Abstract
We have previously localized the core centromere protein-binding domain of a 10q25.2-derived neocentromere to an 80-kb genomic region. Detailed analysis has indicated that the 80-kb neocentromere (NC) DNA has a similar overall organization to the corresponding region on a normal chromosome 10 (HC) DNA, derived from a genetically unrelated CEPH individual. Here we report sequencing of the HC DNA and its comparison to the NC sequence. Single-base differences were observed at a maximum rate of 4.6 per kb; however, no deletions, insertions, or other structural rearrangements were detected. To investigate whether the observed changes, or subsets of these, might be de novo mutations involved in neocentromerization (i.e., in committing a region of a chromosome to neocentromere formation), the progenitor DNA (PnC) from which the NC DNA descended, was cloned and sequenced. Direct comparison of the PnC and NC sequences revealed 100% identity, suggesting that the differences between NC and HC DNA are single nucleotide polymorphisms (SNPs) and that formation of the 10q25.2 NC did not involve a change in DNA sequence in the core centromere protein-binding NC region. This is the first study in which a cloned NC DNA has been compared directly with its inactive progenitor DNA at the primary sequence level. The results form the basis for future sequence comparison outside the core protein-binding domain, and provide direct support for the involvement of an epigenetic mechanism in neocentromerization.
[The sequences in this paper have been submitted to GenBank under accession nos. AF222855 (not yet available) for HC; AF042484 for NCI; AF222854 (not yet available) for NCII; and AF222856 (not yet available) for PnC.]
The centromere is a critical structure found on all eukaryotic chromosomes. It is the site of kinetochore assembly that allows the faithful pairing and segregation of chromosomes during cell division (Choo 1997a). Although the function of the centromere and the proteins that make up the kinetochore are highly conserved, there is no obvious conservation of centromere DNA sequence between species and even among some chromosomes of a single species (for review, see Choo 1997b). Normal human centromeres are composed of large (1–4 Mb) tandem arrays of a 171-bp α-satellite DNA. The discovery of neocentromeres (NCs) on analphoid marker chromosomes indicates that alphoid DNA is not always required for centromere function (Choo 1997b; Barry et al. 1999). Over half of the human chromosomes have now been shown to contain at least one site at which a NC has formed (Choo 1997b; Depinet et al. 1997).
We have characterized a NC that was identified on a chromosome 10-derived marker designated mardel(10), at a region corresponding to 10q25.2 on the normal chromosome 10 (Voullaire et al. 1993; du Sart et al. 1997). This NC is indistinguishable from normal centromeres in terms of protein association and distribution and is 100% mitotically stable (du Sart et al. 1997; Saffery et al. 2000). Detailed Southern hybridization analyses and fingerprinting comparisons demonstrated that the active NC DNA contained an overall similar organization to the inactive, normal 10q25.2 DNA (HC DNA), thereby suggesting the involvement of an epigenetic mechanism in neocentromerization (Choo 2000). Epigenetic modifications have been proposed to account for kinetochore assembly on noncentromeric sequences in the fission yeast (Steiner and Clarke 1994) and Drosophila (Williams et al. 1998) and implicated in phenomena such as position effect variegation (Wakimoto 1998), imprinting (Surani 1998), X chromosome inactivation (Panning and Jaenisch 1998), and the regulation of gene transcription by inducing higher order chromatin folding (Monk 1990; Laurenson and Rine 1992; Sandell and Zakian 1992; Shaffer et al. 1993). DNA sequence analyses of the 80-kb core centromere-protein binding region of the 10q25 NC DNA revealed the complete absence of alphoid sequences (Barry et al. 1999). The NC DNA was unremarkable in its sequence composition when compared to other human genomic DNA, except perhaps for some clustering of AT-rich islands, one of which (the AT28 region) appears to have special structural features that may have implications for centromere function (Koch 2000).
An alternative model to the epigenetic theory of neocentromere activation involves de novo mutational changes to the DNA that allow the nucleation and formation of a kinetochore complex at a previously non centromeric chromosomal region. Earlier restriction mapping comparison of the active (NC) and inactive (HC) regions has not considered the entire sequence nor allowed small sequence rearrangements or single-base substitutions to be detected (du Sart et al. 1997, Cancilla et al. 1998). These studies therefore do not permit a conclusive distinction between the epigenetic and mutational models of neocentromerization. To overcome these shortcomings, we have sequenced the normal HC DNA and compared it directly with the previously obtained NC DNA. In particular, we have also cloned and sequenced the progenitor allele (PnC DNA) from the patient's father from whom the 10q25.2 NC was derived. Comparison of the three DNA sequences has provided further support for the epigenetic mode of neocentromerization.
RESULTS
Quality of the DNA Sequences
DNA sequences were generated primarily from cloned DNA. The cloned DNA was subcloned further and PCR-amplified to allow complete sequencing. Replication errors and misincorporation of nucleotides during PCR were inevitable and resulted in a small but definite number of sequencing errors. At least five-fold coverage of each of the sequences was achieved during this analysis in an attempt to minimize such errors. It should also be noted that for the long PCR used to prepare template fragments for sequencing, a mixture of Taq and Pwo DNA polymerases was used (see Methods). Pwo DNA polymerase contains proofreading ability that enables the correction of replication errors and therefore should minimize errors in the sequences generated from the PCR products.
In a previous study, we have described the sequence for the NC DNA (Barry et al. 1999) (GenBank accession no. AF042484). This sequence was derived entirely from cloned DNA and was redesignated NCI sequence here to distinguish it from the NCII sequence described below. During the course of the present study, it was necessary (see Methods) to resequence regions of the NC DNA using the cloned DNA as template and/or using PCR templates prepared directly from genomic DNA. This allowed the correction of 126 errors in the NCI sequence, caused presumably by cloning and sequencing artifacts. The resulting sequence was designated NCII (GenBank accession no. AF222854) and was used in the following comparative studies. Similarly, the PnC sequence was derived through a combination of the use of cloned DNA (see below) and genomic DNA (see Methods), and was also expected to be of high quality.
Comparison of the HC DNA Sequence to NC DNA Sequence
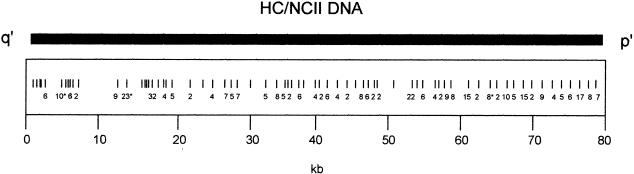
The NC DNA used in this study was obtained directly from the mardel(10) chromosome present in patient (BE) (Cancilla et al. 1998; Barry et al. 1999), whereas the HC DNA originated from an unrelated CEPH individual (du Sart et al. 1997). Previous high-density comparison of HC DNA with NC DNA using RsaI restriction enzyme fingerprinting showed no differences, except within the polymorphic VNTR (Cancilla et al. 1998). In the present study, the HC DNA was sequenced and compared to the NCII DNA by alignment in Sequencher program (Gene Codes Corp.). The HC DNA sequence consists of 80622 nucleotides (GenBank accession no. AF222855) and covers the entire 80202 bp of NCII. Base pair 1 of NCII corresponds to base pair 383 of the HC DNA. When these sequences were compared, no gross deletions, insertions, or other structural rearrangements were detected. A total of 370 single-nucleotide changes were detected with the distribution of change relatively uniform and only a few regions with multiple changes (Fig. 1). These were found to be primarily within regions of high mutability such as poly(A) stretches and the VNTR known as AT28 (Barry et al. 1999). This approximates to 4.6 SNP per kb, which is somewhat higher than the average of 1 per kb, calculated previously from a comparison of DNA from two unrelated individuals (Cooper et al. 1985; Hofker et al. 1986; Kwok et al. 1996). The value of 4.6 is likely to be an overestimation because this has not been corrected for potential cloning/sequencing errors in the HC sequence due to the unavailability of the genomic DNA for this sequence. Given the observed differences, and the difficulty in distinguishing between normal polymorphic variations and phenotype-related mutational changes, it was not possible to conclude whether the changes between the HC and NCII sequences were directly relevant to neocentromerization. Therefore we undertook the cloning and sequence analysis of the progenitor allele from which the NC DNA has directly descended.
Figure 1.
Differences between the ∼80-kb HC and NCII sequences. Vertical bars represent the positions of single nucleotide differences between the two sequences. The numbers below represent the number of single nucleotide differences (where there is more than one difference) over a 1-kb region. Numbers with asterisks represent clusters of differences.
Identification and Cloning of the Progenitor NC Allele
The progenitor allele (designated PnC) refers to the corresponding region of the NC DNA on the pre-rearranged and morphologically normal chromosome 10. Its source has been traced previously to the patient BE's father (CE) by multiloci STS polymorphism analyses at both the NC core region and adjacent domains (du Sart et al. 1997; Cancilla et al. 1998; Barry et al. 1999). These analyses, which identified a VNTR (AT28) polymorphism within the NC DNA, also provided a means of differentiating between the two normal chromosome 10s present in the father (Barry et al. 1999).
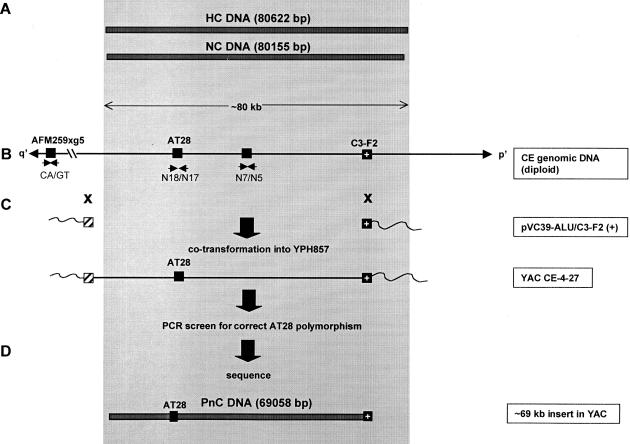
The PnC DNA was cloned from the total genomic DNA of CE using a previously described transformation-associated recombination (TAR) approach (Cancilla et al. 1998). This radial TAR method relies on the high recombination efficiency of yeast and an ARS (autonomously replicating sequence)-negative vector containing a sequence homologous and flanking the DNA of choice. Propagation of the circular YACs is possible provided that the genomic fragment being cloned contains sequences that can act as yeast ARS sequences. These ARS-like sequences occur every 20–40 kb in the human genome (Larionov et al. 1996). Cloning (Fig. 2) was performed with the vector pVC39-Alu/C3-F2(+), previously used successfully to clone the NC DNA (Cancilla et al. 1998).
Figure 2.
TAR cloning and sequencing of PnC DNA. The shaded area represents the region corresponding to the ∼80-kb 10q25.5 NC DNA (du Sart et al. 1997). (A) Sequenced regions of the HC DNA (derived from a CEPH library YAC clone) and NC DNA [derived from the mardel(10) neocentromere]. Total number of nucleotides sequenced is shown in brackets. (B) Structure of the HC/NC region and flanking DNA. Solid boxes represent STSs used in the identification and cloning of the DNA. AFM259xg5 is a (CA)n microsatellite located ∼150 kb (represented by the broken line) from the core region (Cancilla et al. 1998). AT28 (Barry et al. 1999) is a polymorphic VNTR used to identify the progenitor allele. C3-F2 is a 1.4-kb EcoRI fragment that served as the specific TAR “hook”(Cancilla et al. 1998). Small arrows indicate oligonucleotides used in PCR of the STSs. p′ and q′ refer to the short and long arms of mardel(10), respectively. (C) Radial TAR strategy using the vector pVC39-Alu/C3-F2(+) for the direct cloning of the progenitor DNA from the total genomic DNA of CE. The hatched box indicates the position of the Alu consensus sequence hook. Crosses denote the sites of recombination between the TAR vector pVC39-Alu/C3-F2( +) and CE genomic DNA at the C3-F2 and Alu hooks during cloning. The resulting circular YAC, CE-4–27, was shown by the AT28 polymorphism (see Fig. 3) to contain the PnC DNA from the progenitor chromosome 10. (D) The ∼69-kb sequenced portion of PnC DNA, represented by the bar.
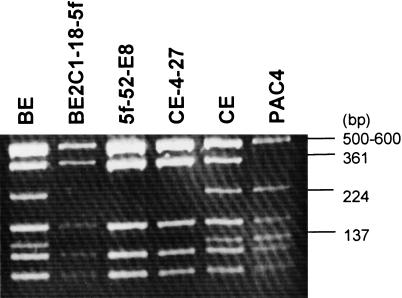
Initial characterization of clones was achieved using a PCR screening strategy (Fig. 2; Methods). One positive His + clone, designated CE-4–27, was further characterized using the polymorphic AT28 region to determine from which of the chromosome 10s of CE the clone was derived. For comparison, DNA from a number of different sources was included in the analysis (Fig. 3). When digested with RsaI, the nonprogenitor chromosome 10 allele in CE could be identified by the presence of two fragments of 224 and 137 bp, whereas the allele from the progenitor chromosome 10 produced a 361-bp band (due to the absence of a RsaI restriction site) (Barry et al. 1999). All three bands could be seen in PCR products derived from the diploid cell lines BE and CE due to the presence of both alleles. The presence of the 361-bp fragment but not the 224- and 137-bp fragments in CE-4–27, BE2C1–18–5f, and 5f-52-E8 confirmed that the CE-4–27 clone carried the progenitor PnC DNA.
Figure 3.
Identification and confirmation of the PnC DNA clone using the AT28 polymorphism. AT28 was amplified by PCR as described in Methods. PCR products were purified and digested with RsaI and electrophoresed on 3% (wt/vol) agarose. (BE) Cell line from mardel(10) patient; (BE2C1–18–5f) a somatic cell hybrid line containing mardel(10) chromosome but not the normal chromosome 10; (5f-52-E8) a BAC clone containing the NC region derived from the BE2C1–18–5f somatic cell hybrid; (CE-4–27) a circular YAC containing the PnC DNA; (CE) a cell line from patient BE's father; (PAC4) a PAC clone containing the HC DNA derived from the normal chromosome 10 of a CEPH individual (du Sart et al. 1997; Cancilla et al. 1998). Note the identical fingerprints of CE-4–27, BE2C1–18–5f and 5f-52-E8.
The CE-4–27 clone has an insert of ∼69 kb, spanning the entire q′ side but missing ∼11 kb at the p′ end of the 80-kb NC DNA (Fig. 2). Previous FISH analysis indicated that the core centromere protein-binding domain of the 80-kb NC DNA resided preferentially towards the q′ end of this DNA (du Sart et al. 1997). Furthermore, the ∼69 kb cloned q′ region of CE-4–27 contains the AT28 repeat that was shown previously to bind a centromere-enriched protein poly(ADP-ribose) polymerase (Earle et al. 2000) and share common structural features with the unrelated primary sequences of both the human α-satellite DNA and the centromere DNA of the budding yeast Saccharomyces cerevisiae (Koch 2000). Based on these observations, we inferred that the critical region of the 10q25.2 NC resided within the ∼69-kb insert of the CE-4–27 clone. This provided the justification for proceeding with the following sequence analysis without further attempting to isolate the missing 11-kb region at the p′ end of the PnC DNA.
Generation of the PnC DNA Sequence
The PnC DNA sequence was initially generated from the CE-4–27 template using PCR primers employed in the sequencing of the HC and NC DNA, in conjunction with specific primers designed for PnC sequencing. Regions of ambiguous sequences were resequenced using the CE genomic DNA as a template (see Methods). The completed PnC DNA sequence consists of 69058 bp (GenBank accession no. AF222856) where nucleotides 1 and 69058 correspond to the same nucleotides from the NC DNA at the q′ and p′ ends of the mardel(10) chromosome, respectively. Direct comparison of the final sequences for the PnC and the NCII DNA showed that they were 100% identical in their primary nucleotide organization. Furthermore, when the HC sequence was used as the normal allele to compare with the NCII sequence within the 11-kb p′ segment that was missing from the PnC DNA, the nature and distribution of base changes were not noticeably different from those of the remaining 69 kb q′ region (Fig. 1), suggesting that such changes were likely to be due to cloning and/or sequencing errors, similar to those shown for the rest of the sequenced region.
DISCUSSION
Previous comparisons using restriction mapping indicated a similar gross sequence organization between the NC DNA and its normal counterpart; however, the sensitivity of these analyses were limited (du Sart et al. 1997, Cancilla et al. 1998). In this study two different normal alleles corresponding to the 10q25.2 NC region were sequenced to allow unequivocal comparison with the NC DNA at the primary nucleotide level. The first allele is the ∼80-kb HC DNA derived from a CEPH YAC library and thus represents a genetically unrelated source to the mardel(10) patient BE. Alignment of this sequence with the NCII sequence revealed approximately 4.6 nucleotide differences per 1000 bp. This value is higher than the 1/1000 average rate of polymorphism previously calculated between two unrelated individuals (Cooper et al. 1985; Hofker et al. 1986; Kwok et al. 1996). Notwithstanding the fact that the average genomic rate does not take into account differences in regional mutability within the human genome that could explain the higher value seen in the HC/NCII region, the possibility that the observed differences might be significant to NC formation was raised. Sequencing of a second allele derived specifically from the progenitor chromosome of mardel(10) was required and undertaken to resolve this possibility.
Although the rate of de novo mutations in the human germ line has never been measured accurately, the rate of mutation is expected to be equal to the rate of substitution over evolutionary time. Human–ape comparisons predict a base substitution/mutation rate of ∼1/50,000,000 per base per gamete, which for a sequence of ∼80 kb, gives an average mutation probability of 0.0016 (i.e., 80,000 of 50,000,000) for a single random mutation per gamete (A. Jeffreys, pers. comm.). However, this calculation also does not take into consideration regional differences in mutation susceptibility within the genome (e.g., higher in regions containing CpG dinucleotides and tandem repeats) (Cooper et al. 1985; Jeffreys et al. 1985). Because of this relatively low predicted rate of germ-line mutation, detection of substantial changes between the progenitor PnC and its descendant NCII DNA could potentially signify an underlying triggering mechanism for neocentromerization. However, direct comparison of the sequences between these two DNA revealed total identity, suggesting that the transition from an inactive to an active state of the mardel(10) NC has not been accompanied by any mutational change in the primary sequence of this core centromere protein-binding region. The differences detected between the NCII and HC sequences are therefore likely to be due to the random accumulation of SNPs.
Epigenetic mechanisms have been proposed to explain the formation of NCs from non-centromeric genomic DNA (Choo 1997b, 2000; Karpen and Allshire 1997; Murphy and Karpen 1998; Wiens and Sorger 1998). These have included mechanisms such as marking a DNA for centromere assembly through the binding of a centromere-specific nucleosomal protein (e.g., the histone H3-like homolog CENP-A) (Shelby et al. 1997), or via chemical modification [e.g., methylation, deacetylation, phosphorylation, poly(ADP)-ribosylation, ubiquitination] (Choo 2000). A model based on the synchronization of centromere DNA replication timing and the expression of a centromere-marking protein such as CENP-A has also been suggested (Csink and Henikoff 1998). A critical criterion upon which the importance of these epigenetic mechanisms is based is the assumption that neocentromerization is not in any way compromised by mutational changes at the primary nucleotide sequence in the DNA undergoing the transformation. For human NCs, this assumption has been based on three observations to date. First, cytogenetic banding has indicated the absence of detectable morphological changes at the chromosomal subregions where NCs have formed (Choo 1997b). Second, fluorescence in situ hybridization (Choo 1997b) and, in one particular case, direct sequencing (Barry et al. 1999), have failed to detect α satellite DNA signals in NCs. Third, restriction map comparison of a cloned NC DNA with its corresponding normal DNA has indicated no major structural changes (du Sart et al. 1997; Cancilla et al. 1998). However, the techniques used in these observations do not have sufficient sensitivity to detect small nucleotide sequence alterations. The present study represents the first comparison of a NC DNA with its corresponding normal DNA at the primary sequence level. Significantly, the comparison has employed the progenitor DNA from which the NC has descended. The result demonstrates no nucleotide change between the progenitor and NC DNA, thus providing direct evidence in support of an unknown epigenetic modification in the neocentromerization process. Although we cannot rule out the possibility that changes in DNA sequences outside the core region may be important, the present study forms the basis for future work extending into these sequences.
METHODS
Sequencing
A series of restriction fragments from overlapping cosmids containing the HC DNA (du Sart et al. 1997) were subcloned into pBluescript-KS(+) and end-sequenced with vector-specific primers or sequenced directly with internal primers. Long PCR products were generated using the Long Template PCR kit (Boehringer Mannheim) from the TAR-cloned PnC DNA using primers designed from HC or NC DNA sequences. Following removal of residual oligonucleotides and buffers from the PCR reaction with the High Pure PCR product purification kit (Boehringer Mannheim), the products were sequenced directly using the appropriate primers as described previously (Barry et al. 1999). Automated sequencing was performed using the ABI PRISM cycle sequencing protocol and electrophoresed on an ABI377 system according to the manufacturer's instructions. All sequences were edited and assembled into contigs using Sequencher software (Gene Codes Corporation).
During the initial comparison between the PnC and NC sequences, some differences were detected. To determine whether the differences were sequencing or cloning artifacts, resequencing of the appropriate regions was performed using the cloned DNA and/or PCR amplified templates prepared directly from genomic DNA. For the NC sequence, genomic DNA was prepared from a mardel(10)-containing somatic hybrid cell line (BE2C1–18–5f) (du Sart et al. 1997). For the PnC DNA, genomic DNA was prepared from the mardel(10) patient's father CE, from which the NC was derived (du Sart et al. 1997; Cancilla et al. 1998; Barry et al. 1999). Because the CE genomic DNA contained two copies of chromosome 10, use of this DNA for the generation of PCR templates could potentially be complicated by polymorphic differences between two alleles. Fortunately, among the relatively small number of differences for which genomic DNA confirmation was required, only a single clear nucleotide peak was observed in every case. Where required, the NC DNA was resequenced using the same method as described previously (Barry et al. 1999). Genomic DNA sequences were generated by amplifying the appropriate regions from total genomic DNA by PCR and sequenced as described above. Automated sequencing was performed using the ABI Prism cycle sequencing protocol and electrophoresed on an ABI377 system according to the manufacturer's instructions. All sequences were edited and assembled into contigs using Sequencher software (Gene Codes Corporation).
TAR Cloning of PnC DNA
PnC DNA was subcloned by TAR in yeast with the pVC39-Alu/C3-F2(+) vector using a previously described method (Cancilla et al. 1998). Approximately 1μg of linearized TAR vector was combined with at least 5μg of high-molecular-weight genomic DNA from the CE cell line and cotransformed into S. cerevisiae YPH857 spheroplasts (∼109 cells). More than 500 His+ colonies were generated and screened by PCR using 2 μl of lysed yeast suspension, 500 ng of each of the primers N7 (5′-TCTGCATAGTGGCTGAAGGC-3′) and N5 (5′-TACTTCGTATCCCATAGGCT-3′), 0.2mm dNTPs, 1× PCR reaction buffer containing 1.5 mm MgCl2 (Perkin Elmer) and 1U AmpliTaq DNA Polymerase containing Gene Amp (Perkin Elmer) in a total reaction mix of 20 μl placed in a thermal cycler for 26 cycles of 94°C for 30 sec, 45°C for 30 sec, and 72°C for 1 min. The positive clone was characterized by PCR using the Long Template PCR kit (Boehringer Mannheim) according to the manufacturer's instructions with 5 μl lysed yeast suspension and the primers N17 (5′- TGCAGGGAGAGAAAGGAACT-3′) and N18 (5′- GAATCGTATGTGCTGCTTGC -3′) in a 50 μl reaction mix, cycling with 2-min extensions, with increments of 20 sec per cycle after the first 10 cycles, and an annealing temperature of 58°C. High-molecular-weight DNA was then prepared from the positive clone (Cancilla et al. 1998), and 1μl used in Long PCR for sequencing (see above).
Acknowledgments
This work was supported by funds from AMRAD, AusIndustry and NHMRC to KHAC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL CHOO@CRYPTIC.RCH.UNIMELB.EDU.AU; FAX 61-3-9348 1391.
REFERENCES
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KHA. Sequence analysis of a 80 kb human neocentromere. Hum Mol Genet. 1999;8:217–227. doi: 10.1093/hmg/8.2.217. [DOI] [PubMed] [Google Scholar]
- Cancilla MR, Tainton KM, Barry AE, Larionov V, Kouprina N, Resnick M, duSart D, Choo KHA. Direct cloning of human 10q25 neocentromere DNA using transformation associated recombination (TAR) in yeast. Genomics. 1998;47:399–404. doi: 10.1006/geno.1997.5129. [DOI] [PubMed] [Google Scholar]
- Choo KHA. The Centromere. Oxford, New York and Tokyo: Oxford University Press; 1997a. [Google Scholar]
- ————— Centromere DNA dynamics: latent centromeres and neocentromere formation. Am J Hum Genet. 1997b;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Centromerization. Trends Cell. Biol. (in press). [DOI] [PubMed]
- Cooper D, Smith B, Cooke H, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Dyke DLV, Willard HF, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KHA. A functional neocentromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earle E, Saxena A, MacDonald A, Hudson DF, Shaffer LG, Saffery R, Cancilla MR, Cutts SM, Howman E, Choo KHA. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum Mol Genet. 2000;9:187–194. doi: 10.1093/hmg/9.2.187. [DOI] [PubMed] [Google Scholar]
- Hofker M, Skraastad M, Bergen A, Wapenaar M, Bakker E, Millington-Ward A, van Ommen G, Pearson P. The X-chromosome shows less genetic variation at restriction sites than the autosomes. Am J Hum Genet. 1986;39:438–451. [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Hypervariable minisatellite regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Koch J. Neocentromeres and alpha satellite: A proposed structural code for functional human centromere DNA. Hum Mol Genet. 2000;9:149–154. doi: 10.1093/hmg/9.2.149. [DOI] [PubMed] [Google Scholar]
- Kwok P-Y, Deng Q, Zakeri H, Taylor S, Nickerson D. Increasing the information content of STS-based genome maps: Identifying polymorphisms in mapped STSs. Genomics. 1996;31:123–126. doi: 10.1006/geno.1996.0019. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci. 1996;93:13925–13930. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing and heritable transcriptional states. Microbiol Rev. 1992;56:543–60. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M. Variation in epigenetic inheritance. Trends Genet. 1990;6:110–114. doi: 10.1016/0168-9525(90)90124-o. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell. 1998;93:317–20. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KHA. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- Sandell L, Zakian V. Telomeric position effect in yeast. Trends Cell Biol. 1992;2:10–14. doi: 10.1016/0962-8924(92)90138-d. [DOI] [PubMed] [Google Scholar]
- Shaffer C, Wallrath L, Elgin S. Regulating genes by packaging domains: Bits of heterochromatin in euchromatin? Trends Genet. 1993;9:35–37. doi: 10.1016/0168-9525(93)90171-D. [DOI] [PubMed] [Google Scholar]
- Shelby R, Vafa O, Sullivan K. Assembly of CENP-A into chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner NC, Clarke, L L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Surani A. Imprinting and the initiation of gene silencing in the germline. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KHA. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Weins GR, Sorger PK. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell. 1998;93:313–316. doi: 10.1016/s0092-8674(00)81157-5. [DOI] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg M L, Karpen GH. Neocentromere activity of structurally acentric minichromosomes in Drosophila. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]