Abstract
Objective:
The aim of this study was to investigate the epidemiology and antibiotic susceptibility profiles of isolated bacterial organisms in relation to empiric treatment of neutropenic fever over a 15-year period.
Methods:
All patients with or at risk for febrile neutropenia and treated in the hematology ward of the Antwerp University Hospital during 1994–2008 were prospectively included. Skin, blood, and urine cultures were taken. Oral quinolone prophylaxis was started in patients with neutropenia without fever. Empiric starting therapy consisted of amikacin in combination with cefepime.
Results:
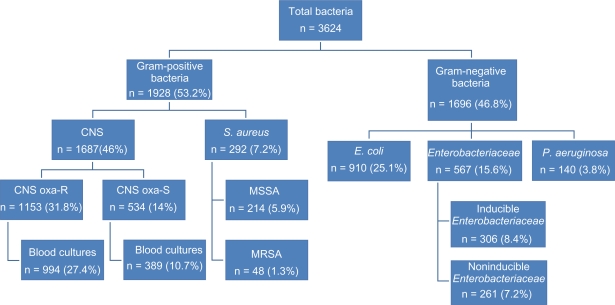
A total of 3624 bacteria were isolated. The most common pathogens were coagulase-negative Staphylococci (46%), followed by Escherichia coli (25%), Enterobacteriaceae (15.6%), Staphylococcus aureus (7.2%), and Pseudomonas aeruginosa (3.8%). The balance between Gram-positive and Gram-negative bacteria remained stable, with a majority of Gram-positive bacteria. A shift from oxacillin-sensitive to oxacillin-resistant coagulase-negative Staphylococci was observed. Regarding susceptibility patterns, no vancomycin resistance was detected in coagulase-negative Staphylococci or in S. aureus. The E. coli susceptibility rates remained stable. However, 66% of bloodstream infections were ciprofloxacin-resistant. A reduced susceptibility of P. aeruginosa strains to meropenem was noticed.
Conclusions:
Improvement in antibiotic susceptibility of inducible Enterobacteriaceae following a switch of empiric antibiotic therapy was maintained 15 years after starting the latter treatment. Further improvement in antibiotic susceptibility of these bacteria to ceftazidime was observed, but continuous vigilance is warranted.
Keywords: bacterial susceptibility, febrile neutropenia, antibiotic change
Introduction
Patients with malignancy are very susceptible to nosocomial infections. Because of this susceptibility, impaired immune response, and the potential for infection with antimicrobial-resistant pathogens, patients suspected of having developed these infections are commonly treated with broad-spectrum antibiotics.1 In this specific group of patients, the most important determinant of successful treatment of infection is prompt institution of effective empiric antibiotics, especially in patients with neutropenic fever. Optimization of empiric therapy requires a knowledge of likely antimicrobial resistance patterns, especially given that neutropenic patients can show different bacterial susceptibilities to those found with other diseases.2,3
Control of emerging resistant organisms remains one of the major challenges for the future. One strategy to achieve this is prospectively monitoring bacterial susceptibility rates. The effect of such intervention strategies on the prevention and control of nosocomial infections has been documented.4
In 1995, after years of using a third-generation cephalosporin (ceftazidime) in combination with a glycopeptide as empiric therapy for neutropenic fever, we were confronted with a high resistance rate of inducible Enterobacteriaceae to ceftazidime.5 Consequently, in May 1995, we introduced a new combination therapy comprising a fourth-generation cephalosporin (cefepime) and an aminoglycoside (amikacin) for the empiric treatment of fever in neutropenic patients in our hematology unit.
The aim of this prospective study was to analyze trends in the bacterial spectrum and antibiotic susceptibilities of bacterial isolates in relation to empiric treatment during a long-term (15-year) follow-up period in our unit.
Materials and methods
This prospective, open-label, epidemiologic study was conducted between 01 January 1994 and 31 December 2008 in the hematology ward at Antwerp University Hospital. This is a tertiary center including units for treatment of acute leukemia and hematopoietic stem cell transplantation. The patient population consisted of adults with acute and chronic leukemias (around 50%), lymphomas (around 25%), or other hematologic malignancies. About 25% of patients were hospitalized for transplantation.
Bacterial cultures
All hospitalized hematology patients with neutropenia were included in the study. Fever was defined as two consecutive axillary temperature recordings of 38°C or higher in a 12-hour period, or as a single recording of 38.5°C or higher. Patients were considered neutropenic if they had fewer than 500 neutrophils/mm3 or if the absolute neutrophil count was greater than 500/mm3 and less than 1000/mm3, but decreasing progressively and expected to be less than 500/mm3 within 24–48 hours. The standard protocol required “inventory” cultures to be taken once a week from all patients with neutropenia (with or without fever) and twice a week from hematopoietic stem cell transplantation patients. The cultures consisted of skin swabs (from nose, throat, ears, axilla, groin, perianal region, vagina, or preputium), a urine culture, and a stool culture. In cases of neutropenic fever, three sets (each set consisting of one aerobic and one anaerobic) of blood cultures were taken, as well as a midstream urine culture, and throat swabs before starting empiric antibiotics, according to the protocol. Suspected foci of infection were also cultured. In cases of persistent fever, one set of blood cultures was taken on each day.
All bacterial isolates were cultured and tested when appropriate for antibiotic susceptibility using Kirby-Bauer disk diffusion according to the standards of the Clinical and Laboratory Standards Institute (CLSI). Breakpoints were based on CLSI guidelines.6 Pathogens with intermediate susceptibility were considered resistant. In the analysis for susceptibility testing, each pathogen was calculated only once per patient and per type of isolate. The total number of hospitalizations per year for neutropenia was retrieved from hospital administration records. All data were collected using dBase IV till 1996, and 1997 Microsoft Access software thereafter. To avoid interference, no major changes in hygiene procedures were made.
Treatment
From February 1991 until the end of April 1995 (before the start of this study), patients were treated with ceftazidime 2 g tid intravenously (IV) in combination with vancomycin 1 g bid IV or teicoplanin 1200 mg/day IV, followed by 800 mg/day IV. At the start of this study in May 1995, the empiric regimen was changed to cefepime 2 g tid IV and amikacin as a loading dose of 25 mg/kg (if renal function was normal) followed by 15 mg/kg/day IV. Vancomycin or teicoplanin were added after 48 hours based on clinical (eg, persistent fever) or microbiologic results. If fever persisted for at least an additional 48 hours, ceftazidime or cefepime were replaced by imipenem-cilastatin or meropenem 1 g tid IV. The oral antibiotic prophylaxis policy was stable, ie, fluoroquinolones (ciprofloxacin 500 mg bid or levofloxacin 500 mg once daily) and/or trimethoprim-sulfamethoxazole (160 mg + 800 mg bid), the latter being for stem cell transplant patients. Oral antibiotic prophylaxis was discontinued after initiation of the empiric antibiotic regimen. The total consumption of antibiotics (in grams) was obtained from the pharmacy department and was converted to defined daily doses (DDDs) per 100 patient days for the years 1994–2008.
Statistical analysis
Because repeated measures were taken for the bacteria, a correlation has to be taken into account when analyzing the data. We used linear mixed models to account for the heterogeneity across the bacteria (and homogeneity within the bacteria). Stepwise model selection was used to identify the statistically significant effects, starting from all two-way interaction effects. P < 0.05 was considered statistically significant. SAS® (version 9.2) was used for all statistical analyses.
Results
Bacterial epidemiology
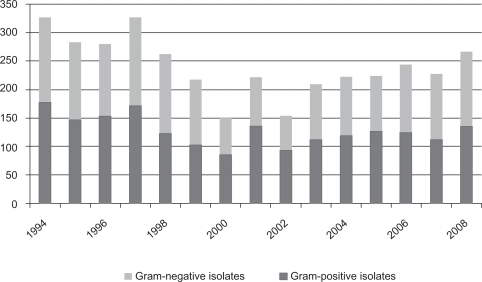
During the 15 years of follow-up, a total of 3624 bacteria (Gram-positive and Gram-negative) were isolated and cultured (Figures 1 and 4). Gram-positive bacteria were more common (n = 1928, 53.2%) than Gram-negative bacteria (n = 1696, 46.8%) throughout the study period. No significant difference was observed between the proportion of Gram-positive and Gram-negative isolates. A decrease in the total number of isolates was observed during the study period (P <0.0001). This was not due to fewer hospitalizations for neutropenia.
Figure 1.
Gram-classification of the isolated bacteria in all cultures (1994–2008).
Figure 4.
Characteristics of the isolated bacteria (1994–2008).
Abbrevations: CNA, coagulase-negative Staphylococci; CNS oxa-R, CNS oxacillin-resistant; CNS oxa-S, CNS oxacillin-sensitive; MSSA, methicillin-sensitive S. aureus; MRSA, methicillin-resistant S. aureus.
The most common Gram-positive isolates were coagulase-negative Staphylococci resistant for oxacillin (n = 1153, 31.8%). Of these bacteria, 994 samples were isolated from blood cultures. Other isolated coagulase-negative Staphylococci were oxacillin-sensitive (n = 534, 14.7%); 389 bacteria were isolated from blood cultures. During follow-up, a statistically significant shift to the more oxacillin-resistant phenotypes occurred (confidence interval [CI] 32.6–49.88).
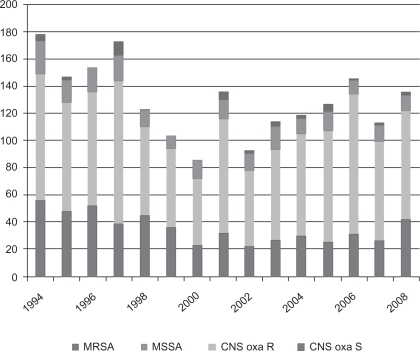
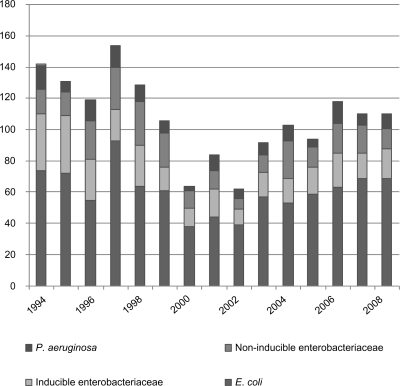
S. aureus was isolated in 262 (7.2%) cases. Only 48 isolates (1.3%) were methicillin-resistant (Figure 2). There was a significant decrease in S. aureus isolates (P <0.0001) throughout the study period. Among the Gram-negative bacteria, the majority were Escherichia coli (n = 910, 25%). A significant decrease in isolates was seen (P = 0.0006). Enterobacteriaceae was isolated in 567 (15.6%) cases. Of these, 306 isolates were inducible Enterobacteriaceae (Enterobacter spp, Serratia spp, Morganella morganii, Citrobacter spp, Providencia spp, and Pantoea agglomerans, 8.4%). A significant decrease over time of inducible Enterobacteriaceae was noted (P = 0.0027). Pseudomonas aeruginosa was isolated in 140 (3.8%) cases (Figure 3).
Figure 2.
Isolated Gram-positive bacteria in all cultures (1994–2008).
Abbreviations: MRSA, Methicillin-resistant S. Aureus; MSSA, Methicillin-sensitive S. Aureus; CNS oxa R, CNS oxacillin-resistant; CNS oxa S, CNS oxacillin-sensitive.
Figure 3.
Isolated Gram-negative bacteria in all cultures (1994–2008).
Antibiotic susceptibility trends
The antibiotics tested and the percentages of isolates with decreased susceptibility are listed in Tables 1 and 2 (for Gram-positive and Gram-negative isolates, respectively). Data are provided for most frequently encountered bacteria for all cultures. Antibiotic susceptibility data for blood cultures are provided separately for oxacillin-sensitive and oxacillin-resistant coagulase-negative Staphylococci.
Table 1.
Behavior of antimicrobial sensitivity of Gram-positive bacteria isolated in the period 1994–2008
| 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSSA, all cultures | |||||||||||||||
| n | 24 | 16 | 18 | 19 | 12 | 10 | 14 | 14 | 12 | 17 | 11 | 14 | 10 | 12 | 11 |
| Penicillin | 83.8 | 87.5 | 88.9 | 89.5 | 100 | 90.0 | 85.7 | 78.6 | 91.7 | 88.2 | 90.0 | 91.7 | 66.7 | 100 | 90.9 |
| Oxacillin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Amikacin | 0 | 0 | 5.6 | 5.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cotrimoxazole | 0 | 0 | 11.1 | 5.3 | 0 | 0 | 14.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ciprofloxacin | – | 12.5 | 33.3 | 15.8 | 0 | 0 | 0 | 0 | 8.3 | 0 | – | – | 0 | 0 | 0 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CNS, oxacillin-sensitive, all cultures | |||||||||||||||
| n | 56 | 48 | 52 | 27 | 45 | 36 | 23 | 32 | 22 | 27 | 30 | 25 | 31 | 26 | 42 |
| Penicillin | 66.1 | 79.2 | 73.1 | 74.1 | 48.9 | 66.7 | 60.9 | 53.1 | 50.0 | 55.6 | 56.7 | 56 | 66.7 | 80.8 | 73.8 |
| Amoxy-Clav | 0 | 2.1 | 7.7 | 0 | 2.2 | 2.8 | 4.3 | 0 | 0 | – | – | – | – | – | – |
| Oxacillin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Meropenem | – | – | – | – | 2.2 | 0 | 4.3 | 0 | 0 | – | – | – | – | – | – |
| Cefepime | – | 12.5 | 7.7 | 3.7 | 2.3 | 8.3 | 4.3 | 0 | 0 | – | – | – | – | – | – |
| Amikacin | 0 | 0 | 3.8 | 0 | 2.2 | 0 | 4.3 | 0 | 0 | 0 | 0 | 0 | 3.2 | 0 | 0 |
| Cotrimoxazole | 17.9 | 18.8 | 11.5 | 11.1 | 13.3 | 16.7 | 13.0 | 3.1 | 0 | 3.7 | 16.7 | 24.0 | 22.6 | 23.1 | 26.2 |
| Ciprofloxacin | – | 29.2 | 15.4 | 11.1 | 15.6 | 11.1 | 17.4 | 3.1 | 27.3 | 16.7 | – | – | 50.0 | 53.8 | 42.9 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 2.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.2 | 0 | 0 |
| CNS, oxacillin-sensitive, blood cultures | |||||||||||||||
| n | 47 | 35 | 38 | 27 | 39 | 30 | 20 | 29 | 16 | 22 | 20 | 17 | 22 | 16 | 26 |
| Penicillin | 68.1 | 77.1 | 76.3 | 74.1 | 48.7 | 63.3 | 65.0 | 51.7 | 43.8 | 59.1 | 55.0 | 47.1 | 71.4 | 87.5 | 84.6 |
| Amoxy-Clav | 0 | 2.9 | 5.3 | 0 | 2.6 | 3.3 | 5 | 0 | 0 | – | – | – | – | – | – |
| Oxacillin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Meropenem | – | – | – | – | 2.6 | 0 | 0 | 0 | 0 | – | – | – | – | – | – |
| Cefepime | – | 10.0 | 5.3 | 3.7 | 2.6 | 10.0 | 0 | 0 | 0 | – | – | – | – | – | – |
| Amikacin | 0 | 0 | 5.3 | 0 | 2.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 | 0 | – |
| Cotrimoxazole | 8.3 | 7.7 | 15.8 | 11.1 | 12.8 | 16.7 | 0 | 3.4 | 0 | 4.5 | 20.0 | 17.6 | 22.7 | 37.5 | 30.8 |
| Ciprofloxacin | – | 15.4 | 21.1 | 11.1 | 15.4 | 13.3 | 0 | 3.4 | 0 | 20.0 | 0 | – | 50.0 | 68.8 | 50.0 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 | 0 | 0 |
| Levofloxacin | |||||||||||||||
| CNS, oxacillin-resistant, all cultures | |||||||||||||||
| n | 93 | 80 | 84 | 105 | 65 | 58 | 49 | 84 | 56 | 66 | 75 | 82 | 103 | 73 | 80 |
| Penicillin | 100 | 97.4 | 100 | 99.0 | 100 | 100 | 100 | 96.4 | 100 | 100 | 98.7 | 96.9 | 98.6 | 94.7 | 98 |
| Amoxy-Clav | 40.0 | 71.4 | 71.4 | 73.3 | 67.7 | 67.2 | 69.4 | 61.9 | 91.1 | – | – | – | – | – | – |
| Oxacillin | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Meropenem | – | – | – | – | 50.8 | 63.8 | 63.3 | 60.7 | 28.6 | – | – | – | – | – | – |
| Cefepime | – | 71.4 | 59.5 | 53.3 | 53.8 | 60.3 | 57.1 | 73.8 | 63.6 | – | – | – | – | – | – |
| Amikacin | 6.7 | 28.8 | 29.8 | 24.8 | 29.2 | 27.6 | 46.9 | 28.6 | 32.1 | 27.3 | 29.7 | 23.2 | 24.5 | 17.8 | – |
| Cotrimoxazole | 53.3 | 60.0 | 67.9 | 79.0 | 64.6 | 56.9 | 71.4 | 44.0 | 51.2 | 74.3 | 78.7 | 70.7 | 69.6 | 61.1 | 70.0 |
| Ciprofloxacin | – | 77.5 | 70.2 | 85.7 | 80.0 | 75.9 | 87.7 | 65.5 | 83.9 | 72.7 | 0 | – | 75.0 | 85.7 | 89.8 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 1.0 | 1.5 | 1.7 | 4.1 | 1.2 | 1.8 | 4.5 | 1.3 | 2.4 | 0 | 1.4 | 1.3 |
| CNS, oxacillin-resistant, blood cultures | |||||||||||||||
| n | 89 | 71 | 79 | 91 | 61 | 48 | 45 | 75 | 47 | 62 | 59 | 71 | 81 | 57 | 58 |
| Penicillin | 100 | 97.1 | 100 | 98.9 | 100 | 100 | 100 | 96.0 | 100 | 100 | 98.3 | 96.5 | 98.5 | 93.3 | 100 |
| Amoxy-Clav | 30.8 | 76.5 | 72.2 | 73.6 | 67.2 | 64.6 | 75.6 | 64.0 | 89.4 | – | – | – | – | – | – |
| Oxacillin | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Meropenem | – | – | – | – | 52.5 | 62.5 | 66.7 | 62.7 | 32 | – | – | – | – | – | – |
| Cefepime | – | 77.8 | 60.8 | 52.7 | 52.5 | 58.3 | 62.2 | 72.0 | 61.9 | – | – | – | – | – | – |
| Amikacin | 0 | 29.6 | 30.4 | 24.2 | 29.5 | 27.1 | 51.1 | 32.0 | 31.8 | 29.0 | 28.8 | 19.7 | 22.2 | 22.9 | – |
| Cotrimoxazole | 46.2 | 57.7 | 68.4 | 81.3 | 62.3 | 62.5 | 75.6 | 44.0 | 51.0 | 74.2 | 76.3 | 70.4 | 67.9 | 69.6 | 60.3 |
| Ciprofloxacin | – | 74.6 | 68.4 | 84.6 | 78.7 | 75.0 | 88.9 | 64.0 | 83.1 | 72.7 | – | - | 66.7 | 88.9 | 90.0 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 1.1 | 1.6 | 2.1 | 4.4 | 1.3 | 2.1 | 4.8 | 0 | 2.8 | 0 | 1.8 | 1.7 |
| Levofloxacin | – | – | – | – | – | – | – | – | – | 78.3 | 83.7 | 82.7 | 76.7 | – | – |
Notes: n, number of isolates; –, not tested; Shown results indicate decreased sensitivity.
Abbreviations: CNS, Coagulase-negative Staphylococci; MRSA, Methicillin-resistant S. aureus; MSSA, Methicillin-sensitive S. aureus.
Table 2.
Behavior of antimicrobial sensitivity of Gram-negative bacteria isolated in the period 1994–2008
| 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli, all cultures | |||||||||||||||
| n | 74 | 72 | 55 | 93 | 64 | 61 | 38 | 44 | 39 | 57 | 53 | 59 | 63 | 69 | 69 |
| Ampicillin | 60.8 | 59.7 | 52.7 | 72.0 | 53.1 | 63.9 | 55.3 | 65.9 | 61.5 | 33.3 | 58.5 | 62.1 | 63.5 | 60.3 | 70.1 |
| Amoxy-clav | 5.4 | 16.7 | 3.6 | 11.8 | 7.8 | 14.8 | 15.8 | 0 | 0 | 3.5 | 9.4 | 6.8 | 7.9 | 17.4 | 13.4 |
| Piperacillin | 40.5 | 40.3 | 41.8 | 61.3 | 37.5 | 45.9 | 36.8 | 56.8 | 46.2 | 63.8 | 53.9 | – | – | – | – |
| Ceftazidime | 5.4 | 4.2 | 0 | 4.3 | 0 | 13.1 | 5.3 | 0 | 2.6 | 5.3 | 3.8 | 1.7 | 0 | 3.2 | 14 |
| Cefepime | 2.7 | 0 | 0 | 0 | 0 | 11.5 | 0 | 0 | 2.6 | 3.5 | 0 | 0 | 0 | 1.6 | 7.3 |
| Meropenem | – | – | – | – | 0 | 1.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Imipenem | 0 | 0 | 0 | 0 | – | – | – | – | – | – | – | – | – | – | – |
| Amikacin | 1.4 | 0 | 0 | 0 | 0 | 1.6 | 0 | 0 | 0 | 0 | 1.9 | 0 | 1.6 | 0 | 1.4 |
| Ciprofloxacin | 31.5 | 35.2 | 29.1 | 38.7 | 26.6 | 42.6 | 28.9 | 36.4 | 38.5 | 26.1 | – | 9.1 | 49.3 | 40.6 | |
| Cotrimoxazole | 44.6 | 40.3 | 49.1 | 53.8 | 40.6 | 57.4 | 39.5 | 47.7 | 48.7 | 35.1 | 34.0 | 51.7 | 52.4 | 56.5 | 50.7 |
| Non-inducible Enterobacteriaceae, all cultures | |||||||||||||||
| n | 16 | 15 | 25 | 27 | 28 | 22 | 11 | 12 | 7 | 11 | 24 | 13 | 19 | 19 | 13 |
| Ampicillin | 68.8 | 80 | 80 | 70.4 | 78.6 | 68.2 | 90.9 | 75 | 85.7 | 54.6 | 75.0 | 84.6 | 78.9 | 81.3 | 69.2 |
| Amoxy-clav | 12.5 | 26.7 | 16.0 | 7.4 | 3.6 | 9.1 | 0 | 8.3 | 14.3 | 0 | 16.7 | 30.8 | 5.3 | 26.3 | 16.7 |
| Piperacillin | 31.3 | 33.3 | 20.0 | 29.6 | 35.7 | 27.3 | 36.4 | 33.3 | 14.3 | 9.1 | 20.8 | – | – | – | – |
| Ceftazidime | 25.0 | 6.7 | 12.0 | 9.0 | 17.9 | 9.1 | 9.1 | 0 | 0 | 0 | 0 | 7.7 | 0 | 0 | 0 |
| Cefepime | 6.7 | 9.1 | 0 | 0 | 3.6 | 4.5 | 0 | 8.3 | 0 | 0 | 0 | 7.7 | 0 | 0 | 0 |
| Meropenem | – | – | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.3 | 5.3 | 0 |
| Imipenem | 0 | 6.7 | 0 | 0 | – | – | – | – | – | – | – | – | – | – | – |
| Amikacin | 0 | 6.7 | 0 | 0 | 0 | 4.5 | 0 | 0 | 0 | 0 | 0 | 7.7 | 5.3 | 5.3 | 0 |
| Ciprofloxacin | 12.5 | 0 | 0 | 38.7 | 10.7 | 4.5 | 36.4 | 25.0 | 14.3 | 0 | – | – | 0 | 10.5 | 7.7 |
| Cotrimoxazole | 12.5 | 6.7 | 12.0 | 30.0 | 28.6 | 13.6 | 45.5 | 33.3 | 14.3 | 9.1 | 16.7 | 7.7 | 15.8 | 15.8 | 23.1 |
| Inducible Enterobacteriaceae, all cultures | |||||||||||||||
| n | 36 | 37 | 26 | 20 | 26 | 15 | 12 | 18 | 10 | 16 | 16 | 17 | 22 | 16 | 19 |
| Ampicillin | 94.4 | 80.6 | 84.6 | 85.0 | 72.7 | 73.3 | 83.3 | 88.9 | 80.0 | 100 | 81.3 | 94.1 | 95.5 | 93.8 | 78.9 |
| Amoxy-clav | 88.9 | 75.0 | 84.6 | 85.0 | 72.7 | 86.7 | 66.7 | 77.8 | 80.0 | 68.8 | 62.5 | 88.2 | 81.8 | 87.5 | 57.9 |
| Piperacillin | 72.2 | 48.6 | 30.8 | 15.0 | 36.4 | 13.3 | 16.7 | 22.2 | 10.0 | 75.1 | 37.5 | – | – | – | – |
| Ceftazidime | 75.0 | 35.1 | 30.8 | 15.0 | 31.8 | 13.3 | 8.3 | 11.1 | 10.0 | 62.8 | 37.5 | 47.1 | 36.4 | 37.5 | 35.5 |
| Cefepime | 13.9 | 0 | 7.7 | 0 | 0 | 0 | 0 | 0 | 0 | 12.5 | 0 | 17.6 | 4.5 | 12.5 | 12.5 |
| Meropenem | – | – | – | – | 4.6 | 0 | 0 | 0 | 0 | 0 | 0 | 5.9 | 0 | 0 | 0 |
| Imipenem | 0 | 0 | 0 | 5.0 | – | – | – | – | – | – | – | – | – | – | – |
| Amikacin | 36.1 | 18.9 | 0 | 5.0 | 18.2 | 6.7 | 0 | 0 | 0 | 18.8 | 6.3 | 11.8 | 13.6 | 0 | 15.8 |
| Ciprofloxacin | 52.8 | 24.3 | 15.4 | 10.0 | 31.8 | 6.7 | 33.3 | 5.6 | 10.0 | 60.0 | - | 33.3 | 25.0 | 33.3 | |
| Cotrimoxazole | 52.8 | 33.3 | 11.5 | 10.0 | 40.9 | 6.7 | 0 | 5.6 | 10.0 | 50.0 | 31.3 | 35.3 | 22.7 | 31.3 | 47.4 |
| Pseudomonas aeruginosa, all cultures | |||||||||||||||
| n | 15 | 7 | 13 | 14 | 11 | 8 | 3 | 10 | 6 | 8 | 10 | 5 | 14 | 7 | 9 |
| Piperacillin | 0 | 14.3 | 0 | 14.3 | 18.2 | 12.5 | – | 0 | 0 | 12.5 | 10.0 | – | – | – | – |
| Ceftazidime | 0 | 0 | 0 | 0 | 9.1 | 12.5 | – | 0 | 0 | 12.5 | 20.0 | 0 | 28.6 | 14.3 | 11.1 |
| Cefepime | 53.3 | 25.0 | 0 | 14.3 | 9.1 | 12.5 | – | 0 | 0 | 25.0 | 10.0 | 20.0 | 21.4 | 14.3 | 11.1 |
| Imipenem | 0 | 14.3 | 15.4 | 21.4 | – | – | – | – | – | – | – | – | – | – | – |
| Meropenem | – | – | – | – | 18.2 | 0 | – | 0 | 0 | 25.0 | 10.0 | 20.0 | 7.1 | 14.3 | 33.3 |
| Amikacin | 0 | 0 | 0 | 7.1 | 0 | 0 | – | 0 | 0 | 25.0 | 10.0 | 0 | 7.1 | 0 | 11.1 |
| Ciprofloxacin | 6.7 | 16.7 | 0 | 7.1 | 18.2 | 0 | – | 0 | 0 | 0 | – | – | 100 | 14.3 | 22.2 |
Notes: n, number of isolates; –, not tested; Shown results indicate decreased sensitivity.
No significant decrease in susceptibility was noted for methicillin-sensitive S. aureus (MSSA). MSSA isolates remained susceptible to ciprofloxacin, cotrimoxazole, and amikacin. None of the 48 methicillin-resistant S. aureus (MRSA) isolates showed true vancomycin resistance (data not shown).
Among the oxacillin-sensitive, coagulase-negative Staphylococci, 60%–70% were not susceptible to penicillin. Some lower susceptibility rates against cefepime were noted at the start of the follow-up period, but this disappeared with time. Vancomycin, teicoplanin, amikacin, meropenem, cefepime, and amoxycillin-clavulanic acid remained equally effective against oxacillin-sensitive, coagulase-negative Staphylococci.
The oxacillin-resistant coagulase-negative Staphylococci showed full susceptibility to vancomycin. Approximately 80% of the oxacillin-resistant, coagulase-negative Staphylococci were susceptible to amikacin. A significant increase in susceptibility to cotrimoxazole was observed (P = 0.0073). No difference was observed between blood and other cultures for these bacteria.
E. coli had a low susceptibility rate to ampicillin of approximately 40%. Additionally, piperacillin showed a very low activity. A major problem existed for prophylactic antibiotics. A low susceptibility rate was noted for ciprofloxacin (60%) and for cotrimoxazole (approximately 50%). Susceptibility of E. coli to cefepime remained higher than to ceftazidime and amoxycillin-clavulanate. Up to 44% of the E. coli isolates in blood cultures showed susceptibility to ciprofloxacin (data not shown). The most active antibiotics against E. coli were meropenem and amikacin.
Cefepime, amikacin, and meropenem remained very active against noninducible Enterobacteriaceae. The susceptibility rate to ceftazidime improved after stopping its use in the empiric antibiotic regimen, rising from 75% to 100%. There was a statistically significant increase in susceptibility for piperacillin (P = 0.0390), a statistically significant decrease in susceptibility to cefepime (P = 0.0298), and a borderline statistically significant decrease in susceptibility to ciprofloxacin (P = 0.0943).
For inducible Enterobacteriaceae, cefepime remained as active as before (after several years of usage). Together with meropenem, cefepime remained the most active antibiotic in this category of bacteria. The susceptibility rate to ceftazidime improved from 30% to 65% (P = 0.0136), but remained lower than that of cefepime. Susceptibility rates to amikacin tended to improve, as did those to meropenem. The susceptibility of inducible Enterobacteriaceae for ciprofloxacin gradually deteriorated (66.7%). Furthermore, cotrimoxazole showed a low susceptibility to these bacteria (42.6%). However, they were still more efficient than ceftazidime.
P. aeruginosa had a low susceptibility rate to cefepime at the start of the follow-up period. Although this low susceptibility rate increased over time, it remained equal to the susceptibility rate of ceftazidime. A decreasing susceptibility rate to meropenem was also apparent.
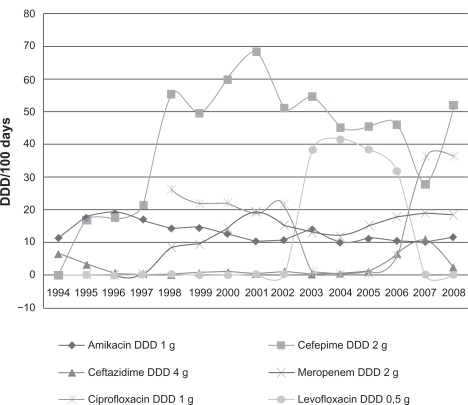
Antibiotic usage
The analysis of antibiotic consumption according to DDDs showed a stable consumption of amikacin, quinolones, and meropenem during the years of the study (Figure 5). There was a statistically significant increase in use of cefepime (P = 0.0001) and a significantly decrease in use of ceftazidime (P = 0.0393).
Figure 5.
Antibiotic consumption in the hematology ward (1994–2008).
Abbrevation: DDD, defined daily doses.
Discussion
After several years of using the new empiric antibiotic therapy, a continuing good profile of decreasing incidence of inducible Enterobacteriaceae was seen in our study. After the change in antibiotic policy, further increasing susceptibility for ceftazidime was observed. This is a long-term confirmation of our own observations and those of others.5,7 Moreover, during the 15 years of follow-up, no significant decrease was seen in susceptibility of these bacteria against the empiric antibiotics used, particularly cefepime and amikacin. To our knowledge, this is the first report showing that long-term use of cefepime and amikacin has no significant effect on susceptibility rates of inducible Enterobacteriaceae in neutropenic patients.
An international, well documented balance of predominantly occurring pathogens has existed since the 1980s. In this balance, Gram-positive organisms continue to be the more common organisms in most hospitals.8,9 In our survey, the relative proportion of Gram-negative and Gram-positive isolates remained stable. Consistent with other international studies, a slight preponderance of Gram-positive organisms was observed in our survey.
However, this is in contrast with data obtained in some other centers, where an increasing number of Gram-negative isolates has been observed in recent years, nearly equaling the Gram-positive isolates.10 This was confirmed in a recent French study in patients with febrile neutropenia.11 In this study, 51% of the documented microbiologic infections were Gram-negative between 2001 and 2003.11 The reason for this change is probably a result of complex interactions involving associated clinical conditions, chemotherapeutic regimens, antibiotic use, prolonged hospitalization, and the quality of care provided.12
Regarding the Gram-positive isolates in our survey, the most common ones were the coagulase-negative Staphylococci, with a significant shift from oxacillin-sensitive to oxacillin-resistant organisms over the years, which occurred in blood cultures as well. In recent years, coagulase-negative Staphylococci has been recognized as one of the leading causes of bacteremia in patients suffering from neutropenia.12–14 A possible explanation for this observation could be the regional use of prophylactic quinolones. The prevalence of MRSA differs markedly among the European countries (from <1% to 80%), with the lowest rate in the Scandinavian countries.14 In our study, only 1.3% of S. aureus isolates were methicillin-resistant, which can be considered favorable. The exact reason for this is unclear. Possible explanations could be the strict infection control measures applied to infected patients, frequent screening cultures, and rigorous hand hygiene procedures. None of the Gram-positive bacterial isolates in our survey was vancomycin-resistant, probably reflecting a low consumption of glycopeptides.
Several reports of reduced ciprofloxacin susceptibility in E. coli and other Gram-negative bacteria have been published.7,14,15 This is confirmed by our results, which show a consistently reduced susceptibility of Gram-negative bacteria to ciprofloxacin. This was noted for E. coli, inducible Enter-obacteriaceae, and P. aeruginosa (approximately 40%, 30%, and 20% reduced susceptibility, respectively). In our survey, only 34% of isolates in blood cultures were susceptible to ciprofloxacin, which is of major concern. One of the possible explanations for this is the prior use of quinolones, which have been identified as a risk factor for infection with resistant Gram-negative bacteria.15
P. aeruginosa was present only in 3.8% of our isolates, which is in accordance with other studies.7,9,13 The susceptibility to the broad-spectrum antibiotics (ceftazidime and cefepime) remained stable during the observation period. However, reduced susceptibility of P. aeruginosa to ceftazidime, cefepime, and meropenem of 11%–33% is a cause for concern. This susceptibility pattern highlights the necessity for continuous surveillance, and can be a reason to start empiric treatment for neutropenic fever with combination therapy targeting Gram-negative infections.16
Our study has some limitations. In particular, it only gives information about local susceptibility patterns. A future investigation could include combined analysis of blood, clinical, and surveillance cultures. However, given that only 15%–25% of patients with neutropenia develop bloodstream infections, we argue that clinicians should consider the entire spectrum of bacterial infections when deciding about empiric antibiotic therapy.
Conclusion
A good susceptibility pattern of inducible Enterobacteriaceae was maintained over 15 years following the introduction of amikacin and cefepime. Although the number of Gram-negative bacteria was rising, the Gram-positive bacteria remained the main pathogens. Coagulase-negative Staphylococci were the most frequently isolated pathogens, with an increase in oxacillin-resistance. No true vancomycin resistance was noted. Another finding was the low susceptibility of ciprofloxacin against E. coli, inducible Enterobacteriaceae, and P. aeruginosa. The decreased susceptibility of P. aeruginosa to most antibiotics is a major concern. Continuous monitoring of susceptibility rates is thus warranted, whereby careful selection of antibiotics could reduce bacterial resistance.
Acknowledgments
The statistical analysis was supported with a grant from the quality fund at Jessa Hospital, Hasselt, Belgium. The authors would like to thank Dr I Gyssens, infectious diseases, for revision of manuscript, and Dr R Saenen and Dr L Luyten, medical registration, for providing patient data.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Viscoli C, Varnier O, Machetti M. Infections in patients with febrile neutropenia: Epidemiology, microbiology, and risk stratification. Clin Infect Dis. 2005;40:S240–S245. doi: 10.1086/427329. [DOI] [PubMed] [Google Scholar]
- 2.Inai K, Iwasaki H, Noriki S, et al. Frequent detection of multidrug-resistant pneumonia-causing bacteria in the pneumonia lung tissues of patients with hematological malignancies. Int J Hematol. 2007;86:225–232. doi: 10.1532/IJH97.07233. [DOI] [PubMed] [Google Scholar]
- 3.Sharma A, Lokeshwar N. Febrile neutropenia in haematological malignancies. J Postgrad Med. 2005;51:42–48. [PubMed] [Google Scholar]
- 4.Jarvis WR, Cookson ST, Robles MB. Prevention of nosocomial bloodstream infections: A national and international priority. Infect Control Hosp Epidemiol. 1996;17:272–275. doi: 10.1086/647294. [DOI] [PubMed] [Google Scholar]
- 5.Mebis J, Goossens H, Bruyneel P, et al. Decreasing antibiotic resistance of Enterobacteriaceae by introducing a new antibiotic combination therapy for neutropenic fever patients. Leukemia. 1998;12:1627–1629. doi: 10.1038/sj.leu.2401158. [DOI] [PubMed] [Google Scholar]
- 6.Wayne PA. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. 16th international supplement document M100-S16 CLSI/NCCLS M100-S15. [Google Scholar]
- 7.Lockhart SR, Abramson MA, Beekmann SE, et al. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007;45:3352–3359. doi: 10.1128/JCM.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanamaru A, Tatsumi Y. Microbiological data for patients with febrile neutropenia. Clin Infect Dis. 2004;39:S7–S10. doi: 10.1086/383042. [DOI] [PubMed] [Google Scholar]
- 9.Wisplinghoff H, Seifert H, Wenzel RP, Edmond MB. Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin Infect Dis. 2003;36:1103–1110. doi: 10.1086/374339. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo C, Quaresmini G, Casari S, et al. Recent changes in bacterial epidemiology and the emergence of fluoroquinolone-resistant Escherichia coli among patients with haematological malignancies: Results of a prospective study on 823 patients at a single institution. J Antimicrob Chemother. 2008;61:721–728. doi: 10.1093/jac/dkm514. [DOI] [PubMed] [Google Scholar]
- 11.Dutronc H, Billhot M, Dupon M, et al. Management of 315 neutropenic febrile episodes in a cancer center. Med Mal Infect. 2009;39:388–393. doi: 10.1016/j.medmal.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Elouennass M, Foissaud V, Trueba F, et al. A 7-year survey of strains identified in blood cultures in a clinical hematology unit. Med Mal Infect. 2004;34:62–69. doi: 10.1016/j.medmal.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Cherif H, Kronvall G, Bjorkholm M, Kalin M. Bacteraemia in hospitalized patients with malignant blood disorders: A retrospective study of causative agents and their resistance profiles during a 14-year period without antibacterial prophylaxis. Hematol J. 2003;4:420–426. doi: 10.1038/sj.thj.6200334. [DOI] [PubMed] [Google Scholar]
- 14.Sigurdardottir K, Digranes A, Harthug S, et al. A multi-centre prospective study of febrile neutropenia in Norway: Microbiological findings and antimicrobial susceptibility. Scand J Infect Dis. 2005;37:455–464. doi: 10.1080/00365540510038497. [DOI] [PubMed] [Google Scholar]
- 15.Paterson DL. “Collateral damage” from cephalosporin or quinolone antibiotic therapy. Clin Infect Dis. 2004;38:S341–S345. doi: 10.1086/382690. [DOI] [PubMed] [Google Scholar]
- 16.Klastersky J. Management of fever in neutropenic patients with different risks of complications. Clin Infect Dis. 2004;39:S32–S37. doi: 10.1086/383050. [DOI] [PubMed] [Google Scholar]