Abstract
Computational methods for automated genome annotation are critical to our community's ability to make full use of the large volume of genomic sequence being generated and released. To explore the accuracy of these automated feature prediction tools in the genomes of higher organisms, we evaluated their performance on a large, well-characterized sequence contig from the Adh region of Drosophila melanogaster. This experiment, known as the Genome Annotation Assessment Project (GASP), was launched in May 1999. Twelve groups, applying state-of-the-art tools, contributed predictions for features including gene structure, protein homologies, promoter sites, and repeat elements. We evaluated these predictions using two standards, one based on previously unreleased high-quality full-length cDNA sequences and a second based on the set of annotations generated as part of an in-depth study of the region by a group of Drosophila experts. Although these standard sets only approximate the unknown distribution of features in this region, we believe that when taken in context the results of an evaluation based on them are meaningful. The results were presented as a tutorial at the conference on Intelligent Systems in Molecular Biology (ISMB-99) in August 1999. Over 95% of the coding nucleotides in the region were correctly identified by the majority of the gene finders, and the correct intron/exon structures were predicted for >40% of the genes. Homology-based annotation techniques recognized and associated functions with almost half of the genes in the region; the remainder were only identified by the ab initio techniques. This experiment also presents the first assessment of promoter prediction techniques for a significant number of genes in a large contiguous region. We discovered that the promoter predictors' high false-positive rates make their predictions difficult to use. Integrating gene finding and cDNA/EST alignments with promoter predictions decreases the number of false-positive classifications but discovers less than one-third of the promoters in the region. We believe that by establishing standards for evaluating genomic annotations and by assessing the performance of existing automated genome annotation tools, this experiment establishes a baseline that contributes to the value of ongoing large-scale annotation projects and should guide further research in genome informatics.
Genome annotation is a rapidly evolving field in genomics made possible by the large-scale generation of genomic sequences and driven predominantly by computational tools. The goal of the annotation process is to assign as much information as possible to the raw sequence of complete genomes with an emphasis on the location and structure of the genes. This can be accomplished by ab initio gene finding, by identifying homologies to known genes from other organisms, by the alignment of full-length or partial mRNA sequences to the genomic DNA, or through combinations of such methods. Related techniques can also be used to identify other features, such as the location of regulatory elements or repetitive sequence elements. The ultimate goal of genome annotation, the functional classification of all the identified genes, currently depends on discovering homologies to genes with known functions.
We are interested in an objective assessment of the state of the art in automated tools and techniques for annotating complete genomes. The Genome Annotation Assessment Project (GASP) was organized to formulate guidelines and accuracy standards for evaluating computational tools and to encourage the development of new models and the improvement of existing approaches through a careful assessment and comparison of the predictions made by current state-of-the-art programs.
The GASP experiment, the first of its kind, was similar in many ways to the CASP (Critical Assessment of techniques for protein Structure Prediction) contests for protein structure prediction (Dunbrack et al. 1997; Levitt 1997; Moult et al. 1997, 1999; Sippl et al. 1999; Zemla et al. 1999), described at http://predictioncenter.llnl.gov. However, unlike the CASP contest, GASP was promoted as a collaboration to evaluate various techniques for genome annotation.
The GASP experiment consisted of the following stages: (1) Training data for the Adh region, including 2.9 Mb of Drosophila melanogaster genomic sequence, was collected by the organizers and provided to the participants; (2) a set of standards was developed to evaluate submissions while the participating groups produced and submitted their annotations for the region; and (3) the participating groups' predictions were compared with the standards, a team of independent assessors evaluated the results of the comparison, and the results were presented as a tutorial at ISMB-99(Reese et al. 1999).
Participants were given the finished sequence for the Adh region and some related training data, but they did not have access to the full-length cDNA sequences that were sequenced for the paper by Ashburner et al. (1999b) that describes the Adh region in depth. The experiment was widely announced and open to any participants. Submitters were allowed to use any available technologies and were encouraged to disclose their methods. Because we were fortunate to attract a large group of participants who provided a wide variety of annotations, we believe that our evaluation addresses the state of art in genome annotation.
Twelve groups participated in GASP, submitting annotations in one or more of six categories: ab initio gene finding, promoter recognition, EST/cDNA alignment, protein similarity, repetitive sequence identification, and gene function. Table 1 lists each participating group, the names of the programs or systems it used, and which of the six classes of annotations it submitted (see enclosed poster in this issue for a graphic overview of all the groups' results). Additional papers in this issue are written by the participants themselves and describe their methods and results in detail.
Table 1.
Participating Groups and Associated Annotation Categories
| Program name | Gene finding | Promoter recognition | EST/c DNA alignment | Protein similarity | Repeat | Gene function | |
|---|---|---|---|---|---|---|---|
| Mural et al. Oakridge, US | GRAIL | X | X | X | |||
| Parra et al. Barcelona, ES | GeneID | X | |||||
| Krogh Copenhagen, DK | HMMGene | X | |||||
| Henikoff et al. Seattle, US | BLOCKS | X | X | ||||
| Solovyev et al. Sanger, UK | FGenes | X | |||||
| Gaasterland et al. Rockefeller, US | MAGPIE | X | X | X | X | X | |
| Benson et al. Mount Sinai, US | TRF | X | |||||
| Werner et al. Munich, GER | CoreInspector | X | |||||
| Ohler et al. Nuremberg, GER | MCPromoter | X | |||||
| Birney Sanger, UK | GeneWise | X | X | ||||
| Reese et al. Berkeley/Santa Cruz, US | Genie | X | X |
It should be noted that the lack of a standard that is absolutely correct makes evaluating predictions problematic. The expert annotations described by the Drosophila experts (Ashburner et al. 1999b) are our best available resource, but their accuracy will certainly improve as more data becomes available. At best, the data we had in hand is representative of the true situation, and our conclusions would be unchanged by using a more complete data set. At worst, there is a bias in the available data that makes our conclusions significantly misleading. We believe that the data is not unreasonable and that conclusions based on it are correct enough to be valuable as the basis for discussion and future development. We do not believe that the values for the various statistics introduced below are precisely what they would be using the extra information, and we emphasize that they should always be considered in the context of this particular annotated data set [for a further detailed discussion of evaluating these predictions, see Birney and Durbin (2000)].
In the next section we describe the target genomic sequence and the auxiliary data, including a critical discussion of our standard sets. Methods gives a short description of existing annotation methods that complements other papers in this issue, including a review article of existing gene-finding methods by Stormo (2000) and papers describing the methods used by the individual participants. Results assesses the individual annotation methods and the Conclusions discusses what the experiment revealed about issues involved in annotating complete genomes. An article by Ashburner (2000) provides a biological perspective on the experiment.
Data: The Benchmark Sequence: The Adh Region in D. melanogaster
The selection of a genomic target region for assessing the accuracy of computational genome annotation methods was a difficult task for several reasons: The genomic region had to be large enough, the organism had to be well studied, and enough auxiliary data had to be available to have a good experimentally verified “correct answer,” but the data should be anonymous so that a blind test would be possible. The Adh region of the D. melanogaster genome met these criteria. D. melanogaster is one of the most important model organisms, and although the Adh region had been extensively studied, the best gene annotations and cDNAs for the region were not published until after the conclusion of the GASP experiment. The 2.9 Mb Adh contig was large enough to be challenging, contained genes with a variety of sizes and structures, and included regions of high and low gene density. It was not a completely blind test, however, because several cDNA and genomic sequences for known genes in the region were available prior to the experiment.
Genomic DNA Sequence
The contiguous genomic sequence of the Adh region in the D. melanogaster genome spans nearly 3 Mb and has been sequenced from a series of overlapping P1 and BAC clones as a part of the Berkeley Drosophila Genome Project (BDGP; Rubin et al. 1999) and the European Drosophila Genome Project (EDGP; Ashburner et al. 1999c). This sequence is believed to be of very high quality with an estimated error rate of <1 in 10,000 bases, based on PHRAP quality scores. A detailed analysis of this region can be accessed through the BDGP Web site (http://www.fruitfly.org/publications/Adh.html) as well as in Ashburner et al. (1999b).
Curated Training Sequences
We provided several D. melanogaster-specific data sets to the GASP participants. This enabled participants to tune their tools for Drosophila and facilitated a comparison of the various approaches that was unbiased by organism-specific factors. The following curated sequence sets, extracted from Flybase and EMBL (provided by the EDGP at Cambridge and provided by the BDGP, were made available and can be found at http://www.fruitfly.org/GASP/data/data.html): (1) A set of complete coding sequences (start to stop codon), excluding transposable elements, pseudogenes, noncoding RNAs, and mitochondrial and viral sequences (2122 entries); (2) nonredundant set of repetitive sequences, not including transposable elements (96 entries); (3) transposon sequences, containing only the longest sequence of each transposon family and excluding defective transposable elements (44 entries); (4) genomic DNA data from 275 multi- and 141 single-exon nonredundant genes together with their start and stop codons and splice sites, taken from GenBank version 109; (5) a set of 256 unrelated promoter regions, taken from the Eukaryotic Promoter Database (EPD; Cavin Périer et al. 1999, 2000) and a collection made by Arkhipova (1995); and (6) an uncurated set of cDNA and EST sequences from work in progress at the BDGP. Five of the 12 participating groups reported making use of these data sets.
Resources for Assessing Predictions: The “Correct” Answer
In a comparative study, the gold standard used to evaluate solutions is the most important factor in determining the usefulness of the study's results. For the results to be meaningful, the standard must be appropriate and correct in the eyes of the study's audience. Because our goal was to evaluate tools that predict genes and gene structure in complex eukaryotic organisms, we drew our standard from a complex eukaryotic model organism, choosing to work with a 2.9-Mb sequence contig from the Adh region of D. melanogaster. Comparing predicted annotations in such a region is only consequential if the standard is believed to be correct, if that correctness has been established by techniques that are independent of the approaches being studied, and if the predictors had no prior knowledge of the standard. Ideally, it would contain the correct structure of all the genes in the region without any extraneous annotations. Unfortunately, such a set is impossible to obtain because the underlying biology is incompletely understood. We built a two-part approximation to the perfect data set, taking advantage of data from the BDGP cDNA sequencing project (http://www.fruitfly.org/EST) and a Drosophila community effort to build a set of curated annotations for this region (Ashburner et al. 1999b). Our first component, known as the std1 data set, used high-quality sequence from a set of 80 full-length cDNA clones from the Adh region to provide a standard with annotations that are very likely to be correct but certainly are not exhaustive. The second component, known as the std3 data set, was built from the annotations being developed for Ashburner et al.(1999b) to give a standard with more complete coverage of the region, although with less confidence about the accuracy and independence of the annotations. We believe that this two-part approximation allows us to draw useful conclusions about the ability to accurately predict gene structure in complex eukaryotic organisms even though the absolutely perfect data set does not exist.
Eukaryotic transcript annotations have complex structures based on the composition of fundamental features such as the TATA box and other transcription factor binding sites, the transcription start site (TSS), the start codon, 5′ and 3′ splice site boundaries, the stop codon, the polyadenylation signal, exon start and end positions, and coding exon start and end positions. Our gene prediction evaluations focused on annotations that are specific to the coding region, from the start codon through the various intron–exon boundaries to the stop codon, and on promoter annotations. Although other types of features are also biologically interesting, we were unable to devise reliable methods for evaluating their predictions. Whenever possible, we relied on unambiguous biological evidence for our evaluations; when that was not available, we combined several types of evidence curated by domain experts.
Our goal for our first standard set, called std1, was to build a set of annotations that we believed were very likely to be correct in their fine details (e.g., exact locations for splice sites), even if we were unable to include every gene in the region. We based std1 on alignments of 80 high-quality, full-length cDNA sequences from this region with the high-quality genomic sequence for the contig. The cDNA sequences are the product of a large cDNA sequencing project at the BDGP and had not been submitted to GenBank at the time of the experiment. Working from five cDNA libraries, the longest clone for each unique transcript was selected and sequenced to a high-quality level. Starting with these cDNA sequences, we generated alignments to the genomic sequence using sim4 (Florea et al. 1998) and filtered them on several criteria. Of the 80 candidate cDNA sequences, 3 were paralogs of genes in the Adh region and 19 appeared to be cloning artifacts (unspliced RNA or multiple inserts into the cloning vector), leaving us with alignments for 58 cDNA clones. These alignments were further filtered based on splice site quality. We required that all of the proposed splice sites include a simple “GT”/“AG” core for the 5′ and 3′ splice sites, respectively, and that they scored highly (5′ splice sites ≥ 0.35 threshold, which gives a 98% true positive rate, and 3′ splice sites ≥ 0.25, which gives a 92% true positive rate) using a neural network splice site predictor trained on D. melanogaster data (Reese et al. 1997). This process left us with 43 sequences from the Adh region for which we had structures confirmed by alignments of high-quality cDNA sequence data with high-quality genomic data and by the fit of their splice sites to a Drosophila splice site model. Of these 43 sequences, 7 had a single coding exon and 36 had multiple coding exons. We added start codon and stop codon annotations to these structures from the corresponding records in the std3 data set.
After the experiment, we recently discovered four inconsistent genes in the std1 data set. For two genes (DS07721.1, DS003192.4), the cDNA clones (CK02594, CK01083, respectively) are likely to be untranscribed genomic DNA that was inappropriately included in the cDNA library. Two other genes from std3 (DS00797.5 and wb) were incorrectly reported in std1 as three partial all incomplete EST alignments (cDNA clones: CK01017, LD33192, and CK02229). In keeping with std1's goal of highly reliable annotations, all four sequences have been removed from the std1 data set that is currently available on the GASP web site. The results reported here use the larger, less reliable, data set as presented at the ISMB-99 tutorial.
The complete set of the original 80 aligned high-quality, full-length cDNA sequences was named std2. This set was never used in the evaluation process because it did not add any further compelling information or conclusions because of the unreliable alignments.
Our goal for the second, used standard set, called std3, was to build the most complete set of annotations possible while maintaining some confidence about their correctness. Ashburner et al. (1999b) compiled an exhaustive and carefully curated set of annotations for this region of the Drosophila genome based on information from a number of sources, included BLASTN, BLASTP (Altschul et al. 1990), and PFAM alignments (Sonnhammer et al. 1997, 1998; Bateman et al. 2000), high scoring GENSCAN (Burge and Karlin 1997) and Genefinder (P. Green, unpubl.) predictions, ORFFinder results (E. Friese, unpubl.), full-length cDNA clone alignments (including those used in std1), and alignments with full-length genes from GenBank. This set included 222 gene structures: 39 with a single coding exon and 183 with multiple coding exons. Of these 222 gene structures, 182 are similar to a homologous protein in another organism or have a Drosophila EST hit. For these structures, the intron–exon boundaries were verified by partial cDNA/EST alignments using sim4 (Florea et al. 1998), homologies were discovered using BLASTX, TBLASTX, and PFAM alignments, and gene structure was verified using a version of GENSCAN trained for finding human genes. Of the 54 remaining genes, 14 had EST or homology evidence but were not predicted by GENSCAN or Genefinder, and 40 were based entirely on strong GENSCAN and Genefinder predictions. All of this evidence was evaluated and edited by experienced Drosophila biolo gists, resulting in a protein coding gene data set that exhaustively covers the region with a high degree of confidence and represents their view of what should or should not be considered an annotated gene. Their gene data set excluded the 17 found transposable elements [6 LINE-like elements (G, F, Doc, and jockey) and 11 retrotransposons with long terminal repeats (LTRs; copia, roo, 297, blood, mdg1-like, and yoyo)], which almost all contain long ORFs. Some of these ORFs code for known and some others for, so far, unknown protein sequences.
Both of these data sets have shortcomings. As mentioned above, std1 only includes a subset of the genes in the region. It also includes a pair of transcripts that represent alternatively spliced products of a single gene. Although this is not incorrect, it confounds our scoring process. Because the cDNA alignments do not provide any evidence for the location of the start and stop codons, we based those annotations in std1 on information from the std3 set. Many of the gene structures in std3 are based on GENSCAN and Genefinder predictions without other supporting evidence, so it is possible that the fine details are incorrect, that the entries are not entirely independent of the techniques used by the predictors in the experiment, and that the set overestimates the number of genes in the region.
See Birney and Durbin (2000) and Henikoff and Henikoff (2000) for further discussion of the difficulties of evaluating these predictions especially in the protein homology annotation category, in which, by training, these programs will recognize protein-like sequences such as the ORFs in transposable elements as genes. They and others (see other GASP publications in this issue) have raised the issues of annotation oversights, transposons, and pseudogenes. In cases where GASP submissions suggest a missed annotation, this information has been passed onto biologists for further research, including screening cDNA libraries. We believe that it would have been biased to retroactively change the scoring scheme used at the GASP experiment based solely on missed annotations discovered by the participant's submissions. See Discussion for an example of an annotation that may be missing in the standard data sets. In the std3 data set we based our standard for what is or is not a Drosophila gene on the expert annotations provided by Ashburner et al. (1999b). It is clear that both transposons and pseudogenes are genuine features of the genome and that gene-finding technologies might recognize them. Because they were not included as coding genes in the expert annotations, we decided against including them in the standard set.
Building a set for the evaluation of transcription start site or, more generally, for promoter recognition proved to be even more difficult. For the genes in the Adh region almost no experimentally confirmed annotation for the transcription start site exists. As the 5′ UTR regions in Drosophila can extend up to several kilobases, we could not simply use the region directly upstream of the start codon. To obtain the best possible approximation, we took the 5′ ends of annotations from Ashburner et al. (1999b) where the upstream region relied on experimental evidence (the 5′ ends of full-length cDNAs) and for which the alignment of the cDNA to the genomic sequence included a good ORF. The resulting set contained 92 genes of the 222 annotations in the std3 set (Ashburner et al. 1999b). This number is larger than the number of cDNAs used for the construction of the std1 set described above because we included cDNAs that were already publicly available. The 5′ UTR of these 96 genes has an average length of 1860 bp, a minimum length of 0 bp (when the start codon was annotated at the beginning, due to the lack of any further cDNA alignment information; this is very likely to be only a partial 5′ UTR and therefore an annotation error), and a maximum length of 36,392 bp.
Data Exchange Format
One of the challenges of a gene annotation study is finding a common format in which to express the various groups' predictions. The format must be simple enough that all of the groups involved can adapt their software to use it and still be rich enough to express the various annotations.
We found that the General Feature Format (GFF) (formerly known as the Gene Feature Finding format) was an excellent fit to our needs. The GFF format is an extension of a simple name, start, end record that includes some additional information about the sequence being annotated: the source of the feature; the type of feature; the location of the feature in the sequence; and a score, strand, and frame for the feature. It has an optional ninth field that can be used to group multiple predictions into single annotations. More information can be found at the GFF web site: http://www.sanger.ac.uk/Software/formats/GFF/. Our evaluation tools used a GFF parser for the PERL programming language that is also available at the GFF web site.
We found that it was necessary to specify a standard set of feature names within the GFF format, for instance, declaring that submitters should describe coding exons with the feature name CDS. We produced a small set of example files (accessible from the GASP web site) that we distributed to the submitters and were pleased with how easily we were able to work with their results.
METHODS
Genome annotation is an ongoing effort to assign functional features to locations on the genomic DNA sequence. Traditionally, most of these annotations record information about an organism's genes, including protein-coding regions, RNA genes, promoters, and other gene regulatory elements, as well as gene function. In addition to these gene features, the following general genome structure features are also commonly annotated: repetitive elements and general A, C, G, T content measures (e.g., isochores).
Genome Annotation Classes
Although the GASP experiment invited and encouraged any class of annotations, most submissions were for gene-related features, emphasizing ab initio gene predictions and promoter predictions. In addition, two groups submitted functional protein domain annotations, and two groups submitted repeat element annotations. In the sections that follow, we categorize and discuss the submitted predictions.
Gene Finding
Protein coding region identification is a major focus of computational biology. A separate article in this issue (Stormo 2000) discusses and compares current methods, whereas an early paper by Fickett and Tung (1992) and a more recent review of gene identification systems by Burge and Karlin (1998) give excellent overviews of the field. Table 2 lists the six groups that predicted protein-coding regions with the corresponding program names. It also categorizes the submissions based on the types of information used to build the model for predictions. Although all groups used statistical information for their models—predominantly coding bias, coding preference, and consensus sequences for start codon, splice sites, and stop codons—only two groups used protein similarity information or promoter information to predict gene structure. More than half of the groups incorporated sequence information from cDNA sequences. In general, state-of-the-art gene prediction systems use complex models that integrate multiple gene features into a unified model.
Table 2.
Gene-Finding Submissions
| Program name | Statistics | Promoter | EST/cDNA alignment | Protein similarity | |
|---|---|---|---|---|---|
| Mural et al. (Oakridge, US) | GRAIL | X | X | ||
| Guigó et al. (Barcelona, ES) | GeneID | X | |||
| Krogh (Copenhagen, DK) | HMMGene | X | X | X | |
| Solovyev et al. (Sanger, UK) | FGenes | X | |||
| Gaasterland et al. (Rockefeller, US) | MAGPIE | X | X | X | |
| Reese et al. (Berkeley/Santa Cruz, US) | Genie | X | X | X | X |
Promoter Prediction
The complicated nature of the transcription initiation process makes computational promoter recognition a hard problem. We define promoter prediction as the identification of TSSs of protein coding genes that are transcribed by eukaryotic RNA polymerase II. A detailed description of the structure of promoter regions and existing promoter prediction systems is beyond the scope of this paper. Fickett and Hatzigeorgiou (1997) provide an excellent review of the field.
We can broadly identify three different approaches to promoter prediction, with at least one GASP submission in each category. The first class consists of “search by signal” programs, which identify single binding sites of proteins involved in transcription initiation or combinations of sites to improve the specificity. The program CoreInspector by Werner's group (M. Scherf, A. Klingenhoff, and T. Werner, in prep.) belongs to this category and searches for co-occurrences of two common binding sites within the core promoter (the core promoter usually denotes the region where the direct contact between the transcription machinery, the holoenzyme of the transcription complex, and the DNA takes place). The second class is often termed “search by content,” as programs within this group do not rely on specific signals but take the more general approach of identifying the promoter region as a whole, frequently based on statistical measures. Sometimes the promoter is split into several regions to obtain more accurate statistics. The MCPromoter program (Ohler et al. 1999) is a member of this second group. In comparison with the signal-based group, the content-based systems usually are more sensitive but less specific. The third class can be described as “promoter prediction through gene finding.” Simply using the start of a gene prediction as a putative TSS can be very successful if the 5′ UTR region is not too large. This approach can be improved by including similarity to EST sequences and/or a promoter module in the statistical systems used for gene prediction. The TSS predictions submitted by the participants of the MAGPIE and the Genie groups belong to this last class.
The notorious difficulty of the problem itself is exacerbated by the limited amount of existing reliably annotated training material. The experimental mapping of a TSS is a laborious process and is therefore not routinely carried out, even if the gene itself is studied extensively. So, both training the models and evaluating the results is a difficult task, and the conclusions we draw from the results must be considered with much caution.
Repeat Finders
Detecting repeated elements plays a very important role in modeling the three-dimensional structure of a DNA molecule, specifically, the packing of the DNA in the cell nucleus. It is believed that the packing of the DNA around the nucleosome is correlated with the global sequence structure produced predominantly by repetitive elements. Repeats also play a major role in evolution (for review, see Jurka 1998). Two groups, Gary Benson [tandem repeats finder v. 2.02 (TRF; Benson 1999)] and the MAGPIE team using two programs Calypso (D. Field, unpubl.) and REPuter (Kurtz and Schleiermacher 1999) submitted repetitive sequence annotations. TRF (Benson 1999) locates approximate tandem repeats (i.e., two or more contiguous, approximate copies of a pattern of nucleotides) where the pattern size is unspecified but falls within the range from 1 to 500 bases. The Calypso program (D. Field, unpubl.) is an evolutionary genomics program. Its primary function is to find repetitive regions in DNA and protein sequences that have higher than average mutation rates. The REPuter program (Kurtz and Schleiermacher 1999) determines repeats of a fixed preselected length in complete genomes.
Protein Homology Annotation
Homologies to gene sequences from other organisms can often be used to identify protein-coding regions in anonymous genomic sequence. In addition to the location, it is often possible to infer the function of the predicted gene based on the function of the homologous gene in the other organism or of a known structural and functional protein element in the gene. Whereas the tools in the gene prediction category and the EST/cDNA alignment category are usually intended to determine the exact structure of a gene, the protein homology-based tools are usually optimized to find conserved parts of the sequence without worrying about the exact gene structure. Traditionally, this area of genome annotations has been dominated by the suite of local alignment search tools of BLAST (Altschul et al. 1990) and more global search tools such as FASTA (Pearson and Lipman 1988). Recent reviews in this area include Agarwal and States (1998), Marcotte et al. (1999), and Pearson (1995).
In the GASP experiment, two groups specializing in functional protein domain or motif identification in genomic DNA submitted annotations. The Henikoff group found hits to the BLOCKS+ database (http://blocks.fhcrc.org), a database consisting of conserved protein motifs (Henikoff and Henikoff 1994; Henikoff et al. 1999a). The second group in this category submitted results from the GeneWise program (Birney 1999). This program searches genomic DNA against a comprehensive hidden Markov model (HMM)-based library (PFAM; Sonnhammer et al. 1997, 1998; Bateman et al. 2000) of protein domains. Both programs look for conserved regions by searching translated DNA against a representation of multiple aligned sequences. Whereas in BLOCKS+ the multiple protein alignments consist of sets of ungapped regions, the GeneWise program searches against a gapped alignment. Both methods will turn up distantly related sequences.
EST/cDNA Alignment
Computational predictions of gene location and structure go hand in hand with EST/cDNA sequencing and alignment techniques for building transcript annotations in genomic sequence. Either can be used as a discovery tool, with the other held in reserve for verification. A researcher can verify the existence and structure of predicted genes by sequencing the corresponding mRNA molecules and aligning their sequences to the original genomic sequence. Alternatively, one can start with an EST or cDNA sequence and build an alignment to the genomic sequence that has been guided and/or verified by tools from the gene prediction arsenal, for example, using likely splice site locations and checking for long ORFs and potential frame shifts.
There are many tools for aligning sequences. Although they have generally been specialized for aligning sequences that are evolutionarily related, some are designed for niche applications such as recognizing overlaps among sequencing runs. Aligning EST/cDNA sequences to the original genomic sequence also presents a unique set of tradeoffs and issues. In some cases (interspecies EST/genomic alignments), these tools must model evolutionary changes in the sequence. Sometimes (e.g., for low-quality EST sequences), they need to model errors in the sequence generated by the sequencing process. For multiexon genes, they need to model the intron regions as cost-free gaps tied to a model for recognizing splice sites. Several tools have been developed for this task: Mott (1997) and Birney and Durbin (1997) describe dynamic programming approaches that include models of splice sites and intron gaps. Florea et al. (1998) describe sim4, a heuristic tool that performs as well as the dynamic programming approaches and is efficient enough to support searching of large databases of genomic sequence.
Using cDNA clones and their sequences to build transcript annotations requires a variety of operations. The tools discussed above align the cDNA sequences to the genomic sequence, but steps must be taken to filter out clones that are merely paralogs of genes in the sequence and to recognize and handle various laboratory artifacts. If the clones represent short ESTs, then a likely annotation can be built by assembling a consistent model from their individual alignments. Longer ESTs or cDNAs might generate several similar alignments, and an automated tool must be able to select the most biologically meaningful variant. Although there are some gene prediction tools that can use information about homologies to known genes or ESTs, and most large-scale sequencing centers have some automated sanity checking for their database search results, there are not any tools that automate the production of transcript annotations from cDNA sequences.
Gene Function
Gene function predictions are the most difficult annotations to produce and to evaluate. Current technologies use similarity to proteins (or protein domains) with known function to predict functional domains in genomic sequence. Although some tools use simple sequence alignments, more powerful tools have developed significantly more sensitive models.
It quickly became apparent that a consistent and correct assessment of function predictions as part of the GASP experiment was not possible because of the incomplete understanding of the protein products encoded by the 222 genes in the Adh region.
Evaluating Gene Predictions
An ideal gene prediction tool would produce annotations that were exactly correct and entirely complete. The fact that no existing tool has these characteristics reflects our incomplete understanding of the underlying biology as well as the difficulty to build adequate gene models in a computer. Although no tool is perfect, each tool has particular strengths and weaknesses, and any performance evaluation should be in the context of an intended use. For example, researchers who are interested in identifying gene-rich regions of a genome for sequencing would be happy with a tool that successfully recognizes a gene's approximate location, even if it incorrectly described splice site boundaries. On the other hand, someone trying to predict protein structures is more interested in getting a gene's structure exactly right than in a tool's ability to predict every gene in the genome.
When assessing the accuracy of predictions, each prediction falls into one of four categories. A true-positive (TP) prediction is one that correctly predicts the presence of a feature. A false-positive (FP) prediction incorrectly predicts the presence of a feature. A true-negative (TN) prediction is correct in not predicting the presence of a feature when it isn't there. A false-negative (FN) prediction fails to predict the existence of a feature that actually exists. The sensitivity (Sn) of a tool is defined as TP / (TP + FN) and can be thought of as a measure of how successful the tool is at finding things that are really there. The specificity (Sp) of a tool is defined as TP / (TP + FP) and can be thought of as a measure of how careful a tool is about not predicting things that aren't really there. Burset and Guigó (1996) also use a correlation coefficient and an average correlation coefficient. We chose not to use these measures because they depend on predictors' TN information, and we recognize that our evaluation sets were constructed in such a way that the TN information is not trustworthy. These Sn and Sp metrics are used for evaluating the submissions in the gene-finding, promoter recognition, and gene identification using protein homology categories. In the gene finding category, they are used for all three levels: base level, exon level, and gene level. In the protein homology category, they are used for base level and gene level only.
In one of the first reviews of gene prediction accuracy, Fickett and Tung (1992) developed a method that measured predictors' ability to correctly recognize coding regions in genomic sequence. They used their method to compare published techniques and concluded that in-frame hexamer counts were the most accurate measure of a region's coding potential. Burset and Guigó (1996) recognized that there are a wide variety of uses for gene predictions and developed measures—including base level, exon level, and gene level Sp and Sn—that describe a predictor's suitability for a particular task.
Base Level
The base level score measures whether a predictor is able to correctly label a base in the genomic sequence as being part of some gene. It rewards predictors that get the broad sweeps of a gene correct, even if they don't get the details such as the splice site boundaries entirely correct. It penalizes predictors that miss a significant portion of the coding sequence, even if they get the details correct for the genes they do predict. We used the Sn and Sp measures defined above as the measures of success in this category.
Exon Level
Exon level scores measure whether a predictor is able to identify exons and correctly recognize their boundaries. Being off by a single base at either end of the exon makes the prediction incorrect. Because we only considered coding exons in our assessment, the first exon is bracketed by the start codon and a 5′ splice site, the last exon is bracketed by a 3′ splice site and the stop codon, and the interior exons are bracketed by a pair of splice sites. As measures of success in this category, we used two statistics in addition to Sn and Sp. The missed exon (ME) score is a measure of how frequently a predictor completely failed to identify an exon (no prediction overlap at all), whereas the wrong exon (WE) score is a measure of how frequently a predictor identifies an exon that has no overlap with any exon in the standard sets. The ME score is the percentage of exons in the standard set for which there were no overlapping exons in the predicted set. Similarly, the WE score is the percentage of exons in the predicted set for which there were no overlapping exons in the standard set.
Gene Level
Gene level Sn and Sp measure whether a predictor is able to correctly identify and assemble all of a gene's exons. For a prediction to be counted as a TP, all of the coding exons must be identified, every intron–exon boundary must be exactly correct, and all of the exons must be included in the proper gene. This is a very strict measure that addresses a tool's ability to perfectly identify a gene. In addition to the Sn and Sp measures based on absolute accuracy, we used the missed genes (MG) score as a measure of how frequently a predictor completely missed a gene (a standard gene is considered missed if none of its exons are overlapped by a predicted coding gene) and the wrong genes (WG) score as a measure of how frequently a predictor incorrectly identified a gene (a prediction is considered wrong if none of its exons are overlapped by a gene from the standard set).
Split and Joined Genes
The exon level scores discussed above measure how well a predictor recognizes exons and gets their boundaries exactly correct. The gene level scores measure how well a predictor can recognize exons and assemble them into complete genes. Neither of these scores directly measures a predictor's tendency to incorrectly assemble a set of predicted exons into more or fewer genes than it should. We developed two new measures, split genes (SG) and joined genes (JG), which describe how frequently a predictor incorrectly splits a gene's exons into multiple genes and how frequently a predictor incorrectly assembles multiple genes' exons into a single gene. Because the coverage of the std1 data set is so incomplete, we have only included SG and JG scores from the comparison with std3. A gene from the standard set is considered split if it overlaps more than one predicted gene. Similarly, a predicted gene is considered joined if it overlaps more than one gene in the standard set. The SG measure is defined as the sum of the number of predicted genes that overlap each standard gene divided by the number of standard genes that were split. Similarly, the JG measure is the sum of the number of standard genes that overlap each predicted gene divided by the number of predicted genes that were joined. A score of 1 is perfect and means that all of the genes from one set overlap exactly one gene from the other set.
Application of These Measures to Correct Answer Data Sets std1/std3
We built the std1 data set in such a way that we believe it is correct in the details of the genes that it describes, though we know that it only includes a small portion of the genes in the region. The std3 data set, on the other hand, is as complete as was possible but does not have rigorous independent evidence for all of its annotations. For the std1 data set, we believe that the TP count (it was predicted, and it exists in the standard) and FN count (it was not predicted, but it does exist in the standard) are reliable because of the confidence that we have in the correctness of the predictions in the set. On the other hand, we do not believe that the TN count (it was not predicted, and it is not in the standard set) and FP count (it was predicted, but is not in the standard set) are reliable because they both assume that the standard correctly describes the absence of a feature and we know that there are genes missing from std1. It follows that we believe that Sn is meaningful for std1 because it only depends on TP and FN but that we are less confident about the Sp score because it depends on TP and FP. A similar logic applies to the std3 data set, where our confidence in the set's completeness but not its fine details suggests that the TP and FP scores are usable but that the TN and FN scores are not. This means that for std3, we believe that the Sp measure can be used to describe a predictor's performance but that Sn is likely to be misleading.
Evaluation of Promoter Predictions
We adopted the measures proposed by Fickett and Hatzigeorgiou (1997). They evaluated the success of promoter predictions by giving the percentage of correctly identified TSSs versus the FP rate. A TSS is regarded as identified if a program makes one or more predictions within a certain “likely” region around the annotated site. The FP rate is defined as the number of predictions within the “unlikely” regions outside the likely regions divided by the total number of bases contained in the unlikely set. As our annotation of the TSS is only preliminary and not experimentally confirmed, we chose a rather large region of 500 bases upstream and 50 bases downstream of the annotated TSS as the likely region. The upstream region is always taken as the likely region, even if it overlaps with a neighboring gene annotation on the same strand. The unlikely region for each gene then consists of the rest of the gene annotation, from base 51 downstream of the TSS to the end of the final exon.
Visualization of the Annotations
Generating “good” annotations generally requires integrating multiple sources of information, such as the results of various sequence analysis tools plus supporting biological information. Visualization tools that display sequence annotations in a browsable graphical framework make this process much more efficient. In this experiment we found that visualization tools are essential to evaluate the genome annotation submissions. When annotations are displayed visually, overall trends become apparent, for example, gene-rich versus gene-poor regions, genes that were predicted by most participants versus those that were predicted by few. Additionally, as we discuss below, a visualization tool that is capable of displaying annotations at multiple levels of detail provides a way to examine individual predictions in detail.
Building genome annotation visualization tools is a daunting task. Many such tools have been developed, starting with ACeDB (Eeckman and Durbin 1995; Stein and Thierry-Mieg 1998). We were fortunate in that the BDGP has built a flexible suite of genome visualization tools (Helt et al. 1999) that could be extended to display the GASP submissions. We adapted the BDGP's annotated clone display and editing tool, CloneCurator (Harris et al. 1999), which is based on a genomic visualization toolkit (Helt et al. 1999), to read the annotation submissions in GFF format and display each team's predictions in a unique color and location.
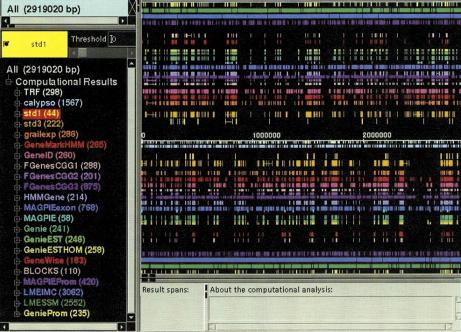
CloneCurator (see Fig. 1) displays features on a sequence as colored rectangles. Features on the forward strand appear above the axis, whereas those on the reverse strand appear below the axis. The display can be zoomed and scrolled to view areas of interest in more detail. A configuration file identifies the feature types that are to be displayed and assigns colors and offsets to each one. For example, the std1 and std3 exons appear in yellow and orange close to the central axis.
Figure 1.
(See facing page.) Screen shot from the CloneCurator program (Harris et al. 1999), featuring the genome annotations of all 12 groups for the 2.9-Mb Adh region. The main panel shows the computational annotations on the forward (above axis) and reverse sequence strands (below axis). Genes located on the top half of each map are transcribed from distal to proximal (with respect to the telomere of chromosome are 2L); those on the bottom are transcribed from proximal to distal. Right below the axis are the two repeat finding results displayed, followed by reference sets from Ashburner et al. (1999b; std1 and std3), followed by the 12 submissions of gene-finding programs, followed by the two protein homology programs, and eventually, farthest away from the axis, the four promoter recognition programs. (Left) The color-coded legend for the program and the number of predictions made by the programs.
RESULTS
The results of an experiment such as GASP are only meaningful if enough groups participate. We were fortunate to have 12 diverse groups involved, and we were very grateful for the speed with which they were able to submit their predictions. We believe that these 12 groups provide a fair representation of the state of the art in annotation system technology. We collected submissions by electronic mail and evaluated them using the std1 and std3 data sets as described above. Before releasing our results at the Intelligent Systems in Molecular Biology conference in August 1999 in Heidelberg, Germany, we assembled a team of independent assessors (Ashburner et al. 1999a) to review our techniques and conclusions. As discussed in the introduction, the accuracy of the various measures discussed below depends heavily on how well our standard sets capture the true set of features in the region. These values should only be considered in the context of the standard data sets.
A detailed description of the results and the evaluation techniques we used can be accessed through the GASP homepage at http://www.fruitfly.org/GASP/.
Gene Finding
Table 3 summarizes the performance of the gene-finding tools using the measures defined above. Three groups submitted multiple submissions. The first group, Fgenes1, Fgenes2, and Fgenes3, submitted three predictions at varying stringency (for details, see Salamov and Solovyev 2000). For the GeneID program, two submitted versions are presented, version 1 (GeneID v1) being the original submission and version 2 (GeneID v2) being a newer submission from a corrected version of the original program (for details, see Parra et al. 2000). The third group with multiple submissions used three versions of the Genie program: the first a pure statistical approach (Genie), the second including EST alignment information (GenieEST), and the third using protein homology information (GenieESTHOM) (for details, see Reese et al. 2000). For all other groups from Table 2, only one submission was evaluated. The following sections discuss the base level, exon level, and gene level performance of these submissions.
Table 3.
Evaluation of Gene-Finding Systems
| FGenes 1 | FGenes 2 | FGenes 3 | GeneID v1 | GeneID v2 | Genie | Genie EST | Genie ESTHOM | HMMGene | MAGPIE exon | GRAIL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base level | Sn std1 | 0.89 | 0.49 | 0.93 | 0.48 | 0.86 | 0.96 | 0.97 | 0.97 | 0.97 | 0.96 | 0.81 |
| Sp std3 | 0.77 | 0.86 | 0.60 | 0.84 | 0.83 | 0.92 | 0.91 | 0.83 | 0.91 | 0.63 | 0.86 | |
| Exon level | Sn std1 | 0.65 | 0.44 | 0.75 | 0.27 | 0.58 | 0.70 | 0.77 | 0.79 | 0.68 | 0.63 | 0.42 |
| Sp std3 | 0.49 | 0.68 | 0.24 | 0.29 | 0.34 | 0.57 | 0.55 | 0.52 | 0.53 | 0.41 | 0.41 | |
| ME (%) std1 | 10.5 | 45.5 | 5.6 | 54.4 | 21.1 | 8.1 | 4.8 | 3.2 | 4.8 | 12.1 | 24.3 | |
| WE (%) std3 | 31.6 | 17.2 | 53.3 | 47.9 | 47.4 | 17.4 | 20.1 | 22.8 | 20.2 | 50.2 | 28.7 | |
| Gene level | Sn std1 | 0.30 | 0.09 | 0.37 | 0.02 | 0.26 | 0.40 | 0.44 | 0.44 | 0.35 | 0.33 | 0.14 |
| Sp std3 | 0.27 | 0.18 | 0.10 | 0.05 | 0.10 | 0.29 | 0.28 | 0.26 | 0.30 | 0.21 | 0.12 | |
| MG (%) std1 | 9.3 | 34.8 | 9.3 | 44.1 | 13.9 | 4.6 | 4.6 | 4.6 | 6.9 | 4.6 | 16.2 | |
| WG (%) std3 | 24.3 | 24.8 | 52.3 | 22.2 | 30.5 | 10.7 | 13.0 | 15.5 | 14.9 | 55.0 | 23.7 | |
| SG | 1.10 | 1.10 | 2.11 | 1.06 | 1.06 | 1.17 | 1.15 | 1.16 | 1.04 | 1.22 | 1.23 | |
| JG | 1.06 | 1.09 | 1.08 | 1.62 | 1.11 | 1.08 | 1.09 | 1.09 | 1.12 | 1.06 | 1.08 |
The evaluation is divided into three categories: base level, exon level, and gene level. The different statistical features reported are Sn, Sp, ME, WE, MG, WG, SG, and JG. std1 and std3 indicate against which standard set the statistics are reported.
Base Level Results
Several gene prediction tools had a Sn of >0.95 at the base level. This suggests that current technology is able to correctly identify >95% of the D. melanogaster proteome. A few tools demonstrated a specificity of >0.90 at the base level, only infrequently labeling a noncoding base as coding. Generally, the tools have a higher Sn than Sp. Two programs, Fgenes2 and GeneID, were designed to be conservative about their predictions and do not follow this trend.
Exon Level Results
There was a great deal of variability in the exon level scores. Several tools had Sn scores ∼0.75, correctly identifying both exon boundaries ∼75% of the time. Their Sps were generally much lower (the highest was 0.68), probably a reflection of the strict definition of exon level scores both splice sites had to be predicted correctly and possible inaccuracies in the std3 data set. The low ME scores (several <0.05) combined with the fairly high Sn suggest that several tools were successful at identifying exons but had trouble finding the correct exon boundaries. Programs that incorporate EST alignment information, such as GenieEST and HMMGene, had sensitivity scores that were up to 10% better than the other tools. The high WE scores suggest either that the tools are overpredicting or that there are genes that are missing even from std3.
Gene Level Results
All of the predictors had considerable difficulty correctly assembling complete genes. The best tools were able to achieve Sns between 0.33 and 0.44, meaning that they are incorrect over half of the time. This value seems to be very similar in Drosophila and human sequences, based on a recent analysis of the BRCA2 region in human (T.J. Hubbard, pers. comm.). Even on the more complete std3 data set, the programs tended to incorrectly predict many genes. The very low MG score (as low as 4.6%) is reassuring because it suggests that several tools are able to recognize a gene, even if they have difficulty figuring out the exact details of its structure. Comparing the WG and MG measures suggests that existing tools tend to predict genes that do not exist more often than they miss genes that do exist. Because it is almost certain that there are real genes that are missing from both standard sets, this conclusion must be viewed with some skepticism. Although there were several tools with good SG or JG scores, none of them performed well in both categories.
Promoter Prediction
Table 4 shows the performance of the promoter prediction systems, grouped by approach: search-by-signal, search-by-region, and gene prediction programs.
Table 4.
Evaluation of Promoter Prediction Systems
| System name | Sensitivity | Rate of false-positive predictions in regiona (853,180 bases) | Rate of predictions in regionb (2,570,232 bases) |
|---|---|---|---|
| CoreInspector | 1 (1%) | 1/853,180 | 1/514,046 |
| MCPromoter v1.1 | 26 (28.2%) | 1/2,633 | 1/2,537 |
| MCPromoter v2.0 | 31 (33.6%) | 1/2,437 | 1/2,323 |
| GeniePROM | 25 (27.1%) | 1/14,710 | 1/28,879 |
| GenieESTPROM | 30 (32.6%) | 1/16,729 | 1/29,542 |
| MAGPIE | 33 (35.8%) | 1/14,968 | 1/16,370 |
We show the Sn for identified TSSs in comparison with the FP rate for non-TSS regions and general gene regions: athe unlikely region defined as the rest of the gene starting 51 bases downstream from its annotated TSS; bthe general gene region, spanning from half the distance to the previous and next annotated genes including the annotated TSS (taken from the std3 annotation).
Gene-finding programs that include a prediction of the TSS obtained the best results. The number of false predictions made by the region-based programs is very high (giving them a low Sp), and because the signal-specific programs only identify one promoter, their Sn is very low. The high Sp of the gene finders is obviously due to the context information: All promoter predictions within gene predictions are ruled out in advance, and the location of the possible start codon provides the system with a good initial guess of where to look for a promoter. The MAGPIE system also uses EST alignments to obtain information on 5′ UTRs, which mirrors the way the std sets were constructed: Roughly one-third of the putative TSS assignments rely on cDNAs that were publicly available in GenBank. A closer look at the results reveals that the region-based programs have a Sn that is comparable with the gene finders and the signal based program had only a single FP, showing that both types of tools can be used for different applications.
Our data set, and the evaluation based on it, relies on the assumption that the 5′ ends of the full-length cDNAs are reasonably close to the TSS. This makes it very hard to draw strong conclusions from the presented results. Even the most sensitive systems could identify only roughly one third of the start sites. This could of course be caused by the fact that the existing annotation is only an approximation and some of the true TSSs may be located further upstream. It also hints at the diversity of promoter regions that mirrors the possibilities for gene regulation and at the existing bias toward housekeeping genes in the current data sets used for the training of the models.
Gene Identification Using Protein Homology
Gene-finding evaluation statistics, such as those described above, can be used to summarize the ability of a program to identify complete and accurate gene structures in genomic DNA. In Table 5 we have applied the same evaluation statistics to the homology-based search programs GeneWise and BLOCKS+. Because these programs are not optimized to deal with exact exon boundary assignments, Table 5 only shows the performance for the base level and the MG and WG.
Table 5.
Evaluation of Similarity Searching
| BLOCKS | GeneWise | MAGPIE cDNA | MAGPIE EST | Grail Similarity | ||
|---|---|---|---|---|---|---|
| Base level | Sn std1 | 0.04 | 0.12 | 0.02 | 0.31 | 0.31 |
| Sp std3 | 0.80 | 0.82 | 0.55 | 0.32 | 0.81 | |
| Gene level | MG (%) std1 | 62.7 | 69.7 | 95.3 | 27.9 | 41.8 |
| WG (%) std3 | 12.9 | 14.1 | 0.0 | 44.3 | 7.4 |
Base and gene level statistics are shown. The base level is described using Sn and Sp, and the statistics for the gene level are given as MG and WG.
The very low Sns at the base level are not surprising, because the programs identify only conserved protein motifs or particular domains and make no effort to predict complete genes. Sp, which should be high given that only conserved protein motifs are scored, was lower than expected. Detailed studies of these predictions (see Birney and Durbin 2000; Henikoff and Henikoff 2000) show that most of the FP predictions were hits to transposable elements or to possible genes that are missing in the standard sets. Both programs use a database of protein domains or conserved protein motifs. Both databases are large and are believed to contain at least 50% of the existing protein domains. The high number of MG, 62.7% for BLOCKS and 69.7% for GeneWise, means that these programs will miss a significant number of Drosophila genes when used to search genomic DNA directly. The WG scores of 12.9% BLOCKS and 14.1% for GeneWise are lower than the gene finding programs discussed in the previous section.
Gene Identification Using EST/cDNA Alignments
It is believed that some cDNA information exists for approximately half of the genes in the D. melanogaster genome. This cDNA database (available as the EST data set at the GASP web site) was used as a basis for the cDNA/EST alignment category. The Sn of 31% for MAGPIEEST and GrailSimilarity (Table 5) implies that the coding portion of the available EST data currently covers one-third of the genome's coding sequence. The low Sp is very surprising and suggests that the EST/cDNA alignment problem is not a trivial one. The only program that tried to align complete cDNAs to genomic DNA, MAGPIEcDNA, could find complete cDNAs for only 2.4% of the genes. EST alignments also resulted in high numbers of missed genes, suggesting that the EST libraries are biased toward highly expressed genes. The high WG scores suggest that some genes are missing even from std3.
Selected Gene Annotations
The summary statistics discussed above only provide a global view of the predicting programs' characteristics. A much better understanding of how the various approaches behave can be obtained by looking at individual gene annotations. Such a detailed examination can also help identify issues that are not addressed by current systems.
In the following paragraphs we will discuss a few interesting examples. Figure 1 shows the color codes of the participating groups that are used throughout this section. Genes located on the top of each map are transcribed from distal to proximal (with respect to the telomere of chromosome arm 2L); those on the bottom are transcribed from proximal to distal. std1 and std3 are the expert annotations described in Ashburner et al.(1999b). Just below the axis, you can see the annotations for the two repeat finding programs. These have no sequence orientation and are therefore only shown on one side. Farther away from the axis, after std1 and std3, we grouped all of the ab initio gene-finding programs together. Next to the gene finders are the homology-based annotations. On the bottom and the top of the figure we show the three promoter annotations, but for clarity we did not include these annotations in the subsequent figures. (On the front page and in the legend of Fig. 1, you can see the full set of annotations of all programs, which are also accessible from the GASP web site.)
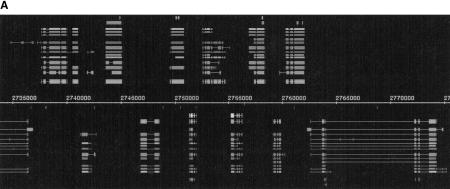
Our first example is a “busy” region with 12 complete genes and 1 partial gene in a stretch of only 40 kb (Fig. 2A). This region is located at the 3′ end of the Adh region from base 2,735,000 to base 2,775,000. Genes exist on both strands, and it is striking that in this region the genes tend to alternate between the forward and the reverse strands. We selected this region for its gene density and because it has characteristics that are typical of the complete Adh region. Figure 2A vividly demonstrates that all of the gene-finding programs' predictions are highly correlated with the annotated genes from std1/std3. In the past, gene finders had often mistakenly predicted a gene on the noncoding strand opposite of a real gene, leading to FP predictions known as “shadow exons.” Figure 2A makes it clear that gene finders have overcome this problem, because there are almost no shadow exon predictions for any of the genes in std3. Another characteristic, captured in the high base level sensitivity and the low missing genes statistics, is that every gene in the std3 set was predicted by at least a few groups and that most of these predictions agree with each other. Except for the second and third genes [DS02740.5, I(2)35Fb] on the forward strand (2,740,000–2,745,000), which seem to be single exon genes, all of the genes in this region are multiexon genes with between two and eight exons. The exon size varies widely. There are genes that consist of only two large exons, some that consist of a mix of large and small exons, and some that are made up exclusively of many small exons. The distribution seems to be almost random. Except for the long final intron in the last gene on the reverse strand (cact), the region consists exclusively of short introns.
Figure 2.
(A) Annotations for the following known genes described in Ashburner et al. (1999b) are shown for the region from 2,735,000 to 2,775,000 (from the left to the right of the map): crp (partial, reverse (r)), DS02740.4 (forward (f)), DS02740.5 (f), I(2)35Fb (f), heix (r), DS02740.8 (f), DS02740.9 (r), DS02740.10 (f), anon-35Fa (r), Sed5 (f), cni (r), fzy (f), cact (r). (B) Annotations for the following known gene described in Ashburner et al. (1999b) are shown for the region from 600,000 to 635,000 (left to right): DS01759.1 (r).
Predictions on the reverse strand indicate a possible gene from base 2,741,000 to base 2,745,000. Most of the gene finders agree on this prediction, but neither std1 nor std3 describes a gene at this location. This could be a real gene that was missed by the expert annotation pathway described in Ashburner et al (1999b). Neither BLOCKS+ nor GeneWise found any homologies in this region, but we can see from the table in the previous section that many real genes do not have any homology annotations. Interestingly, this is the only area in the region where two gene finders predicted a possible gene that likely consists of shadow exons.
The fifth gene on the forward strand (DS02740.10, bases 2,752,500–2,755,000) shows that long genes with multiple exons are much harder to predict than single exon genes or genes with only a few exons. In this region splitting and joining genes does not seem to be a problem. Repeats occur sparsely and mostly in noncoding regions, predominantly in introns.
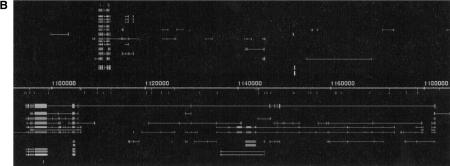
In contrast to the busy region in Figure 2A, Figure 2B highlights a region of almost equal size in which only one gene (DS01759.1) is present in both std1 and std3. There are very few FP predictions by any group, but there is one case where the “false” predictions by different programs are located at very similar positions (on the reverse strand near base 620,000). This suggests a real gene that is missing from both standard sets.
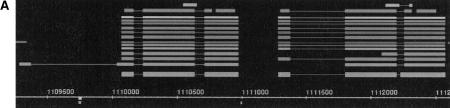
Figure 3, A–D, depicts selected genes that illustrate some interesting challenges in gene finding. Figure 3A shows the Adh and the Adhr genes that occur as gene duplicates. The encoded proteins have a sequence identity of 33%. The positions of the two introns interrupting the coding regions are conserved and give additional evidence to tandem duplication. Both genes are under the control of the same regulatory promoter, the Adhr gene does not have a TSS of its own, and its transcript is always found as part of an Adh–Adhr dicistronic mRNA. Gene duplications occur very frequently in the Drosophila genome—estimates show that at least 20% of all genes occur in gene family duplications. In an additional twist, Adh and Adhr are located within an intron of another gene, outspread (osp), that is found on the opposite strand (for details, see Fig. 3B). The Adh gene is correctly predicted by most of the programs, although one erroneously predicts an additional first exon. Most of the programs also predict the structure of Adhr correctly; one program misses the initial exon and shortens the second exon. Both Adh and Adhr show hits to the protein motifs in BLOCKS+ as well as alignments to a PFAM protein domain family through GeneWise. Both genes hit two different PFAM families, and the order of these two domains is conserved in the gene structure.
Figure 3.
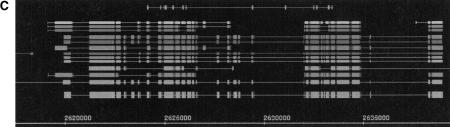
(A) Annotations for the following known genes described in Ashburner et al. (1999b) are shown for the region from 1,109,500 to 1,112,500 (forward strand only) (left to right): Adh, Adhr. (B) Annotations for the following known genes described in Ashburner et al. (1999b) are shown for the region from 1,090,000 to 1,180,000 (left to right): osp (r), Adh (f), Adhr (f), DS09219.1 (r), DS07721.1 (f). (C) Annotations for the following known gene described in Ashburner et al. (1999b) are shown for the region from 2,617,500 to 2,640,000 (forward strand only) (left to right): Ca-α1D. (D) Annotations for the following known genes described in Ashburner et al. (1999b) are shown for the region from 2,894,000 to 2,904,000 (forward strand only) (left to right): idgf1, idgf2, idgf3.
Figure 3B highlights the osp gene region. This is an example of a gene with exceptionally long (>20 kb) introns, making it hard for any gene finder to predict the entire structure correctly. In addition, there are a number of smaller genes [including the Adh and Adhr genes discussed above, DS09219.1 (r.) and DS07721.1 (f.)] within the introns of osp. No current gene finder includes overlapping gene structures in its model; as a consequence, none of the GASP gene finders were able to predict the osp structure without disruption. This is clearly a shortcoming of the programs because genes containing other genes are often observed in Drosophila (Ashburner et al. 1999b report 17 cases for the Adh region). However, it should be noted that most of the gene finders predict the 3′ end of osp correctly and therefore get most of the coding region right. The region that includes the 5′ end of osp shows a lot of gene prediction activity, but there is no consistency among the predictions. One program (FGenesCCG3) does correctly predict the DS09219.1 gene.
Figure 3C shows the entire gene structure of the Ca-α1D gene. This gene is the most complex gene in the Adh region, with >30 exons. This is a very good example for studying gene splitting. Several predictors break the gene up into several genes, but some groups make surprisingly close predictions. This shows the complex structure that genes can exhibit and that extent to which this complexity has been captured in the state-of-the-art prediction models. It is interesting to note that most of the larger exons are predicted, whereas the shorter exons are missed. Such a large complex gene is a good candidate for alternative splicing, which can ultimately be detected only by extensive cDNA sequencing.
Figure 3D shows the triple duplication of the idgf gene (idgf1, idgf2, and idgf3) on the forward strand. Two programs mistakenly join the first two genes into a single gene; all the others correctly predict all three genes.
DISCUSSION
The goal of the GASP experiment was to review and assess the state of the art in genome annotation tools. We believe that the noncompetitive framework and the community's enthusiastic participation helped us achieve that goal. By providing all of the participants with an unprecedented set of D. melanogaster training data and using unreleased information about the region as our gold standard, we were able to establish the level playing field that made it possible to compare the performance of the various techniques. The large size of the Adh contig and the diversity of its gene structures provided us with an opportunity to compare the capabilities of the annotation tools in a setting that models the genome-wide annotations currently being attempted. However, the lack of a completely correct standard set means that our results should only be considered in the context of the std1 and std3 data sets.
Assessing the Results
The most difficult part of the assessment was developing a benchmark for the predicted annotations. By dividing the predictions into different classes and developing class-specific metrics that were based on the best available standards, we feel that we were able to make a meaningful evaluation of the submissions. Although most of the information that was used to evaluate the submissions was unreleased, some cDNA sequences from the region were in the public databases. As sequencing projects move forward, it will become increasingly difficult for future experiments to find similarly unexplored regions. This makes it very different from the CASP protein structure prediction contests, which can use the three-dimensional structure of a novel target protein that is unknown to the predictors.
As discussed in the introduction, the lack of an absolutely correct standard against which to evaluate the various predictions is a troubling issue. Although we believe that the standard sets sufficiently represent the true nature of the region and that conclusions based on them are interesting, it must be remembered that the various results can only be evaluated in the context of these incomplete data sets. This also makes GASP more difficult and less clear cut than CASP, where the three-dimensional protein structure is experimentally solved at least to some degree of resolution.
It should also be noted that the gene-finding tools with the highest Sp have a great deal in common with GENSCAN, the gene prediction tool used in the development of the std3 data set. This suggests that std3's origins might have led to a bias favoring GENSCAN-like predictors. Because std1 was exclusively created using full-length cDNA alignments, this set might be biased towards highly expressed genes, because the cDNA libraries were not normalized.
Progress in Genome-Wide Annotation
The rapid release of completed genomes, including the imminent release of the D. melanogaster and human genomes, has driven significant developments in genome annotation and gene-finding tools. Problems that have plagued gene-finding programs, such as predicting shadow exons, restricting predictions to a single strand, recognizing repeats, and accurately identifying splice sites, have been overcome by the current state of the art. In this section, we discuss some of the remaining issues in genome annotation that the GASP experiment highlighted.
Successful gene prediction programs use complex models that integrate information from statistical features that are driven by the three-dimensional protein–DNA/RNA interactions. They make integrated predictions on both strands and have been tuned to predict all the genes in gene-rich regions and avoid overpredicting genes in gene-poor regions (Fig. 2A,B). Although most of the programs identify almost all the existing genes (as evidenced by the Sn and MG statistics), there is significant variation in their ability to accurately predict precise gene structures (see the Sp statistics, particularly at the exon level). If any global performance conclusion can be drawn, it is that the probabilistic gene finders (mostly HMM based) seem to be more reliable. The integration of EST/cDNA sequence information into the ab initio gene finders [see HMMGene, GenieEST, and GRAIL (Fig. 2A,B and Fig. 3A–D)] significantly improves gene predictions, particularly the recognition of intron–exon boundaries. Some groups submitted multiple annotations of the Adh region using programs that were tuned for different tasks. The suite of Fgenes programs shows very nicely the results of such a three-part submission. The first Fgenes submission (Fgenes1) is a version adjusted to weight Sn and Sp equally. The second submission (Fgenes2) is very conservative and only annotates high-scoring genes. This results in a high Sp but a low Sn. The third submission (Fgenes3) tries to maximize Sn and to avoid missing any genes, at the cost of a loss in Sp. These differently tuned variants may be useful for different types of tasks.
A comparison (data not shown) to a gene-finding system that was trained on human data showed that it did not perform as well as the programs that were trained on Drosophila data.
None of the gene predictors screened for transposable elements, which have a protein-like structure. As described in Ashburner et al. (1999b), the Adh region has 17 transposable element sequences. Eliminating transposons from the predictions or adding them to the standard sets would have reduced the FP counts, raising the Sp and lowering the WE and WG scores. Although this accounts for a portion of the high FP scores, we believe that there may also be additional genes in this region not annotated in std3. Future biological experiments (Rubin 2000) to identify and sequence the predicted genes that were not included in std3 should improve the completeness and accuracy of the final annotations.
There were fewer submissions of homology-based annotations than those by ab initio gene finders, and their results were significantly affected by their FP rates. A significant portion of those FPs were matches to transposable elements, some appear to be matches to pseudogenes, and others are likely to be real, but as yet unannotated, genes. The homology-based approaches seem to be the most promising techniques for inferring functions for newly predicted genes.
Even using EST/cDNA alignments to predict gene structures is not as simple as expected. Paralogs, low sequence quality of mRNAs, and the difficulty of cloning infrequently expressed mRNAs make this method of gene finding more complex than believed, and it is difficult to guarantee completeness with this method. Normalized cDNA libraries and other more sophisticated technologies to purify genes with low expression levels, along with improved alignment and annotation technologies, should improve predictions based on EST/cDNA alignments.
Lessons for the Future
To fully assess the submitted annotations, the correct answer must be improved. Only extensive full-length cDNA sequencing can accomplish this. A possible approach would be to design primers from predicted exons and/or genes in the genomic sequence and then use hybridization technologies to fish out the corresponding cDNA from cDNA libraries. For promoter predictions, another way to improve the correct answer is to make genome-to-genome alignments with the DNA of related species (e.g., Caenorhabditis briggsae vs. Caenorhabditis elegans; D. melanogaster vs. D. virilis). More detailed guidelines, including how to handle ambiguous features such as pseudogenes and transposons, will make the results of future experiments even more useful.
A successful system to identify all genes in a genome should consist of a combination of ab initio gene finding, EST/cDNA alignments, protein homology methods, promoter recognition, and repeat finding. All of the various technologies have advantages and disadvantages, and an automated method for integrating their predictions seems ideal.
Beyond the identification of gene structure is the determination of gene functions. Most of the existing prototypes of such systems are based on sequence homologies. Although this is a good starting point, it is definitely not sufficient. The state of the art for predicting function in protein sequences uses the protein's three-dimensional structure, but the difficulty of accurately predicting three-dimensional structure from primary sequences makes applying these techniques on complete genomes problematic. The new field of structural genomics will hopefully give more answers in these areas.
Another approach to function classification is the analysis of gene expression data. Improvements in TSS annotations, along with correlation in expression profiles, should be very helpful in identifying regulatory regions.
Conclusions
The GASP experiment succeeded in providing an objective assessment of current approaches to gene prediction. The main conclusions from this experiment are that current methods of gene predictions are tremendously improved and that they are very useful for genome scale annotations but that high-quality annotations also depend on a solid understanding of the organism in question (e.g., recognizing and handling transposons).
Experiments like GASP are essential for the continued progress of automated annotation methods. They provide benchmarks with which new technologies can be evaluated and selected.
The predictions collected in GASP showed that for most of the genes, overlapping predictions from different programs existed. Whether or not a combination of overlapping predictions would do better than the best performing individual program was not explicitly tested in this experiment. For such a test, additional experiments such as cDNA library screening and subsequent full-length cDNA sequencing in this selected Adh test bed region would be necessary. These experiments are currently under way, and it would be interesting to perform a second GASP experiment when more cDNAs have been sequenced.
We believe that existing automated annotation methods are scalable and that the ultimate test will occur when the complete sequence of the D. melanogaster genome becomes available. This experiment will set standards for the accuracy of genome-wide annotation and improve the credibility of the annotations done in other regions of the genome.
URLs
Gene Finding
HMMGene, http://www.cbs.dtu.dk/services/HMMGene/; GRAIL, http://compbio/ornl.gov/droso; Fgenes, http://genomic/sanger.ac.uk/gf/gf.shtml; GeneID, http://www1/imim.es/∼rguigo/AnnotationExperiment/index.html; Genie, http://www.neomorphic.com/genie.
Promoter Prediction
MCPromoter, http://www5.informatik.uni-erlangen.de/HTML/English/Research/Promoter; CoreInspector, http://www.gsf.de/biodv.
Protein Homology
BLOCKS+, http://blocks.fhcrc.org and http://blocks.fhcrc.org/blocks-bin/getblock.sh?<block name>; GeneWise, http://www.sanger.ac.uk/Software/Wise2/.
Repeat Finders
Acknowledgments
We thank all of the participants who submitted their annotations, without which the project would not have been such a success, for their original contributions, their publication, and their patience with the organizers during this very intense project. We also thank the Drosophila Genome Sequencing Center at LBNL, headed by Sue Celniker, for providing such high-quality sequence; the annotation team at the Berkeley Drosophila Genome Sequencing Center and especially Sima Misra, Gerry Rubin, and Michael Ashburner; and the entire Drosophila community for producing such a thoroughly studied genomic region. Special thanks go to the independent assessor team, consisting of Michael Ashburner, Peer Bork, Richard Durbin, Roderic Guigó, and Tim Hubbard, who critiqued our evaluation. Thanks goes also to the organizers of ISMB-99 Heidelberg, especially Thomas Lengauer and Reinhard Schneider, for encouraging our tutorial and the tremendous support in the preparation process and during the conference. We also thank Richard Durbin, David Haussler, Tim Hubbard, and Richard Bruskiewich for developing and maintaining the GFF format and their associated tools. Last but not least, a big thank you goes to Gerry Rubin for making the Drosophila Genome Project such a success. This work was supported by NIH grant HG00750.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mgreese@lbl.gov; FAX (510) 486-6798.
REFERENCES
- Agarwal P, States DJ. Comparative accuracy of methods for protein sequence similarity search. Bioinformatics. 1998;14:40–47. doi: 10.1093/bioinformatics/14.1.40. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arkhipova I R. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–1369. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. 2000. A biologist's view of the Drosophila genome annotation assessment. Genome Res. (this issue). [DOI] [PubMed]
- Ashburner M, Bork P, Durbin R, Guigó R, Hubbard TJ. GASP1 assessment meeting. Heidelberg, Germany: EMBL; 1999a. [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of drosophila melanogaster. The adh region. Genetics. 1999b;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. et al. 1999c. European Drosophila Genome Project (EDGP). http://edgp.ebi.ac.uk/.
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam Protein Families Database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. 1999. Wise2. http://www.sanger.ac.uk/Software/Wise2/.
- Birney E, Durbin R. Dynamite: A flexible code generating language for dynamic programming methods used in sequence comparison. Intell Syst Mol Biol. 1997;5:56–64. [PubMed] [Google Scholar]
- ———. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- ————— Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Burset M, Guigó R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- Cavin Périer R, Junier T, Bonnard C, Bucher P. The Eukaryotic Promoter Database (EPD): Recent developments. Nucleic Acids Res. 1999;27:307–309. doi: 10.1093/nar/27.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavin Périer R, Praz V, Junier T, Bonnard C, Bucher P. The Eukaryotic Promoter Database (EPD) Nucleic Acids Res. 2000;28:302–303. doi: 10.1093/nar/28.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack RL, Jr, Gerloff DL, Bower M, Chen X, Lichtarge O, Cohen FE. Meeting review: The Second meeting on the critical assessment of techniques for protein structure prediction (CASP2), Asilomar, California, December 13–16, 1996. Folding Design. 1997;2:R27–R42. doi: 10.1016/S1359-0278(97)00011-4. [DOI] [PubMed] [Google Scholar]
- Eeckman FH, Durbin R. ACeDB and macace. Methods Cell Biol. 1995;48:583–605. [PubMed] [Google Scholar]
- Fickett JW, Tung CS. Assessment of protein coding measures. Nucleic Acids Res. 1992;20:6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett JW, Hatzigeorgiou AG. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaasterland T, Sensen CW. MAGPIE: Automated genome interpretation. Trends Genet. 1996;12:76–78. doi: 10.1016/0168-9525(96)81406-5. [DOI] [PubMed] [Google Scholar]
- Guigó R, Knudsen S, Drake N, Smith T. Prediction of gene structure. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- Harris, N.L., G. Helt, S. Misra, and S.E. Lewis. 1999. CloneCurator. http://www.fruitfly.org/displays/CloneCurator.html.
- Helt G, Blossom E, Morris J, Fineman D, Cherritz S, Shaw S, Harmon CL. Neomorphic Genome Software Development Toolkit (NGSDK). Berkeley, CA: Neomorphic, Inc.; 1999. http://www.neomorphic.com . [Google Scholar]
- Henikoff S, Henikoff JG. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Genomic sequence annotation based on translated searching of the Blocks+ Database. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Henikoff JG, Henikoff S, Pietrokovski S. New features of the Blocks Database servers. Nucleic Acids Res. 1999a;27:226–228. doi: 10.1093/nar/27.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Blocks+: A non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999b;15:471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repeats in genomic DNA: Mining and meaning. Curr Opin Struct Biol. 1998;8:333–337. doi: 10.1016/s0959-440x(98)80067-5. [DOI] [PubMed] [Google Scholar]
- Krogh A. Two methods for improving performance of an HMM and their application for gene finding. Ismb. 1997;5:179–186. [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. Integrating database homology in a probabilistic gene structure model. Pac Symp Biocomput. 1997;2:232–244. [PubMed] [Google Scholar]
- Kurtz S, Schleiermacher C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- Levitt M. Competitive assessment of protein fold recognition and alignment accuracy. Proteins (Suppl.) 1997;1:92–104. doi: 10.1002/(sici)1097-0134(1997)1+<92::aid-prot13>3.3.co;2-1. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comp Appl Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Moult J, Hubbard T, Bryant SH, Fidelis K, Pedersen JT. Critical assessment of methods of protein structure prediction (CASP): Round II. Proteins (Suppl.) 1997;1:2–6. [PubMed] [Google Scholar]
- Moult J, Hubbard T, Fidelis K, Pedersen JT. Critical assessment of methods of protein structure prediction (CASP): Round III. Proteins (Suppl.) 1999;3:2–6. [PubMed] [Google Scholar]
- Ohler U, Harbeck S, Niemann H, Noth E, Reese MG. Interpolated markov chains for eukaryotic promoter recognition. Bioinformatics. 1999;15:362–369. doi: 10.1093/bioinformatics/15.5.362. [DOI] [PubMed] [Google Scholar]
- Ohler U, Stommer G, Harbeck S. Stochastic segment models of eukaroyotic promoter regions. Pac Symp Biocomput. 2000;5:377–388. doi: 10.1142/9789814447331_0036. [DOI] [PubMed] [Google Scholar]
- Parra, G., E. Blanco, and R. Guigó. 2000. GeneID in Drosophila. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Pearson WR. Comparison of methods for searching protein sequence databases. Protein Sci. 1995;4:1145–1160. doi: 10.1002/pro.5560040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG. “Genome annotation in Drosophila melanogaster.” Ph.D. thesis. Germany: University of Hohenheim; 2000. [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Reese MG, Harris NL, Hartzell G, Lewis SE. The 7th conference on Intelligent Systems in Molecular Biology (ISMB'99), Heidelberg, Germany. 1999. http://www.fruitfly.org/GASP , http://www.fruitfly.org/GASP. . [Google Scholar]
- Reese, M.G., D. Kulp, H. Tammana, and D. Haussler. 2000. Genie–Gene finding in Drosophila melanogaster. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Rubin, G.M. 2000. Full-length cDNA project. http://www.fruitfly.org/EST
- Rubin GM, et al. Berkeley Drosophia Genome Project (BDGP). 1999. http://www.fruitfly.org http://www.fruitfly.org. . [Google Scholar]
- Salamov, A.A. and V.V. Solovyev. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Sippl MJ, Lackner P, Domingues FS, Koppensteiner WA. An attempt to analyse progress in fold recognition from CASP1 to CASP3. Proteins (Suppl.) 1999;3:226–230. doi: 10.1002/(sici)1097-0134(1999)37:3+<226::aid-prot29>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- Solovyev VV, Salamov AA, Lawrence CB. Identification of human gene structure using linear discriminant functions and dynamic programming. Ismb. 1995;3:367–375. [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Thierry-Mieg J. Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res. 1998;8:1308–1315. doi: 10.1101/gr.8.12.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo, G.D. 2000. Gene-finding approaches for eukaryotes. Genome Res. (this issue). [DOI] [PubMed]
- Uberbacher EC, Mural RJ. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A, Venclovas C, Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins (Suppl.) 1999;3:22–29. doi: 10.1002/(sici)1097-0134(1999)37:3+<22::aid-prot5>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]