Abstract
A differentially methylated region (DMR) and endoderm-specific enhancers, located upstream and downstream of the mouse H19 gene, respectively, are known to be essential for the reciprocal imprinting of Igf2 and H19. To explain the same imprinting patterns in non-endodermal tissues, additional enhancers have been hypothesized. We determined and compared the sequences of human and mouse H19 over 40 kb and identified 10 evolutionarily conserved downstream segments, 2 of which were coincident with the known enhancers. Reporter assays in transgenic mice showed that 5 of the other 8 segments functioned as enhancers in specific mesodermal and/or ectodermal tissues. We also identified a conserved 39-bp element that appeared repeatedly within the DMR and formed complexes with specific nuclear factors. Binding of one of the factors was inhibited when the target sequence contained methylated CpGs. These complexes may contribute to the presumed boundary function of the unmethylated DMR, which is proposed to insulate maternal Igf2 from the enhancers. Our results demonstrate that comparative genomic sequencing is highly efficient in identifying regulatory elements.
[The sequence data described in this paper have been submitted to GenBank under accession nos. AF087017 and AF049091.]
The mouse Igf2 and H19 genes lie 70 kb apart (Fig. 1A) within a large imprinted domain on distal chromosome 7 (Kato et al. 1999). These two genes are reciprocally imprinted, with Igf2 being paternally expressed and H19 being maternally expressed. The mechanisms of the reciprocal imprinting of the two genes have been studied extensively by introducing germ-line mutations into this imprinted domain. The first such study by Leighton et al. (1995a) showed that a 13-kb deletion of the H19 region caused disruption of the imprinted expression of Igf2 (and Ins2) indicating that Igf2 imprinting is dependent on the H19 region. This deletion contained the entire H19 transcription unit of 3 kb and its upstream region of 10 kb. The result was consistent with the enhancer competition model (Bartolomei and Tilghman 1992; Sasaki et al. 1992), in which the imprinting of the two genes is explained by their competition for common enhancers.
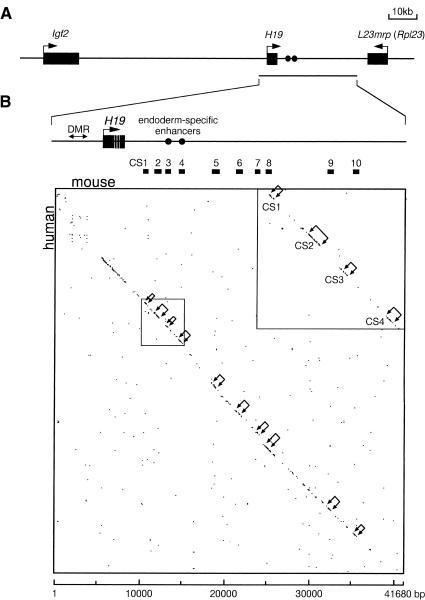
Figure 1.
Comparison of the human and mouse H19 region sequences. (A) Map of the Igf2/H19 region. The region analyzed is indicated by a horizontal line below the map. (B) Dot matrix alignment. (Top) Lower magnification of the H19 region. The homology plot program DNASIS v3.0 (Hitachi Software Engineering) was used to align the mouse (x axis) and human (y axis) H19 sequences with a criterion of 22 matching bases in a window of 30. Brackets with arrows in the matrix and bars between the map and matrix indicate conserved segments (CS1–CS10). (Inset) Higher magnification of the CS1–CS4 region. The GenBank sequences used are as follows: mouse, AF049091 (bases 1–41680); human, AF087017 (bases 1–40558) and AC004556 (bases 82896–88682).
Subsequent studies in mice revealed that two regulatory regions are essential for the imprinting of Igf2 and H19. First, two endoderm-specific enhancers located 5–7 kb downstream of H19 (Yoo-Warren et al. 1988; Fig. 1A,B) were indispensable for the expression and imprinting of both genes in endodermal tissues (Leighton et al. 1995b). Second, a 2-kb differentially methylated region (DMR) located 2-kb upstream of H19 was essential for the imprinting of an H19 transgene (Elson and Bartolomei 1997) or both endogenous Igf2 and H19 (Thorvaldsen et al. 1998). This DMR is unmethylated when maternally derived and methylated when paternally derived. Several lines of evidence from independent experiments suggested that the DMR plays dual roles in imprinting: A methylated DMR on the paternal chromosome appears to act as an inactivation center that methylates and silences the neighboring H19 whereas an unmethylated DMR on the maternal chromosome may serve as a boundary element or a chromatin insulator that blocks the interaction between Igf2 and the downstream enhancers (Thorvaldsen et al. 1998; Webber et al. 1998; Schmidt et al. 1999).
In the present study, we attempted to identify new regulatory sequences involved in the imprinting of the two genes by comparative genomic sequencing. Although both Igf2 and H19 are expressed and imprinted in many mesodermal tissues, only endoderm-specific enhancers have been identified. Therefore, we first looked for the hypothetical nonendodermal enhancers. Then, we attempted to identify sequence elements within the DMR that are conserved through evolution and bound by nuclear factors. Our results showed that comparative sequencing is a highly efficient approach to identify potential regulatory elements located several kilobases to several tens of kilobases away from genes.
RESULTS
To look for hypothetical enhancers of the imprinted domain and functional elements within the DMR, we decided to take a comparative sequencing approach. Previous studies with transgenic mice carrying a YAC transgene suggested that the hypothetical nonendodermal enhancers are located within a 130-kb region extending from promoter 1 of Igf2 to 35 kb downstream of H19 (Ainscough et al. 1997). A more recent study showed that the location of the enhancers is essential for the imprinting of both Igf2 and H19 (Webber et al. 1998), suggesting that the additional enhancers may be located downstream of H19. Therefore, we cloned and sequenced a 41-kb human genomic region extending from 8 kb upstream to 30 kb downstream of H19. The obtained sequence (GenBank accession no. AF087017) plus an additional 6-kb sequence from a human PAC clone (GenBank accession no. AC004556) was compared with the corresponding mouse sequence, which we had reported previously (GenBank accession no. AF049091; Ishihara et al. 1998).
Evolutionarily Conserved Downstream Segments
A homology plot analysis of the human and mouse sequences using DNASIS software (v3.0, Hitachi Software Engineering) revealed that a 28-kb region downstream of the H19 gene contained a number of conserved stretches at homologous positions (Fig. 1B). Higher magnification analyses (e.g., see Fig. 1B, inset) identified 10 discrete conserved segments (Fig. 1B, CS1–CS10) spaced with gap regions consisting of nonhomologous sequences. The conserved segments were 200–500 bp in size and their sequence identity between human and mouse ranged from 68% to 85%, on the basis of maximum matching alignments by DNASIS (Table 1). Unlike protein coding regions, these segments lacked the third base wobble and contained many deletions and insertions. Notably, this analysis identified the two known enhancers as CS3 and CS4, raising the possiblity that the other conserved segments may also be enhancers.
Table 1.
Evolutionarily Conserved Segments Downstream of H19 and the Summary of Transgenesis Experiments
| Segment | Positiona | Size (bp)a | Sequence identity (%)b | No. of embryo | Major sites of expression | Tissue-specific enhancer activity | |
|---|---|---|---|---|---|---|---|
| with integration | with expression | ||||||
| CS1 | 10344–10571 | 228 | 78 | 15 | 5 | ganglionic placode of cranial nerves | + |
| CS2 | 11605–12076 | 472 | 78 | 9 | 5 | various | − |
| CS3 | 12846–13089 | 244 | 72 | 9 | 4 (+1)c | sclerotome | + |
| CS4 | 14280–14664 | 385 | 68 | 16 | 2 (+3)c | liver, gut | + |
| CS5 | 18151–18535 | 385 | 70 | 4 | 3 | ectoderm of limb buds, neural tube floor plate | + |
| CS6 | 21142–21501 | 360 | 68 | 6 | 5 | myotome, rib primordia, intercostal muscles | + |
| CS7 | 23733–23963 | 231 | 74 | 10 | 7 | mesenchymal condensation of limb buds | + |
| CS8 | 24834–25111 | 278 | 85 | 11 | 4 | various | − |
| CS9 | 32288–32773 | 486 | 71 | 8 | 6 | myotome, rib primordia, intercostal muscles | + |
| CS10 | 35463–35748 | 286 | 79 | 9 | 0 | — | − |
Nucleotide position and segment size are given according to the mouse sequence (GenBank accession no. AF049091).
Nucleotide sequence identity between the human and mouse sequences is shown.
Number of embryos that showed ectopic expression only is indicated in parentheses.
Enhancer Assay in Transgenic Mice
To test whether the newly identified conserved segments can function as enhancers in transgenic mice, mouse DNA fragments (0.47–1.54 kb in size) containing the respective segments were linked to a lacZ reporter gene (Fig. 2A; Hatano et al. 1998) and introduced into fertilized eggs. Embryos that developed from the injected zygotes were recovered at 12–12.5 days postcoitum (dpc) and examined for lacZ expression. No embryo was allowed to develop to term to give a transgenic line. Thus, in this assay, DNA fragments directing a consistent tissue-specific expression pattern in multiple independent embryos were judged as possessing enhancer activity.
Figure 2.
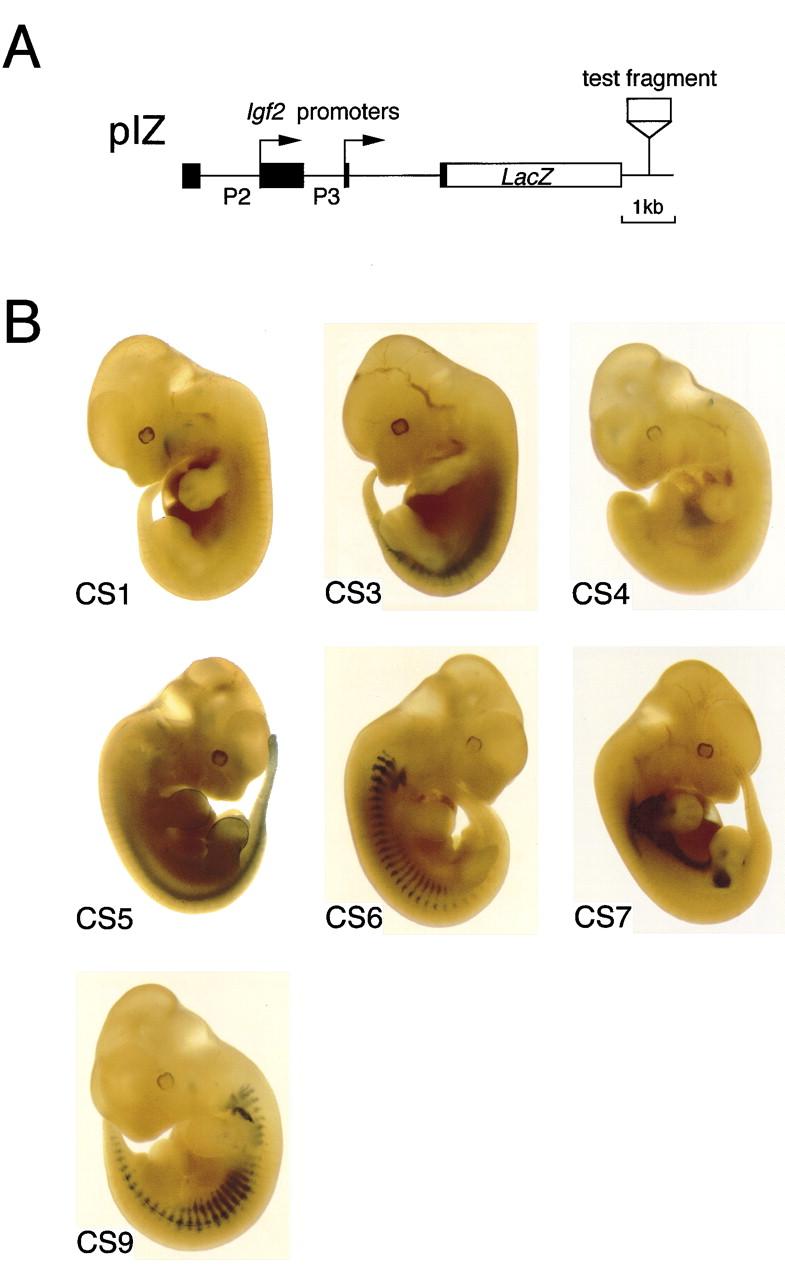
Expression of the lacZ reporter gene driven by the conserved downstream segments in transgenic mice. (A) Structure of the reporter construct. Each transgene construct was made by insertion of one of the conserved segments into plasmid pIZ (Hatano et al. 1998). (B) Representative patterns of lacZ staining in 12- to 12.5-dpc embryos. CS1 transgenes reproducibly stained the ganglionic placodes of the facial nerve (near the first branchial groove) and the inferior edge of the maxillary prominences. CS3 transgenes were expressed in the sclerotome, and CS4 transgenes in the liver and the epithelial layer of the gut. CS5 directed lacZ expression in the neural tube floor plate as well as in the ectoderm of the limb buds. Both CS6 and CS9 transgenes expressed lacZ in the myotome and the rib primordia. CS7 transgenes were expressed in the mesenchyme condensations at the bases of the limb buds. The expression patterns were confirmed by histological examinations.
We first examined whether CS3 and CS4, corresponding to the known enhancers, function as enhancers in endodermal tissues. Four of the five embryos carrying CS3 transgenes were stained for lacZ activity in the sclerotome (a mesodermal tissue) while the last one showed an ectopic expression pattern (Fig. 2B; Table 1). This result is consistent with the recent observation that transgenes containing both CS3 and CS4 were expressed in the sclerotome (Hatano et al. 1998; Brenton et al. 1999). However, little, if any, enhancer activity was detectable in endodermal tissues such as the liver or gut. Two embryos carrying CS4 transgenes showed specific staining in the liver and gut although three others were stained in various patterns (Fig. 2B; Table 1). This result suggested that the endoderm-specific enhancer activity of CS4 was easily influenced by chromosomal environment.
Then, we examined the eight newly identified segments in embryos and found that five (CS1, CS5–CS7 and CS9) exhibited enhancer activity in nonendodermal tissues (Fig. 2B; Table 1). For example, CS1 transgenes reproducibly stained the ganglionic placodes of the facial nerve and the maxillary prominences (ectoderm origin). CS5 directed lacZ expression in the ectoderm of the limb buds and in the neural tube floor plate (Fig. 2B), and CS6 and CS9 showed almost identical staining patterns in the myotome and primordia of ribs (mesoderm origin, Fig. 2B). Also, CS7 transgenes were expressed in masses of mesenchymal (mesoderm) cells. (For details, see Fig. 2 legend and Table 1.) No enhancer activity was detected for CS2, CS8, and CS10 at this stage of development because they did not drive expression of lacZ (CS10) or only gave position-dependent variable expression (CS2 and CS8; Table 1). Thus at least seven tissue-specific enhancers (CS1, CS3-CS7, and CS9), including the two previously known ones, form a large regulatory region reminiscent of the locus control region of the β-globin gene cluster (Li et al. 1999).
Conserved Upstream Elements
Previous studies in mice showed that a 2-kb DMR located upstream of H19 (Figs. 1B and 3B) is essential not only for the methylation and silencing of paternal H19 but also for the prevention of the use of the downstream enhancers by maternal Igf2 (Thorvaldsen et al. 1998). This DMR is located just upstream of a 461-bp G-rich repeat region containing 32 copies of a consensus sequence (G)GGGGTATA (Tremblay et al. 1995; Fig. 3B). Although human H19 also has an upstream DMR, its structure is completely different from that of the mouse DMR (Jinno et al. 1996). For example, the G-rich repeat found adjacent to the mouse DMR is not present in human H19 (Fig. 3B). Furthermore, the human DMR contains a duplication of a region containing a 400-bp sequence and three copies of another 400-bp sequence with one duplication unit lacking a small part of the other (Jinno et al. 1996; Frevel et al. 1999a; Fig. 3B). These 400-bp sequences are not present in the mouse genome or in any other part of the human genome (Jinno et al. 1996).
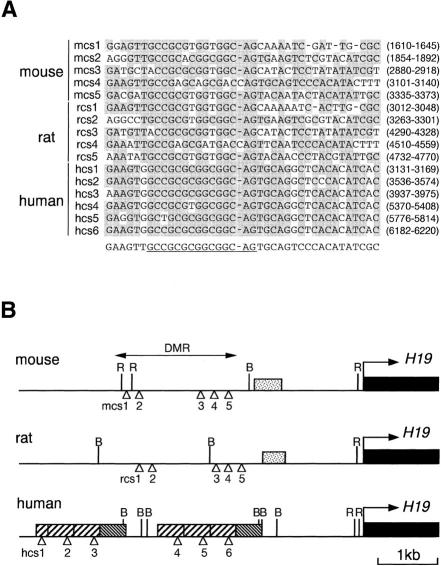
Figure 3.
Evolutionarily conserved 39-bp sequence elements in the upstream DMR of H19. (A) Sequence alignment of the elements conserved in human (hcs1–hcs6), mouse (mcs1–mcs5) and rat (rcs1–rcs5). The bases identical to the consensus sequence (bottom) are shaded. The highly conserved core of the consensus sequence is underlined. Nucleotide positions (in parentheses) follow numbering schemes for AF087017 (human), AF049091 (mouse), and AF043428 (rat). (B) Map of the upstream DMR of human, mouse, and rat H19. (Stippled boxes) G-rich repeat region found in rodents; (hatched boxes) two types of the human 400-bp repeat sequences; (open arrowheads) locations of the conserved 39-bp sequences. B, BamHI; R, EcoRI.
Although no cross-species homology was revealed initially (Jinno et al. 1996), our closer examination of the DMR by homology plot analysis identified a 39-bp sequence that had a high degree of similarity between human, mouse, and rat (Fig. 3A). This sequence element was repeated six times in human (as part of a 400-bp direct repeat, Jinno et al. 1996; Frevel et al. 1999a) and five times in mouse and rat (Fig. 3B). Two recent papers pointed out the presence of some, but not all, of these elements (mcs1, mcs2, mcs3, and mcs5, Stadnick et al. 1999; mcs2, Frevel et al. 1999b). Interestingly, the highly conserved 15-bp core region of the consensus sequence contains four methylatable CpG sites (Fig. 3A). Moreover, four (mcs1–mcs3 and mcs5) of the five mouse elements were associated respectively with a major maternal-specific DNase I hypersensitive site (Hark and Tilghman 1998; Khosla et al. 1999), suggesting that they might bind a regulatory protein(s).
Electrophoretic Mobility Shift Assay
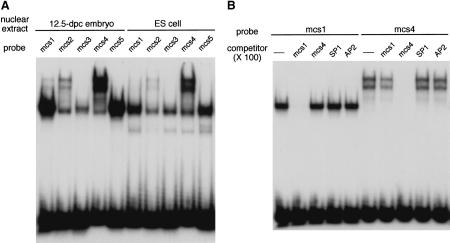
To examine whether the conserved mouse elements mcs1–mcs5 form complexes with specific nuclear factors, electrophoretic mobility shift assays (EMSAs) were carried out with duplexed oligonucleotides containing the core regions. Nuclear extracts from 12.5-dpc mouse embryos and ES cells contained several factors that formed complexes with the duplexes (Fig. 4A). The major complex formed with mcs1, mcs3, and mcs5 showed very similar, if not identical, mobilities (Fig. 4A), suggesting that identical or very similar factors were bound. This suggestion was confirmed by cross competition assays in which excess unlabeled mcs1, for example, competed with the mcs3 or mcs5 duplexes for formation of the complexes (data not shown). Similarly, mcs2 and mcs4 appeared to form multiple complexes with identical or similar proteins as determined by their electrophoretic mobilities (Fig. 4A) and cross competition (data not shown). The mcs1 duplex was also capable of binding these multiple factors with low efficiency (Fig. 4A). Lack of cross competition between mcs1 and mcs4 indicates that the factors binding to the two sequence categories are different (Fig. 4B).
Figure 4.
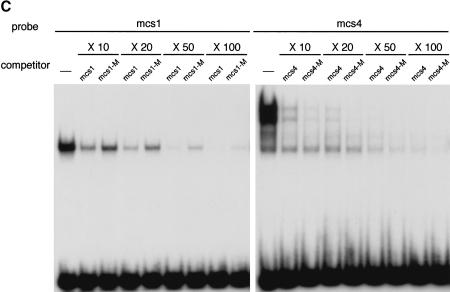
Formation of complexes between the conserved upstream elements and nuclear factors revealed by EMSA. (A) The five mouse elements, mcs1–mcs5, form complexes with specific nuclear proteins in embryos and ES cells. Nuclear extracts prepared from 12.5-dpc mouse embryos and ES cells were incubated with 32P-labeled mcs1–mcs5 oligonucleotide duplexes. The affinity between the factors and elements differed greatly. The patterns obtained with the two different extracts appeared identical except that the ES cell extracts gave one or two additional faint bands with higher mobilities. (B) Lack of cross-competition between mcs1, mcs4, SP1, and AP2 duplexes. (C) Methylation of CpG in mcs1 reduces complex formation. Effects of methylation were not evident for mcs4.
Two transcription factors, SP1 and AP2, are known to bind to CpG-containing sequences. However, excess unlabeled SP1 or AP2 duplexes gave no competition with the mcs1 and mcs4 duplexes (Fig. 4B). This result indicates that these factors do not contribute to the complexes and suggests that the factors binding to the conserved elements are novel.
Finally, we examined the effect of CpG methylation of the target sequences by adding excess in vitro methylated duplexes as competitors. The results showed that methylated mcs1 was a less effective competitor for complex formation than unmethylated mcs1 (Fig. 4C). This result indicates that the binding affinity of the factor(s) to the target is greatly reduced by CpG methylation. In contrast, the factors binding to mcs4 appeared insensitive to methylation (Fig. 4C).
DISCUSSION
We have used a comparative genomic sequencing approach combined with functional assays such as a reporter assay in transgenic mice and EMSA to look for regulatory sequences in the imprinted mouse Igf2/H19 domain. We identified five novel tissue-specific enhancers and five novel nuclear factor binding elements in a 40-kb H19 region. This success clearly demonstrates the effectiveness of the approach. The identified sequences are likely to be involved in the reciprocal imprinting of Igf2 and H19 although the formal proof for this awaits germ-line deletion experiments in mice.
The expression pattern of the lacZ transgene driven by each enhancer was in most cases consistent with that of a 130-kb YAC transgene containing a lacZ reporter at the Igf2 locus (Ainscough et al. 1997) and that of endogenous Igf2 (Stylianopoulou et al. 1988; Lee et al. 1990). However, we observed some minor differences, and the full expression pattern of endogenous Igf2 could not be obtained even if all patterns were put together. For example, none of the enhancer segments showed strong expression in the limb mesenchyme, where the YAC transgene and endogenous Igf2 are highly expressed. It is possible that there exists an unidentified enhancer, or a specific enhancer set may produce this pattern as a combinatorial effect. Because we examined only 12- to 12.5-dpc embryos, it is also possible that there are additional enhancers that act at other developmental stages. In fact, three conserved downstream segments did not show enhancer activity in 12- to12.5-dpc embryos. However, an alternative explanation may be that these segments encode other regulatory functions such as a chromatin insulator or a silencer.
The discovery of a 28-kb regulatory region containing a cluster of at least seven enhancers is reminiscent of the β-globin LCR, which contains five DNase I-hypersensitive sites in a 16-kb region upstream of the β-globin gene cluster (Li et al. 1999). Among the five hypersensitive sites, three are known to be erythroid-specific enhancers. Unlike the β-globin LCR, however, most of the enhancers of the H19 downstream region have distinctive tissue-specificity and those of only CS6 and CS9 overlap. In general, LCRs are defined as DNA regions that confer high level, tissue-specific, site-of-integration-independent, copy number-dependent expression on linked genes in ectopic chromatin sites (Li et al. 1999). It will be interesting to test whether the identified region has these properties.
The identification of the binding sites for methylation-sensitive nuclear factors within the DMR has implications for mechanistic models of the Igf2/H19 imprinting. The original enhancer competition model predicted that the DMR works as an inactivation center when methylated (on the paternal chromosome), but does not play any positive role when unmethylated (on the maternal chromosome). However, on the basis of observations made with mutated mice, a modified version of the model was proposed. In this model, the maternally derived unmethylated DMR works as a chromatin boundary or insulator element that blocks the engagement of the downstream enhancers to maternal Igf2 (Thorvaldsen et al. 1998; Webber et al. 1998; Schmidt et al. 1999). It is conceivable that the complexes formed on the conserved elements, especially those containing the methylation-sensitive factors, are part of the methylation-regulated insulator complex. In this regard, it is interesting that the first example of vertebrate insulator protein was recently identified (Bell et al. 1999), although its binding sequences did not show significant homology to our elements.
It is well known that important regulatory sequences are highly conserved through evolution. With the progress of the Human Genome Project, large-scale comparative sequencing has become a practical means to look for potential regulatory regions. A number of recent works described the presence of conserved noncoding sequences at orthologous loci from different species such as humans, rodents, Fugu (pufferfish), and nematodes. Some studies provided further functional evidence that the identified sequences were indeed regulatory elements (e.g., Oeltjen et al. 1997; Thacker et al. 1999). In the present study, we demonstrated that large-scale comparative sequencing is extremely efficient in identifying elements such as tissue-specific enhancers and nuclear factor binding sites when combined with appropriate functional assays. This approach appears especially useful in studying organization and regulation of large chromatin domains such as imprinted domains.
METHODS
Cosmid Cloning and DNA Sequencing
A human fetal brain genomic library carried by the SuperCos 1 cosmid vector (Stratagene) was screened with a mixture of two PCR-amplified genomic fragments from the human H19 region by colony hybridization. One probe fragment was from the exon 1 region (sense primer H19-h1, 5′-cacttttggttacaggacgtggc-3′; antisense primer H19-h2, 5′-atacagcgtcaccaagtccactg-3′) and the other was from a region 13 kb downstream from H19 (sense primer H19-E1, 5′-tctggaatgtggggaggcaaaca-3′; anti-sense primer H19-E2, 5′-ctggcgaaagcatgaggcaagat-3′). The primers were designed on the basis of published sequence information (Brannan et al. 1990) and our partial sequence data obtained from a phage clone (K. Miura and Y. Jinno, unpubl.). Ten positive cosmid clones were picked up, and seven of them were verified to contain H19 by Southern blotting. Two overlapping clones cHH1 and cHH6 were used to produce a detailed restriction map. Restriction fragments from the two cosmids were subcloned into pBluescript plasmids and sequenced by use of the Dye Primer Cycle Sequencing Kit and an ABI PRISM 377 Sequencer (Perkin Elmer). The obtained sequences were analyzed by use of DNASIS software (v3.0; Hitachi Software Engineering).
Transgenic Mice
Plasmids containing the transgenes were constructed by insertion of DNA fragments to be assayed into the pIZ vector (previously called pIZ-A, Hatano et al. 1998). Transgene fragments were liberated by restriction digestion, fractionated in agarose gels, and recovered by electroelution. Manipulation of mouse embryos, genotyping by PCR, and staining for lacZ activity were done as described previously (Hatano et al. 1998), except that the staining reaction was done at 30°C to reduce background signals. Histological sections were prepared and observed by standard procedures.
EMSA
Cell nuclei from 12.5-dpc mouse embryos were isolated as described (Sasaki et al. 1992). Nuclei from ES cells were prepared as follows. ES cells were suspended in five packed cell volumes of buffer A [10 mm HEPES-KOH (pH 7.6), 10 mm KCl, 0.5 mm EDTA, 1 mm dithiothreitol (DTT), 0.2 mm phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, 2 μg/ml pepstatin A, 2 μg/ml leupeptin], homogenized with an all-glass Dounce homogenizer (20 strokes) and layered over an equal volume of buffer B (buffer A containing 0.5 M sucrose). Nuclei were then pelleted at 5,000 rpm for 15 min in a Beckman SW41Ti rotor. Nuclei from both embryos and ES cells were suspended in an equal volume of buffer C [10 mm HEPES-KOH (pH 7.6), 0.4 M KCl, 0.1 mm EDTA, 3 mm MgCl2, 1 mM DTT, 0.2 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml pepstatin A, 2 μg/ml leupeptin], mixed gently for 30 min, and centrifuged for 30 min at 15,000g. The supernatant was dialyzed overnight against buffer D [20 mm HEPES-KOH (pH 7.6), 100 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, 20% glycerol, 0.2 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml pepstatin A] and centrifuged for 10 min at 15,000g. The supernatant, designated the nuclear extract, was frozen as aliquots and stored at −80°C. EMSA was carried out using the Gel Shift Assay Core System (Promega) according to the manufacturer's protocol. Oligonucleotide duplexes used were: mcs1, 5′-ggagttgccgcgtggtggcagcaa-3′; mcs2, 5′-agggttgccgcacggcggcagtga-3′; mcs3, 5′- gatgctaccgcgcggtggcagcat-3′; mcs4, 5′-gaagttgccgagcagcgaccagtgc-3′; mcs5, 5′-gacgatgccgcgtggtggcagtac-3′; SP1, 5′-attcgatcggggcggggcgagc-3′; AP2, 5′-gatcgaactgaccgcccgcggcccgt-3′. Methylation of duplexes was carried out by use of SssI methylase (New England Biolabs) according to the supplier's instructions. The methylation reaction was monitored by digestion of the duplex with the methylation-sensitive restriction enzyme AciI (CCGC), whose recognition site was present in all duplexes but mcs4.
Acknowledgments
We are grateful to Nao Aoki for maintaining the mouse colonies, Kazue Hatanaka for histological preparations, and Akiko Iwaki, Yasuyuki Fukumaki, and Kenshi Hayashi for discussion and encouragement. K.I. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan, and the Ministry of Health and Welfare of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hisasaki@lab.nig.ac.jp; FAX: 81-(0)559-81-6800.
REFERENCES
- Ainscough JF-X, Koide T, Tada M, Barton SC, Surani MA. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124:3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM. Parental imprinting of mouse chromosome 7. Semin Dev Biol. 1992;3:107–117. [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton JD, Drewell RA, Viville S, Hilton KJ, Barton SC, Ainscough JF-X, Surani MA. A silencer element identified in Drosophila is required for imprinting of H19 reporter transgenes in mice. Proc Natl Acad Sci USA. 1999;96:9242–9247. doi: 10.1073/pnas.96.16.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson DA, Bartolomei MS. A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol Cell Biol. 1997;17:309–317. doi: 10.1128/mcb.17.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frevel MAE, Sowerby SJ, Petersen GB, Reeve AE. Methylation sequencing analysis refines the region of H19 epimutation in Wilms Tumor. J Biol Chem. 1999a;247:29331–29340. doi: 10.1074/jbc.274.41.29331. [DOI] [PubMed] [Google Scholar]
- Frevel MAE, Hornberg JJ, Reeve AE. A potential imprint control element: Identification of a conserved 42-bp sequence upstream of H19. Trends Genet. 1999b;15:216–218. doi: 10.1016/s0168-9525(99)01752-7. [DOI] [PubMed] [Google Scholar]
- Hark AT, Tilghman SM. Chromatin conformation of the H19 epigenetic mark. Hum Mol Genet. 1998;7:1979–1985. doi: 10.1093/hmg/7.12.1979. [DOI] [PubMed] [Google Scholar]
- Hatano N, Eversole-Cire P, Ferguson-Smith AC, Jones PA, Surani MA, Sasaki H. Enhancer-dependent, locus-wide regulation of the imprinted mouse insulin-like growth factor II gene. J Biochem. 1998;123:984–991. doi: 10.1093/oxfordjournals.jbchem.a022034. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Kato R, Furuumi H, Zubair M, Sasaki H. Sequence of a 42-kb mouse region containing the imprinted H19 locus: Identification of a novel muscle-specific transcription unit showing biallelic expression. Mamm Genome. 1998;9:775–777. doi: 10.1007/s003359900863. [DOI] [PubMed] [Google Scholar]
- Jinno Y, Sengoku K, Nakao M, Tamate K, Miyamoto T, Matsuzaka T, Sutcliffe JS, Anan T, Takuma N, Nishiwaki K, Ikeda Y, Ishimaru T, Ishikawa M, Niikawa N. Mouse/human sequence divergence in a region with a paternal-specific methylation imprint at the human H19 locus. Hum Mol Genet. 1996;5:1155–1161. doi: 10.1093/hmg/5.8.1155. [DOI] [PubMed] [Google Scholar]
- Kato R, Shirohzu H, Yokomine T, Mizuno S, Mukai T, Sasaki H. Sequence-ready 1-Mb YAC, BAC and cosmid contigs covering the distal imprinted region of mouse chromosome 7. DNA Res. 1999;6:401–405. doi: 10.1093/dnares/6.6.401. [DOI] [PubMed] [Google Scholar]
- Khosla S, Aitchison A, Gregory R, Allen ND, Feil R. Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Mol Cell Biol. 1999;19:2556–2566. doi: 10.1128/mcb.19.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Pintar J, Efstratiadis A. Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development. 1990;110:151–159. doi: 10.1242/dev.110.1.151. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995a;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995b;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- Li Q, Harju S, Peterson KR. Locus control regions. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Jones PA, Chaillet JR, Ferguson-Smith AC, Barton SC, Reik W, Surani MA. Parental imprinting: Potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 1992;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Levorse JM, Tilghman SM. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc Natl Acad Sci USA. 1999;96:9733–9738. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadnick MP, Pieracci FM, Cranston MJ, Taksel E, Thorvaldsen JL, Bartolomei MS. Role of a 461-bp G-rich repetitive element in H19 transgene imprinting. Dev Genes Evol. 1999;209:239–248. doi: 10.1007/s004270050248. [DOI] [PubMed] [Google Scholar]
- Stylianopoulou F, Efstratiadis A, Herbert J, Pintar J. Pattern of the insulin-like growth factor II gene expression during rat embryogenesis. Development. 1988;103:497–506. doi: 10.1242/dev.103.3.497. [DOI] [PubMed] [Google Scholar]
- Thacker C, Marra MA, Jones A, Baillie DL, Rose AM. Functional genomics in Caenorhabditis elegans: An approach involving comparisons of sequences from related nematodes. Genome Res. 1999;9:348–359. [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran, K.L. KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nature Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Webber AL, Ingram RS, Levorse JM, Tilghman SM. Location of enhancers is essential for the imprinting of H19 and Igf2. Nature. 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- Yoo-Warren H, Pachnis V, Ingram RS, Tilghman SM. Two regulatory domains flank the mouse H19 gene. Mol Cell Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]