triple therapy fuel hopes for major improvements in treatment outcomes increasing the cure rate by more than 30%.75,79,80
To improve the pharmacokinetic profile, dosing intervals, and perhaps some advances in safety and tolerability,80 second-generation protease inhibitors taken once a day (at present TPR and BPR have to be taken orally three times a day) are in Phase II trials. Some of these new-wave NS3 protease inhibitors include the following: BI-201335 (Boehringer Ingelheim, Ingelheim am Rhein, Germany), BMS-650032 (Bristol-Myers Squibb, New York, NY), GS-9256 (Gilead, Foster City, CA), Danoprevir/R7227/ITMN191 (InterMune, Brisbane, CA/Roche) and NS3/NS4 A inhibitors ABT-450 (Abbott, Abbott Park, IL/Novartis), and Vaniprevir/MK7009 (Merck) among many others.
A conclusion extracted from the clinical trials using DAAs has been that in order to improve efficacy, reduce duration of treatment, and facilitate accomplishment of treatment, response-guided therapies should be faced by using predictive values. Virologic response rates have been shown to depend on various baseline host and viral factors such as age, weight, gender, race, liver enzymes, stage of fibrosis, HCV genotype, and HCV RNA concentration at baseline10,11,81–85 and also on treatment factors at the time of HCV RNA clearance. Among chronically infected patients, response to treatment differs, even among cases with similar HCV RNA levels and identical viral genotypes,10,11,86,87 suggesting that other factors should be taken into consideration. A recent genome-wide association study with 1671 individuals infected with HCV genotype 1 showed that genetic variation in the IL28B gene, which encodes IFN-λ, is associated with spontaneous HCV clearance88 and therapy response to SOC treatment.89–91 Particularly, the presence of a C/C genotype in the SNP rs12979860 located on chromosome 19q13 was strongly associated with an SVR. The corresponding SVR rates being ∼80% for the C/C genotype, ∼40% for the T/C genotype, and ∼30% for the T/T genotype aimed at identifying genetic contributions to anti-HCV response.89,92 It has been recently shown that rs12979860 genotype is a significant predictor of SOC response in CHC patients independent of HCV genotype and other covariates.93 Interestingly, in a random multiethnic population of 455 patients, the C-allele frequency was ∼90% in East Asian patients, ∼55% in European American patients, and just ∼25% in African–Americans. This finding finally sheds some light on the precise mechanisms that are behind the significantly lower SVR rates obtained in African–Americans compared with Caucasians94,95 and the higher SVR rates consistently reported in trials of anti-HCV therapy conducted in Asia.96 The association between SNPs in the IL28B region and SVR in HCV-1-infected patients has been confirmed by different studies.20,88–90,95 A more fine mapping revealed seven SNPs associated with NVR: rs8105790, rs11881222, rs8103142, rs28416813, rs4803219, rs8099917, and rs724866891 that need further analysis.
NS3 helicase
The carboxy terminal, two-thirds of NS3, constitutes the helicase domain, which is also essential for the replication of viral RNA. The helicase domain might stimulate the activity of NS5B polymerase, resolve RNA secondary structure immediately prior to replication by the polymerase, and/or separate newly synthesized double-stranded RNA into positive and negative strands. Two tropolone derivatives, BTN10 and BTN11,97–100 and trixsalen101 have significant antiviral activity in the HCV replicon system.
NS4A
NS4A is part of the NS3 protease structure and acts as a membrane anchoring the replication complex. No structural information is known, except that the central portion binds to NS3. It may be a ‘druggable’ target.
NS4B
NS4B possibly has an important activity in the formation of membranous web structure and assembly of replication complex and/or NTPase. It is among the key players in the HCV replication cycle and hypothesized to bind RNA. A nucleotide binding motif was identified within NS4B to bind and hydrolyze GTP,102 and it is suggested that it may also bind polynucleotide structures. It is a hydrophobic and poorly structured protein and may represent a challenge for biochemical analysis and antiviral targeting. In fact, ATPases and GTPases have previously been targeted successfully. Recently, high-throughput screening for inhibitors of the RNA binding function has identified an H1 histamine receptor antagonist (clemizole hydrochloride) that substantially inhibits HCV replication in cell culture. This approach for drug discovery seems useful when dealing with membrane-bound protein targets.103
NS5A
NS5A is a multifunctional protein with critical viral key functions, regulation of HCV RNA replication, virus assembly, multiple viral–viral and viral–host protein interactions, modulation of cellular signaling pathways, and IFN response.104 It contains a putative IFN sensitivity-determining region and may play a role in resistance to IFN-α.105 As a multifunctional protein required for in vivo and in vitro replication with unknown human homologues, NS5A represents an attractive target for therapeutic intervention. Proof of principle for NS5A as a viable target has been demonstrated in early clinical trials in HCV-infected subjects. Two potent NS5A DAAs compounds, AZD-7295 (AstraZeneca, Madrid, Spain) and BMS-790052 (Bristol-Myers Squibb), have been evaluated in Phase II clinical trials. BMS-790052 has shown strong activity against several genotypes106 in replicon and JFH-1 systems, and it has demonstrated a rapid and robust HCV RNA decline (∼3.6 log 10) in clinical trials with no signs of adverse effects.107 Promising results from clinical trials provide the basis for designing second-generation NS5A polymerase inhibitors.108
NS5B, RNA-dependent RNA polymerase
Although protease inhibitors are expected to reach the market first, investigational agents that block other HCV enzymes, such as NS5B protein, are of special interest because NS5B specifically catalyzes viral RNA synthesis and genome replication. Proof of principle for NS5B as viable target has been demonstrated in clinical trials. Inhibitors of HCV NS5B RNA-dependent RNA polymerase (RdRp) are divided into two classes: nucleoside and nonnucleoside inhibitors (NI and NNI, respectively). Nucleoside polymerase inhibitors in particular appear to have a high genetic barrier to resistance, but the nonnucleosides also offer great promise in combination with other drugs.
NI:
In their active triphosphate forms, NI metabolites act as nonobligate chain terminators while competing with the natural substrate nucleotides for the HCV NS5B RdRp, thereby reducing the efficiency of further RNA elongation through steric resistance. To date, many NI polymerase inhibitors are under early stages of clinical investigation,32,80 and the most advanced ones, which are in Phase II, include RG-7128 (Pharmasset, Princeton, NJ/Roche), PSI-7977 (Pharmasset/Roche), Tegobuvir (Gilead), and IDX184 (Idenix Pharmaceuticals, Cambridge, MA).
NNI:
The primary mechanism of action for NNI is specific targeting of different and less-conserved allosteric sites of the HCV NS5B polymerase. Multiple NS5B HCV inhibitors are under clinical investigation, and the ones that are in Phase II include GS-9190 (Gilead), ABT-333 (Abbott Laboratories), ANA598 (Anadys Pharmaceuticals, San Diego, CA), BI-207127 (Boehringer Ingelheim), ABT-072 (Abbott Laboratories), Filibuvir/PF-00868554 (Pfizer, New York, NY), VCH-916, and VX-222 (Vertex Pharmaceuticals).32,80
SOC-free treatments
Side effects caused by SOC contraindicated treatment of many patients. It is clear that monotherapy with a DAA is not possible because resistance mutants appear in a few days of treatment. In this scenario, researchers are looking at using combinations of DAAs targeting different viral functions for which there is no cross-resistance, as has been successfully applied at preventing resistance in HIV treatment. The basic knowledge for this approach is that to get resistance, a combination of two drugs needs to be developed, which is obviously much less likely than with just one drug.109 At this moment, whether SOC-free regimens are possible and, if they are, what will be the best combinations of DAAs, needs further study.
The first IFN/RBV-free regimen trial (INFORM-1) was performed by combining the HCV protease inhibitor RG7227 (also known as ITMN-191) with the polymerase inhibitor RG7128 (Roche/Genentech, San Francisco, CA) and results showed that over the course of a 14-day study, there was an HCV viral load reduction below the limit of quantification in 63% of subjects enrolled,110 opening the possibility to treat patients with contraindications to IFN and/or RBV treatments. Other combinations that are under study include BMS-650032 (NS3 inhibitor) with BMS-790052 (NS5A inhibitor) by Bristol-Myers Squibb or GS9256 (protease inhibitor) with tegobuvir (NI NS5B inhibitor) by Gilead80 (Table 3).
Table 3.
SOC-free DAA combinations
| Agent 1 | Agent 2 | Phase of development |
|---|---|---|
| RG7227 (ITMN-191) (Danoprevir) (protease inhibitor) | RG7128 (polymerase inhibitor) | Phase I |
| BMS-650032 (protease inhibitor) | BMS-790052 (NS5A inhibitor) | Phase I |
| GS9256 (protease inhibitor) | Tegobuvir (polymerase inhibitor) | Phase II |
| Telaprevir (protease inhibitor) | VX-222 (polymerase inhibitor) | Phase II |
Antiviral drug resistance
Antiviral drug resistance mutations, especially to new DAAs, cause an important drawback in the treatment of chronic infection. The primary factor involved in viral resistance is the quasispecies nature of HCV, a consequence of the high viral turnover via the error-prone polymerase. HCV has a half-life of only 2–5 h, with the production and clearance of 1010–1012 virions/day in an infected patient.111,112 The enzyme used for the HCV for replication (RdRp codified by the NS5B genetic region) has poor fidelity and lacks an exonucleolytic proofreading mechanism; replication is, therefore, inherently error-prone, with an error-rate in the order of 10−3–10−4 mutations/site and genomic replication cycle (corresponding to a natural evolutionary rate of 1.5 × 10−3 base substitutions/site/year).113,114 As a consequence of high mutation rate and high level of replication, the virus circulates as a complex mixture of different but closely related genomes known as quasispecies in the infected host.115 Quasispecies represents a highly dynamic structure with mutants continuously arising, all competing but interacting (they may cooperate) and displaying different phenotypes and fitness.116 Single mutations at each position in the genome arise at least once every day, and in consequence, potentially drug-resistant mutants are constantly being generated. Since antiviral-resistant mutants are often less replication-competent than the corresponding wild-type (wt) viruses, they represent minority mutants in the absence of drug therapy. During antiviral treatment, such mutants are selected and arise as major components of the viral quasispecies contributing to treatment failure. Besides, compensatory mutations selected in the same genome contribute to fitness recovery.117,118 On stopping the treatment, if such mutations are less competitive than others, they might be negatively selected and relegated to a minority population, but remaining at a higher proportion than before, generating a memory in the quasispecies. It has been recently reported that a resistant variant may persist for at least 3 years following a 14-day trial with DAA monotherapy.119 If the patient is then retreated with the same drug or with a cross-reactive one, the HCV-resistant variant rapidly emerges, within days, thus contributing to a rapid treatment failure.120 Therefore, the use of any DAAs in monotherapy should not be ever considered since resistance develops within the first day of treatment with the risk of selecting mutants that may cross-react with other inhibitors. For instance, genotype 1b replicons subjected to a single protease inhibitor (ie, TPR or BPR) selected NS3 protease resistant variants such as substitutions A156T/A156V and R109 K to make the virus resistant to all other inhibitors. Some authors argue that in most cases, mutation A156T significantly reduces NS3/4A catalytic efficiency, polyprotein processing, and replicon fitness, but RNA viruses like HCV having an enormous capacity of adaptation and second-site mutations such as P89 L, Q86R, and G162R have been described to partially reverse A156T-associated defects in polyprotein processing and/or replicon fitness, without significantly reducing resistance to the protease inhibitor.121–124 As resistance is a major problem, the FDA has restricted the use of monotherapy to 3 days in early studies and then SOC has to be added.
Key issues relevant to the development of resistance include the number of mutations needed to introduce the required coding change(s), the replication capacity of the variant virus, the prevalence of the variant within the quasi-species, the level of resistance that it confers to the virus, and the potency and bioavailability of the antiviral agent.125 Other viral factors include viral quasispecies heterogeneity and, to a less extent, specific mutations within the core, E2, and NS5 A coding regions. The availability of recently developed platforms for massive parallel high-throughput sequencing of both viral populations and host polymorphisms (ie, IL28 and MHC haplotypes for HCV), especially those capable of clonally sequencing long fragments (ie, 454/Roche), should facilitate optimization of treatment.
Conclusion
Persistent HCV infection is a leading cause of chronic hepatitis, cirrhosis, and HCC and the major indication for liver transplantation in adults. In contrast to HBV and HIV, HCV can be permanently eradicated from the infected host. Current SOC with pegylated IFN-α 2 and RBV for 48 weeks eradicates infection in ∼50% of HCV genotype 1-infected patients and is associated with significant side effects, which limit its effectiveness in many cases (ie, dialysis and HIV-infected patients, transplant recipients, etc.), and are frequently associated with poor compliance or treatment discontinuation, requiring specialized and frequent monitoring.
In recent years, host (IL-28B polymorphisms, MHC haplotypes, and several polymorphisms in cytokine/chemokines and IFN signaling pathways) and viral factors (genotype, viral load, quasispecies complexity, and mutations in viral-encoded proteins) have been strongly associated with SOC treatment outcome. Hence, accurately predicting response to SOC (and to any available DAA) will require identification of host and viral factors independently associated with treatment outcome to design cost-effective, individualized treatment strategies.
To overcome the limited efficacy of SOC, more than 50 DAAs, designed to target viral-encoded proteins essential in the HCV life cycle (ie, NS3 protease, NS5B polymerase, NS5A, etc.), are currently under development. Phase III trials have just been completed for two protease inhibitors (TPR and BPR) and have shown a highly significant increase in viral eradication rates when given in association with SOC treatment. The rapid selection of resistant mutants associated with the high mutation rate of HCV have clearly shown that DAAs cannot be given in monotherapy and SOC–DAA combinations will be required in the near future.
Nonetheless, the ultimate goal of HCV treatment (HCV eradication in every patient within the shortest time interval and minimum side effects) will imply the design of IFN-free specifically targeted anti-HCV treatment (STAT-C) strategies with a combination of DAAs targeting different steps of the HCV life cycle.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.WHO Hepatitis B. Fact sheet 204. 2000Available from: http://www.who.int/mediacentre/factsheets/fs204/en/. Accessed Sep 23 2010.
- 2.Quer J, Esteban JI. Epidemiology. In: Thomas HC, Lemon S, Zuckerman AJ, editors. Viral Hepatitis. 3rd ed. Oxford, UK: Blackwell Publishing; 2005. pp. 407–425. [Google Scholar]
- 3.Willems M, Metselaar HJ, Tilanus HW, Schalm SW, de Man RA. Liver transplantation and hepatitis C. Transpl Int. 2002;15(2–3):61–72. doi: 10.1007/s00147-002-0382-4. [DOI] [PubMed] [Google Scholar]
- 4.NIH National Institutes of Health Consensus Development conference statement: management of hepatitis C 2002 (2002 Jun 10–12) Gastroenterology. 2002;123(6):2082–2099. doi: 10.1053/gast.2002.1232082. [DOI] [PubMed] [Google Scholar]
- 5.Buti M, San Miguel R, Brosa M, et al. Estimating the impact of hepatitis C virus therapy on future liver-related morbidity, mortality and costs related to chronic hepatitis C. J Hepatol. 2005;42(5):639–645. doi: 10.1016/j.jhep.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Pawlotsky JM. Therapy of hepatitis C: from empiricism to eradication. Hepatology. 2006;43(2 Suppl 1):S207–S220. doi: 10.1002/hep.21064. [DOI] [PubMed] [Google Scholar]
- 7.Poynard T, McHutchison J, Manns M, et al. Impact of pegylated interferon alfa-2b and ribavirin on liver fibrosis in patients with chronic hepatitis C. Gastroenterology. 2002;122(5):1303–1313. doi: 10.1053/gast.2002.33023. [DOI] [PubMed] [Google Scholar]
- 8.Bruno S, Stroffolini T, Colombo M, et al. Sustained virological response to interferon-alpha is associated with improved outcome in HCV-related cirrhosis: a retrospective study. Hepatology. 2007;45(3):579–587. doi: 10.1002/hep.21492. [DOI] [PubMed] [Google Scholar]
- 9.WHO Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat. 1999;6(1):35–47. [PubMed] [Google Scholar]
- 10.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 11.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 12.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361(6):580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 13.Rumi MG, Aghemo A, Prati GM, et al. Randomized study of peginterferon-alpha2a plus ribavirin vs peginterferon-alpha2b plus ribavirin in chronic hepatitis C. Gastroenterology. 2010;138(1):108–115. doi: 10.1053/j.gastro.2009.08.071. [DOI] [PubMed] [Google Scholar]
- 14.Bjornsson E, Verbaan H, Oksanen A, et al. Health-related quality of life in patients with different stages of liver disease induced by hepatitis C. Scand J Gastroenterol. 2009;44(7):878–887. doi: 10.1080/00365520902898135. [DOI] [PubMed] [Google Scholar]
- 15.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 16.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 17.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 18.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79(6):3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 21.Bain VG, Kaita KD, Yoshida EM, et al. A phase 2 study to evaluate the antiviral activity, safety, and pharmacokinetics of recombinant human albumin-interferon alfa fusion protein in genotype 1 chronic hepatitis C patients. J Hepatol. 2006;44(4):671–678. doi: 10.1016/j.jhep.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 22.de Leede LG, Humphries JE, Bechet AC, van Hoogdalem EJ, Verrijk R, Spencer DG. Novel controlled-release Lemna-derived IFN-alpha2b (Locteron): pharmacokinetics, pharmacodynamics, and tolerability in a phase I clinical trial. J Interferon Cytokine Res. 2008;28(2):113–122. doi: 10.1089/jir.2007.0073. [DOI] [PubMed] [Google Scholar]
- 23.Buckwold VE, Wei J, Huang Z, et al. Antiviral activity of CHO-SS cell-derived human omega interferon and other human interferons against HCV RNA replicons and related viruses. Antiviral Res. 2007;73(2):118–125. doi: 10.1016/j.antiviral.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Rohloff CM, Alessi TR, Yang B, Dahms J, Carr JP, Lautenbach SD. DUROS technology delivers peptides and proteins at consistent rate continuously for 3 to 12 months. J Diabetes Sci Technol. 2008;2(3):461–467. doi: 10.1177/193229680800200316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witthoft T. Review of consensus interferon in the treatment of chronic hepatitis C. Biologics. 2008;2(4):635–643. doi: 10.2147/btt.s1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perales C, Agudo R, Domingo E. Counteracting quasispecies adaptability: extinction of a ribavirin-resistant virus mutant by an alternative mutagenic treatment. PLoS One. 2009;4(5):e5554. doi: 10.1371/journal.pone.0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J Virol. 2002;76(17):8505–8517. doi: 10.1128/JVI.76.17.8505-8517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanford RE, Guerra B, Lee H, et al. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J Virol. 2003;77(2):1092–1104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanda T, Yokosuka O, Imazeki F, et al. Inhibition of subgenomic hepatitis C virus RNA in Huh-7 cells: ribavirin induces mutagenesis in HCV RNA. J Viral Hepat. 2004;11(6):479–487. doi: 10.1111/j.1365-2893.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- 30.Kearney KR, Thornton JJ, Navarro VJ. Taribavirin for the treatment of chronic hepatitis C. Expert Opin Pharmacother. 2008;9(18):3243–3249. doi: 10.1517/14656560802594459. [DOI] [PubMed] [Google Scholar]
- 31.Poordad F, Lawitz E, Chun E. Treatment week 12 results of weight-based taribavirin versus weight-based ribavirin, both with peginterferon alfa-2b, in naive chronic hepatitis C, genotype 1 patients. 43rd Annual Meeting of the European Asociation for the Study of the Liver (EASL); Milan, Italy. 2008. Abst. 996. [Google Scholar]
- 32.Schinazi RF, Bassit L, Gavegnano C. HCV drug discovery aimed at viral eradication. J Viral Hepat. 2010;17(2):77–90. doi: 10.1111/j.1365-2893.2009.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73(10):8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luquin E, Larrea E, Civeira MP, Prieto J, Aldabe R. HCV structural proteins interfere with interferon-alpha Jak/STAT signalling pathway. Antiviral Res. 2007;76(2):194–197. doi: 10.1016/j.antiviral.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Melen K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73(4):536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- 36.Elazar M, Liu M, McKenna SA, et al. The anti-hepatitis C agent nitazoxanide induces phosphorylation of eukaryotic initiation factor 2alpha via protein kinase activated by double-stranded RNA activation. Gastroenterology. 2009;137(5):1827–1835. doi: 10.1053/j.gastro.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol JF, Elfert A, Keeffe EB. Treatment of chronic hepatitis C using a 4-week lead-in with nitazoxanide before peginterferon plus nitazoxanide. J Clin Gastroenterol. 2010;44(7):504–509. doi: 10.1097/MCG.0b013e3181bf9b15. [DOI] [PubMed] [Google Scholar]
- 38.Kaul A, Stauffer S, Berger C, et al. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5(8):e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borel JF, Feurer C, Gubler HU, Stahelin H. Biological effects of cyclosporin A: a new antilymphocytic agent. Agents Actions. 1976;6(4):468–475. doi: 10.1007/BF01973261. [DOI] [PubMed] [Google Scholar]
- 40.Paeshuyse J, Kaul A, de Clercq E, et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43(4):761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 41.Watashi K. Alisporivir, a cyclosporin derivative that selectively inhibits cyclophilin, for the treatment of HCV infection. Curr Opin Investig Drugs. 2010;11(2):213–224. [PubMed] [Google Scholar]
- 42.Ma S, Boerner JE, TiongYip C, et al. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob Agents Chemother. 2006;50(9):2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flisiak R, Horban A, Gallay P, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47(3):817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 44.Coelmont L, Kaptein S, Paeshuyse J, et al. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob Agents Chemother. 2009;53(3):967–976. doi: 10.1128/AAC.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49(5):1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 47.Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 49.Andre P, Perlemuter G, Budkowska A, Brechot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25(1):93–104. doi: 10.1055/s-2005-864785. [DOI] [PubMed] [Google Scholar]
- 50.Burlone ME, Budkowska A. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol. 2009;90(Pt 5):1055–1070. doi: 10.1099/vir.0.008300-0. [DOI] [PubMed] [Google Scholar]
- 51.Cocquerel L, Voisset C, Dubuisson J. Hepatitis C virus entry: potential receptors and their biological functions. J Gen Virol. 2006;87(Pt 5):1075–1084. doi: 10.1099/vir.0.81646-0. [DOI] [PubMed] [Google Scholar]
- 52.Diedrich G. How does hepatitis C virus enter cells? FEBS J. 2006;273(17):3871–3885. doi: 10.1111/j.1742-4658.2006.05379.x. [DOI] [PubMed] [Google Scholar]
- 53.Evans MJ, von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 54.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80(4):1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanchard E, Belouzard S, Goueslain L, et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koutsoudakis G, Kaul A, Steinmann E, et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80(11):5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Hahn T, Rice CM. Hepatitis C virus entry. J Biol Chem. 2008;283(7):3689–3693. doi: 10.1074/jbc.R700024200. [DOI] [PubMed] [Google Scholar]
- 58.Ploss A, Evans MJ, Gaysinskaya VA, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quer J, Cos J, Murillo P, Esteban JI, Esteban R, Guardia J. Improved attachment of natural HCV isolate to Daudi cells upon elimination of immune complexes and close pH control. Intervirology. 2005;48(5):285–291. doi: 10.1159/000085096. [DOI] [PubMed] [Google Scholar]
- 60.Cheng G, Montero A, Gastaminza P, et al. A virocidal amphipathic {alpha}-helical peptide that inhibits hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2008;105(8):3088–3093. doi: 10.1073/pnas.0712380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meuleman P, Hesselgesser J, Paulson M, et al. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48(6):1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 62.Huang H, Sun F, Owen DM, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104(14):5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82(5):2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durantel D, Alotte C, Zoulim F. Glucosidase inhibitors as antiviral agents for hepatitis B and C. Curr Opin Investig Drugs. 2007;8(2):125–129. [PubMed] [Google Scholar]
- 65.Romero-Lopez C, Diaz-Gonzalez R, Barroso-delJesus A, Berzal-Herranz A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J Gen Virol. 2009;90(Pt 7):1659–1669. doi: 10.1099/vir.0.008821-0. [DOI] [PubMed] [Google Scholar]
- 66.Pawlotsky JM. More sensitive hepatitis C virus RNA detection: what for? J Hepatol. 2010;52(6):783–785. doi: 10.1016/j.jhep.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 68.Han DS, Hahm B, Rho HM, Jang SK. Identification of the protease domain in NS3 of hepatitis C virus. J Gen Virol. 1995;76(Pt 4):985–993. doi: 10.1099/0022-1317-76-4-985. [DOI] [PubMed] [Google Scholar]
- 69.de Francesco R, Pessi A, Steinkuhler C. The hepatitis C virus NS3 proteinase: structure and function of a zinc-containing serine proteinase. Antivir Ther. 1998;3(Suppl 3):99–109. [PubMed] [Google Scholar]
- 70.Vallet S, Gouriou S, Nousbaum JB, Legrand-Quillien MC, Goudeau A, Picard B. Genetic heterogeneity of the NS3 protease gene in hepatitis C virus genotype 1 from untreated infected patients. J Med Virol. 2005;75(4):528–537. doi: 10.1002/jmv.20302. [DOI] [PubMed] [Google Scholar]
- 71.Foy E, Li K, Wang C, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300(5622):1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 72.Foy E, Li K, Sumpter R, Jr, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360(18):1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 74.Hezode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360(18):1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 75.Kwo P, Lawitz EJ, McCone J, Schiff E, Vierling JM, Pound D. HCV SPRINT-1 final results: SVR 24 from a phase 2 study of boceprevir plus PegIntron (peginterferon alfa-2b)/ribavirin in treatment-naive subjects with genotype 1 chronic hepatitis C. J Hepatol. 2009;50:S4. [Google Scholar]
- 76.Reesink HW, Zeuzem S, Weegink CJ, et al. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology. 2006;131(4):997–1002. doi: 10.1053/j.gastro.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Sarrazin C, Rouzier R, Wagner F, et al. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology. 2007;132(4):1270–1278. doi: 10.1053/j.gastro.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 78.Forestier N, Reesink HW, Weegink CJ, et al. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology. 2007;46(3):640–648. doi: 10.1002/hep.21774. [DOI] [PubMed] [Google Scholar]
- 79.Terrault N, Khalili M. A glimpse of future hepatitis C virus treatment paradigms. Hepatology. 2010;51(2):707–710. doi: 10.1002/hep.23523. [DOI] [PubMed] [Google Scholar]
- 80.Opar A. Excitement grows for potential revolution in hepatitis C virus treatment. Nat Rev Drug Discov. 2010;9(7):501–503. doi: 10.1038/nrd3214. [DOI] [PubMed] [Google Scholar]
- 81.Berg T, Sarrazin C, Herrmann E, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37(3):600–609. doi: 10.1053/jhep.2003.50106. [DOI] [PubMed] [Google Scholar]
- 82.Hadziyannis SJ, Sette H, Jr, Morgan TR, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 83.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20(1):23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeffers LJ, Cassidy W, Howell CD, Hu S, Reddy KR. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39(6):1702–1708. doi: 10.1002/hep.20212. [DOI] [PubMed] [Google Scholar]
- 85.Myers RP, Patel K, Pianko S, Poynard T, McHutchison JG. The rate of fibrosis progression is an independent predictor of the response to antiviral therapy in chronic hepatitis C. J Viral Hepat. 2003;10(1):16–22. doi: 10.1046/j.1365-2893.2003.00387.x. [DOI] [PubMed] [Google Scholar]
- 86.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 87.Chung RT, Andersen J, Volberding P, et al. Peginterferon alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351(5):451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 90.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 91.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 92.Asselah T. Genetic polymorphism and response to treatment in chronic hepatitis C: the future of personalized medicine. J Hepatol. 2010;52(3):452–454. doi: 10.1016/j.jhep.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 93.McCarthy JJ, Li JH, Thompson A, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138(7):2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muir AJ, Bornstein JD, Killenberg PG, Altantic Coast Hepatitis Treatment Group Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350(22):2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- 95.Conjeevaram HS, Fried MW, Jeffers LJ, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131(2):470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 96.Yu ML, Dai CY, Huang JF, et al. Rapid virological response and treatment duration for chronic hepatitis C genotype 1 patients: a randomized trial. Hepatology. 2008;47(6):1884–1893. doi: 10.1002/hep.22319. [DOI] [PubMed] [Google Scholar]
- 97.Borowski P, Lang M, Haag A, Baier A. Tropolone and its derivatives as inhibitors of the helicase activity of hepatitis C virus nucleotide triphosphatase/helicase. Antivir Chem Chemother. 2007;18(2):103–109. doi: 10.1177/095632020701800206. [DOI] [PubMed] [Google Scholar]
- 98.Boguszewska-Chachulska AM, Krawczyk M, Najda A, et al. Searching for a new anti-HCV therapy: synthesis and properties of tropolone derivatives. Biochem Biophys Res Commun. 2006;341(2):641–647. doi: 10.1016/j.bbrc.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 99.Krawczyk M, Wasowska-Lukawska M, Oszczapowicz I, Boguszewska-Chachulska AM. Amidinoanthracyclines – a new group of potential anti-hepatitis C virus compounds. Biol Chem. 2009;390(4):351–360. doi: 10.1515/BC.2009.040. [DOI] [PubMed] [Google Scholar]
- 100.Beaulieu PL. Recent advances in the development of NS5B polymerase inhibitors for the treatment of hepatitis C virus infection. Expert Opin Ther Pat. 2009;19(2):145–164. doi: 10.1517/13543770802672598. [DOI] [PubMed] [Google Scholar]
- 101.Huang Z, Peng J, Cai Z, Huang C, Murray MG. 15th International Symposium on Hepatitis C Virus and Related Viruses. San Antonio, TX: 2010. Novel molecular as a potent HCV helicase inhibitor. Abstract # 278:156. [Google Scholar]
- 102.Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78(20):11288–11295. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Einav S, Gerber D, Bryson PD, et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26(9):1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85(Pt 9):2485–2502. doi: 10.1099/vir.0.80204-0. [DOI] [PubMed] [Google Scholar]
- 105.Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology. 2001;284(1):1–12. doi: 10.1006/viro.2001.0885. [DOI] [PubMed] [Google Scholar]
- 106.Heathcote J, Main J. Treatment of hepatitis C. J Viral Hepat. 2005;12(3):223–235. doi: 10.1111/j.1365-2893.2005.00600.x. [DOI] [PubMed] [Google Scholar]
- 107.Nettles RE, Chung E, Persson A. BMS-790052 is a first-in-class potent hepatitis C virus (HCV) NS5A inhibitor for patients with chronic HCV infection: results from a proof-of-concept study. Hepatology. 2008;48(Suppl 4) Abstract # LB12. [Google Scholar]
- 108.Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465(7294):96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Domingo E.RNA virus evolution and the control of viral disease Jucker E.Progress in Drug ResearchVol. XXXIIIBasel, Switzerland: Birkhauser Verlag; 198993–133. [DOI] [PubMed] [Google Scholar]
- 110.Gane EJ, Stedman C. First-in-man demonstration of potent antiviral activity with a nucleoside polymerase (R7128) and protease (R7227/ITMN-191) inhibitor combination in HCV: safety, pharmacokinetics, and virologic results from INFORM-1. J Hepatol. 2008;50(Suppl 1) Abstract # 1046, S380. [Google Scholar]
- 111.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 112.Herrmann E, Neumann AU, Schmidt JM, Zeuzem S. Hepatitis C virus kinetics. Antivir Ther. 2000;5(2):85–90. [PubMed] [Google Scholar]
- 113.Quer J, Esteban JI, Cos J, et al. Effect of bottlenecking on evolution of the nonstructural protein 3 gene of hepatitis C virus during sexually transmitted acute resolving infection. J Virol. 2005;79(24):15131–15141. doi: 10.1128/JVI.79.24.15131-15141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Okamoto H, Kojima M, Okada S, et al. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190(2):894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 115.Domingo E, Holland JJ. Mutation rates and rapid evolution of RNA viruses. In: Morse SS, editor. Evolutionary Biology of Viruses. New York: Raven Press; 1994. pp. 161–184. [Google Scholar]
- 116.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Villet S, Billioud G, Pichoud C, et al. In vitro characterization of viral fitness of therapy-resistant hepatitis B variants. Gastroenterology. 2009;136(1):168–176. doi: 10.1053/j.gastro.2008.09.068. [DOI] [PubMed] [Google Scholar]
- 118.Nijhuis M, van Maarseveen NM, Boucher CA. Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. Handb Exp Pharmacol. 2009;189:299–320. doi: 10.1007/978-3-540-79086-0_11. [DOI] [PubMed] [Google Scholar]
- 119.Forestier N, Susser S, Welker M, Karey U, Zeuzem S, Sarrazin C. Long term follow up of patients previously treated with telaprevir. Hepatology. 2008;48(4 Suppl 1) 760A (1011). [Google Scholar]
- 120.Domingo E, Ruiz-Jarabo CM, Sierra S, et al. Emergence and selection of RNA virus variants: memory and extinction. Virus Res. 2002;82(1–2):39–44. doi: 10.1016/s0168-1702(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 121.Susser S, Welsch C, Wang Y, et al. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology. 2009;50(6):1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 122.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70(2):28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 123.Yi M, Tong X, Skelton A, et al. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem. 2006;281(12):8205–8215. doi: 10.1074/jbc.M510246200. [DOI] [PubMed] [Google Scholar]
- 124.Lin C, Gates CA, Rao BG, et al. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem. 2005;280(44):36784–36791. doi: 10.1074/jbc.M506462200. [DOI] [PubMed] [Google Scholar]
- 125.Thompson AJ, McHutchison JG. Antiviral resistance and specif ically targeted therapy for HCV (STAT-C) J Viral Hepat. 2009;16(6):377–387. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]