Abstract
Background and Aims
Clianthus maximus is a leguminous perennial with an unusual order of floral organ insertion, and inflorescences produced year round that nearly all abort except during a limited time in autumn. This study aimed to determine at what point in floral organ differentiation abortion occurred and whether the expression of the floral identity genes underlies this cessation in flower development.
Methods
Inflorescences were harvested across an annual cycle and flower development was examined by light and scanning electron microscopy. Expression of the C. maximus-equivalents of LEAFY (LFY), APETALA1 (AP1), PISTILLATA (PI) and AGAMOUS (AG) was monitored simultaneously by quantitative, reverse transcriptase PCR.
Key Results
Only those inflorescences formed in autumn proceeded to anthesis. Organogenesis had not begun in inflorescences that aborted. The C. maximus-equivalents of AP1, PI and AG were expressed in sepals, petals, carpels and stamens, as expected from the ABC model of floral organ identity specification; furthermore, the order of expression of the three genes reflected the unusual pattern of organ differentiation. Low expression of LFY and AP1 was observed during inflorescence abortion.
Conclusions
Predictions of gene expression based on the ABC model were upheld despite the unusual mass abortion of inflorescences and the non-standard pattern of organ formation. The lack of expression of LFY and AP1 in inflorescences may have been the cause of inflorescence abortion.
Keywords: ABC model, Clianthus maximus, floral development, floral identity genes, gene expression, inflorescence abortion, LEAFY, APETALA1, PISTILLATA, AGAMOUS, qRT-PCR
INTRODUCTION
Clianthus maximus (kowhai ngutukaka or kakabeak) is endemic to New Zealand and is listed as nationally vulnerable (de Lange et al., 2004). It is a leguminous shrub and as such it is palatable to introduced deer and possums in its natural region, where it is found almost exclusively within 50 km of the east coast of the North Island of New Zealand. With its showy flowers it is also of outstanding horticultural value and is commonly cultivated. However, we have shown that the genetic diversity that remains in the wild is not retained in cultivation – and that which is in cultivation no longer exists in the wild (Song et al., 2008a).
Despite its horticultural value and its endangered status, little is known of the floral ontogeny of Clianthus and details of flower development from inflorescence initiation to organ differentiation and development have not been published, nor has there been any expression analysis of the floral organ identity genes. The development of floral organs in the eudicots is widely accepted to be controlled by the combinatorial action of a set of homeotic genes – the Class A, Class B and Class C genes. Class A genes specify sepals, A and B combined specify petals, B and C combined specify stamens, and C genes alone specifies carpels (Bowman et al., 1991; Coen and Meyerowitz, 1991). This designation of activities is commonly referred to as the ABC model of floral organ identity specification.
Quantitative reverse-transcriptase PCR (qRT-PCR) allows for the simultaneous analysis of multiple genes both during development and in specific organs. However, although the ABC model has been shown to be applicable to a wide range of species (Ng and Yanofsky, 2001; Becker and Theissen, 2003), simultaneous investigation of the expression of these genes during an entire annual cycle in natural conditions is limited to a few studies, including that of Sophora tetraptera, which is also a leguminous species native to New Zealand (Song et al., 2008b).
We have reported contrasting programmes of floral development in S. tetraptera and Metrosideros excelsa, another woody species native to New Zealand. In S. tetraptera, inflorescence initiation started in late spring with all floral organs initiated by mid- to late summer. However, at this stage, development ceased and remained paused for several months. Then, in a rapid phase of development, floral organs were differentiated with full flowering occurring by October (Song et al., 2008b).
In contrast, in M. excelsa, inflorescence development was only detectable by late autumn (May), and proceeded as far as cymule primordia in early winter (June) before a pause of several months through the winter. After this, organ differentiation proceeded rapidly with all floral organs fully differentiating within 4–6 weeks by the end of September (Sreekantan et al., 2001, 2004). We have shown (Henriod et al., 2000) that several weeks of low temperatures are required during winter for cymules of M. excelsa to differentiate floral organs and not abort.
Here we show that the time frame of floral development in C. maximus differs from that of both S. tetraptera and M. excelsa. Inflorescences develop year-round, with only those inflorescences developing during a short period in late autumn proceeding to anthesis. A bimodal pattern of expression of LEAFY (LFY)- and APETALA1 (AP1)-equivalents occurs in both S. tetraptera (Song et al., 2008b) and M. excelsa (Sreekantan et al., 2004), with a period of strong expression during initial inflorescence development, low expression of both genes during the winter ‘pause’ in floral development, followed by an increase during the spring flush of flower development. In this study, we used qRT-PCR to monitor the expression of the Clianthus equivalents of LFY, AP1, PISTILLATA (PI) and AGAMOUS (AG) during inflorescence and floral organ development, with the expectation that expression of the ABC genes in the individual floral organs of Clianthus would match the expression of the ABC genes in the floral organs of Arabidopsis, but that expression of the LFY- and AP1-equivalents might be reduced in those inflorescences destined to be aborted, with a consequent loss of the bimodal expression pattern exhibited by several woody species.
MATERIALS AND METHODS
Plant materials
Vegetatively propagated 3-year-old plants of Clianthus maximus ‘Kaka King’ were obtained from commercial nurseries and were grown in Palmerston North, New Zealand, under prevailing climatic conditions. Inflorescences and floral buds from different developmental stages were harvested at intervals of between 2 to 4 weeks (Supplementary Data Table S1, available online) from spring, over a complete annual growth cycle of vegetative and reproductive development. Monthly average temperature ranged from 15 to 20 °C in summer and from 5 to 10 °C in winter. For histological analysis, harvested samples were fixed initially in formalin–acetic acid–alcohol. Samples were then further prepared for light microscopy or scanning electron microscopy as detailed in Song et al. (2008b). For RNA isolation, harvested samples were frozen immediately in liquid nitrogen and stored at –80 °C until used.
Floral identity gene isolation and sequence analysis
Isolation of putative orthologues from C. maximus was carried out through direct sequencing of RT-PCR products using tissue-specific cDNA templates. For each gene of interest (GOI), degenerate primers were designed based on the sequence conservation of orthologues from Arabidopsis thaliana, Antirrhinum majus, all the legume sequences available in the NCBI database and 2–3 representative dicot species. For the MADS-box gene orthologues/paralogues, degenerate primers were designed to minimize the false isolation of any other coexisting paralogues or MADS-box genes from other subfamilies. Protein sequences were first aligned using the Clustal X program (Thompson et al., 1997) to identify the conserved sequence segments. Corresponding DNA sequences of each conserved protein sequence segment were then aligned and degenerate primers were designed using Primer Premier 5·0 software. These primers were then searched against the NCBI database to verify their specificity; they are listed in Table 1.
Table 1.
Degenerate primers used for floral identity gene isolation from Clianthus (F, forward, R, reverse)
| Genes | Sequences (5′ to 3′) |
|---|---|
| LEAFY | F: CAAGGCTGCHRTHMGRGC |
| R: GCTTKGTDGGNACRTACC | |
| APETALA1 | F: GGTAGRGTNCARYTGAAGMG |
| R: GAGTCAGDTCVAGMTCRTTCC | |
| PISTILLATA | F: GGMAAGATHGAGATMAAGMRG |
| R: GCAGATTKGGCTCVAWNGG | |
| AGAMOUS | F: GAGATHAAGMGVATHGARAAYAC |
| R: CARMTCMAYYTCCCTYTTYTGC | |
| β-Actin | F: GTGAAGGAAAAACATGCSTAYAT |
| R: KGAACCACCACTCAAMACAATG | |
| GAPDH | F: GACARTGGAARMACSAYGA |
| R: TTCCACCTCTCCAGTCCTT | |
| 18S rRNA | F: TACCGTCCTAGTCTCAACCATAA |
| R: AGAACATCTAAGGGCATCACA |
Total RNA was extracted using a modification of the TRI reagent method (Molecular Research Centre, Cincinnati, OH, USA) as described in Song et al. (2008b). The quantity and purity of the RNA was measured using a NanoDrop ND-100 spectrophotometer (Nanodrop Technologies Inc., Wilmington, DE, USA). Its integrity was checked by visualization of ethidium-stained RNA separated on a 2 % (w/v) agarose gel containing 2 % formaldehyde. cDNA was synthesized using Expand Reverse Transcriptase (Roche Diagnostics, Mannheim, Germany) and used as a template for PCR as described in Song et al. (2008b). All bands of approximately the expected size were purified and sequenced on an ABS 3730 sequencer (Applied Bioscience) using the same degenerate primers as for the PCR. The raw sequences from four to eight sequencing reactions (at least two forward and two reverse directions) were aligned using Clustal X software to correct the sequencing errors and to generate a consensus sequence for each GOI.
For phylogenetic analysis, 13–20 representative orthologues/paralogues for each GOI (Supplementary Data Table S2) were used to construct both a neighbour-joining (NJ) phylogentic tree using Clustal X software and a maximum-likelihood (ML) tree using PhyML3·0 software (Guindon and Gascuel, 2003; Guindon et al., 2010), with 1000 bootstrap replicates. Each tree was rooted with an orthologue/paralogue from the gymnosperm species Pinus radiata.
The nucleotide sequences reported in this paper were submitted to the GenBank database with the following accession numbers: CmLFY, DQ418756; CmAP1, DQ418757; CmPI, DQ418758; CmAG, DQ418759.
qRT-PCR
For qRT-PCR analysis, specific primers were designed for each gene as described in Song et al. (2008b) and are listed in Supplementary Data Table S3. Primers were designed such that they were of similar size (within the range 100–350 nucleotides) and spanning at least one intron to eliminate amplification from genomic DNA. qRT-PCR analyses were performed using a LightCyclerTM2·0 and SYBR® Green DNA dye (Roche Diagnostics) as described in Song et al. (2008b).
To determine the PCR amplification efficiency of each GOI and housekeeping gene, a serial dilution of 10-, 100-, 1000-, 10 000- and 100 000-fold of the same cDNA was used for qRT-PCR amplification. A standard curve was obtained by plotting the threshold cycle (CT) value versus the logarithm of the concentration. The PCR efficiency (E) was calculated according to the formula: E = 10 (–1/slope) – 1 (Pfaffl, 2001).
Data analysis
As described in Song et al. (2008b), experimental data were analysed as a block experiment by analysis of variance, where PCR run was the block factor, the stage of development was the treatment factor and relative abundance was the response variable.
RESULTS
Floral ontogeny and development
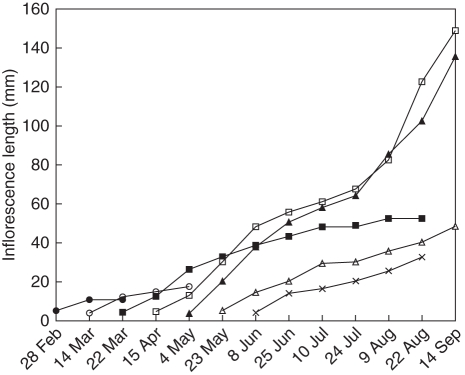
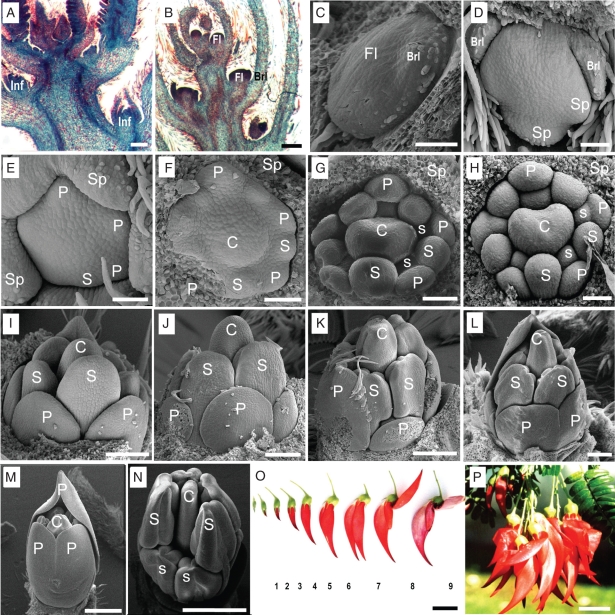
Shoot growth (elongation and node number increase) and leaf emergence occurred continuously throughout the year. Vegetative growth slowed during the flowering season in early spring (September), and resumed in October after flowering. Inflorescences emerged from axils of undeveloped leaves within the shoot tip almost continuously throughout the year, from shortly after the previous flowering season in October until a short time before the next flowering season in August. The early-emerged inflorescences normally aborted during the development period. Furthermore, the inflorescences that emerged after May rarely reached the flowering stage and aborted before or during the flowering season. Only those inflorescences that emerged in autumn (April to early May) developed to maturity, with individual flowers developing to anthesis (Fig. 1). Inflorescences reached the flowering stage about 4 months after their emergence, with individual flower buds undergoing a series of developmental stages before opening in mid-September (Fig. 2O). There were 10–20 flowers in each inflorescence (Fig. 2P).
Fig. 1.
Development of inflorescences of C. maximus emerging at different times of the year. Values are means of 25 inflorescences from five different plants. All inflorescences not elongating to >60 mm aborted.
Fig. 2.
Floral ontogeny and development in C. maximus using SEM (C–N) and light microscopy (A and B counter-stained with safranin and fast green). (A) Inflorescences initiated from the axil of the shoot tip; (B) inflorescence with floral primordia; (C) single floral meristem before organ initiation; (D) sepal initiation; (E) petal initiation; (F) outer stamen and carpel initiation; (G) mass increase of petals, outer stamens and carpel, with similar height; (H) inner stamen initiation; (I–K) organ differentiation and rapid enlargement; (L) petal expansion; (M) ovule formation; (N) pollen formation; (O) flower maturation; (P) inflorescences showing flowers in bloom. Abaxial side is at base of figure in (C–L). Sepals were removed in (F–L). Abbreviations: Brl, bracteole; C, carpel; Fl, flower buds; Inf, inflorescence; O, ovule; P, petal; S, outer stamen; s, inner stamen; Sp, sepal. Scale bars: (A, B, I, J) = 200 µm, (C–F) = 50 µm, (G, H) = 100 µm, (K, L) = 400 µm, (M, N) = 2 mm, (O, P) = 20 mm.
Microscopy studies confirmed that the inflorescences emerged from leaf axils (Fig. 2A) from November to August. Flower primordia were initiated within the inflorescence shortly thereafter (Fig. 2B). Although sepals could occasionally be distinguished between November and March, no further organogenesis was observed in flower primordia from inflorescences harvested before mid-April (Fig. 2C). Organogenesis progressed slowly from late April, with sepals (Fig. 2D) initiated first. This was followed by petal (Fig. 2E), outer stamen and carpel (Fig. 2F) initiation, which occurred sequentially but overlapped within a broadly similar time frame. The inner stamens initiated at the inner side of the petal primordia much later, when the mass of petals, outer stamens and carpel had increased substantially (Fig. 2G). After all the organs within a floral bud had initiated, the floral organs enlarged and differentiated rapidly in June and early July. However, the increase in the mass of the carpel was substantially advanced over that of petals and outer stamens. The inner stamens remained much smaller than the floral organs in the other whorls (Fig. 2H–J). In late July, the size of the petals increased rapidly and further differentiated into distinct vexillum, wing and keel, with the height of the vexillum exceeding that of the carpel (Fig. 2K, L). At this stage, the height of the gynoecium still exceeded that of the androecium. By August, the volume of floral organs, particularly petals, had expanded further, with obvious formation of stigma, ovary and anthers. The androecium then enlarged so that its height now exceeded that of the gynoecium (Fig. 2M, N). At this time, the length of the flower bud was about 5–7 mm, equivalent to stage 1 of flower development (Fig. 2O).
Isolation of floral identity genes
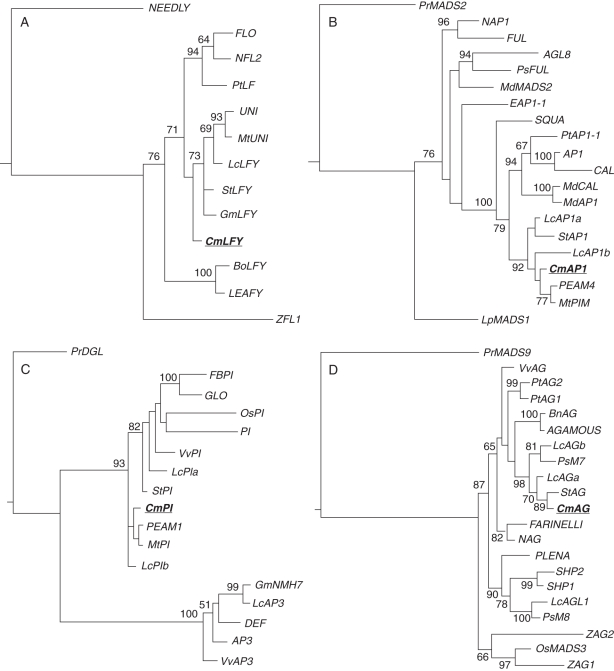
Using the PCR product direct sequencing strategy, one single PCR product was obtained for each GOI. Results of multiple alignments of four to eight raw sequences confirmed that only one copy of each GOI was isolated from C. maximus. A single 401-bp putative LFY orthologue was obtained and was named CmLFY. All positive hits resulting from the BLAST search against the NCBI database were LFY/FLO orthologues from a large range of angiosperm and gymnosperm species. Multiple alignment of the deduced amino acid with related LFY/FLO orthologues showed that CmLFY has characteristics typical of a LFY and FLO orthologue and was most similar to the UNIFOLIATA (UNI) protein from the legume Pisum sativum (Supplementary Data Fig. S1, available online). Unexpectedly, the residue on position 20 of the CmLFY fragment differed from that of all other sequences, with the glutamic acid changed to aspartic acid. This is in agreement with our previous study in comparing the sequencing variability of LEAFY intron 2 for over 70 individual plants from cultivars and wild populations of the same species (Song et al., 2008b). Similar tree topologies were obtained for the ML phylogenetic tree and the NJ phylogenetic tree using the same set of LFY/FLO orthologues, which showed that CmLFY grouped together with LFY/FLO orthologues of other eudicot species, and particularly with those of other leguminous species. It was well separated from the monocot Zea mays and the gymnosperm Pinus radiata (Fig. 3A, Supplementary Data Fig. S2).
Fig. 3.
The maximum-likelihood (ML) phylogentic trees of representative floral identity gene proteins generated with 1000 bootstrap replicates. Values are percentage bootstrap support. Genes isolated in this study are in bold font and are underlined. (A) LFY/FLO proteins. BoLFY, Brassica oleracea; CmLFY, Clianthus maximus, isolated in this study; FLO, Antirrhinum majus; LFY, Arabidopsis thaliana; LcLFY, Lotus corniculatus var. japonicus; MtUNI, Medicago truncatula; NEEDLY, Pinus radiata; NFL2, Nicotiana tabacum; PtLF, Populus trichocarpa; StLFY, Sophora tetraptera; UNI, Pisum sativum; GmLFY, Glycine max; ZFL1, Zea mays. Refer to Supplementary Data Table S2 (available online) for NCBI accession numbers. (B) Class A MADS-box proteins. AGL8, AP1, CAL, Arabidopsis thaliana; CmAP1, Clianthus maximus; EAP1-1, Eucalyptus globulus; LcAP1a, b, Lotus corniculatus var. japonicus; LpMADS1, Lolium perenne; MdAP1, MdCAL, MdMADS2, Malus × domestica; MtPIM, Medicago truncatula; NAP1, Nicotiana tabacum; PEAM4, PsFUL, Pisum sativum; PrMADS2, Pinus radiata; PtAP1-1, Populus trichocarpa; FRUITFUL, SQUA, Antirrhinum majus; StAP1, Sophora tetraptera. Refer to Supplementary Data Table S2 for NCBI accession numbers. (C) Class B MADS-box proteins. AP3, PI, Arabidopsis thaliana; CmPI, Clianthus maximus; DEF, GLO, Antirrhinum majus; FBPI, Petunia hybrida; GmNMH7, Glycine max; LcPIa, b, LcAP3, Lotus corniculatus var. japonicus; MtPI, Medicago truncatula; OsPI, Oryza sativa; PEAM1, Pisum sativum; PrDGL, Pinus radiata; StPI, Sophora tetraptera; VvAP3, VvPI, Vitis vinifera. Refer to Supplementary Data Table S2 for NCBI accession numbers. (D) Class C MADS-box proteins. AG, SHP1, SHP2, Arabidopsis thaliana; BnAG, Brassica napus; CmAG, Clianthus maximus; FARINELLI, PLE, Antirrhinum majus; LcAGa, LcAGb, LcAGL1, Lotus corniculatus var. japonicus; NAG1, Nicotiana tabacum; OsMADS3, Oryza sativa; PsM7, PsM8, Pisum sativum; PtAG1, PtAG2, Populus trichocarpa; PrMADS9, Pinus radiata; StAG, Sophora tetraptera; VvAG, Vitis vinifera; ZAG1, ZAG2, Zea mays. Refer to Supplementary Data Table S2 for NCBI accession numbers.
The possible structure was deduced by comparison of the cDNA sequences and amino acid sequences with those of Arabidopsis. CmLFY spans the second intron, with 129 bp at the end of exon 1 and the rest of the sequence at the starting part of exon 3 (Supplementary Data Fig. S3).
The RT-PCR using degenerate AP1 primers yielded a cDNA fragment of 629 bp. The fragment was named CmAP1. All of the sequences yielded by BLAST searching the GenBank amino acid database were AP1 and SQUA orthologue proteins. Multiple alignment of the deduced amino acid with related Class A MADS-box proteins showed that CmAP1 has characteristics typical of an AP1 orthologue and the most similar protein was PEAM4 from the legume Pisum sativum (Supplementary Data Fig. S4). To elucidate the relationship of CmAP1 with other orthologues in a broad taxonomic range, 20 Class A MADS-box proteins from eudicot and one monocot species were compared to generate phylogenetic trees. Both the ML and the NJ phylogenetic trees clearly showed the eudicot AP1/SQUA orthologues grouped together and well separated from the monocot species. CmAP1 showed closest relationship to other leguminous species (Fig. 3B, Supplementary Data Fig. S5). In comparing the cDNA and amino acid sequences, the segment sequenced covered exons 1–8, including the major part of the MADS-box, and the entire sequence of the K-domain (Supplementary Data Figs S3 and S5).
A 329-bp putative PI orthologue for Clianthus was obtained through RT-PCR and direct sequencing and was named CmPI. BLAST searches for similar sequences in the GenBank amino acid database with CmPI yielded only PI/GLO sequences. Multiple alignment of the deduced amino acid with related Class B MADS-box proteins showed that CmPI has characteristics typical of a PI/GLO orthologue and the most similar protein was PEAM1 from the legume Pisum sativum (Supplementary Data Fig. S6). Phylogenetic trees generated using 15 Class B MADS-box gene proteins were compared to determine the relationship of CmPI with orthologues/paralogues in other species. Both rooted NJ and ML trees showed that CmPI was grouped most closely to the other leguminous species of the PI/GLO clade and was well separated from those of the AP3/DEF clade (Fig. 3C, Supplementary Data Fig. S7). The CmPI fragment consisted of the entire sequences for exons 1–3 and the partial sequence of exon 4 and covered the entire MADS-box and partial sequence of the K-domain (Supplementary Data Figs S3 and S6).
Similarly, a 297-bp putative AG orthologue was isolated and named CmAG. All of the sequences found by BLAST searches of the CmAG amino acid sequence against the GenBank amino acid database were AG/PLE orthologue proteins. Multiple alignment of the deduced amino acid with related Class C MADS-box proteins showed that CmAG has characteristics typical of an AG orthologue and the most similar protein was LcAGa from the legume Lotus corniculatus var. japonicus (Supplementary Data Fig. S8). The rooted ML and NJ phylogenetic trees of the representative Class C MADS-box proteins showed that CmAG clustered with other leguminous species, and that it was more closely related to the AG/FAR orthologues than the PLE/SHP orthologues (Fig. 3D, Supplementary Data Fig. S9). The CmAG fragment consisted of exons 2–5 and contained a short sequence of the MADS-box and almost the entirety of the K-domain (Supplementary Data Figs S3 and S8).
Housekeeping genes and qRT-PCR optimization
Orthologues of housekeeping genes 18S, β-actin and GAPDH from Clianthus were isolated using the same strategy as for the GOIs and were used as internal controls in the gene expression studies. Optimization of the qRT-PCR is described in detail in Song et al. (2008b). The PCR efficiencies are shown in Table 2 for both floral identity genes and housekeeping genes.
Table 2.
PCR efficiencies of floral identity genes and selected housekeeping genes
| Gene | Slope | Efficiency |
|---|---|---|
| CmAP1 | –3·646 | 0·881 |
| CmPI | –3·540 | 0·916 |
| CmAG | –3·497 | 0·932 |
| CmLFY | –3·662 | 0·875 |
| Cm18S | –3·389 | 0·973 |
| CmGAPDH | –3·663 | 0·875 |
| CmACT | –3·566 | 0·907 |
PCR efficiencies were calculated according to the formula: Efficiency = 10(–1/slope) –1 (Pfaffl, 2001). The slope was calculated based on the standard curve generated using a serial dilution of 10-, 100-, 1000-, 10 000- and 100 000-fold of the same cDNA template.
Expression of floral identity genes in vegetative and reproductive tissues
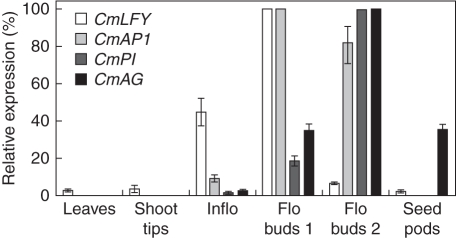
Expression of the four floral identity genes was analysed in shoot tips, adult leaves and inflorescences harvested in late autumn (May), and from early-stage flower buds, mid-stage flower buds and seed pods sampled in July, September and October, respectively. A distinct expression profile was obtained for each gene (Fig. 4). Relative to the expression of CmLFY in early-stage flower buds (set at 100 %), expression decreased to 7 % in mid-stage flower buds. Expression was low in leaves and vegetative shoot tips (2–3 %) and was barely detectable in seed pods (1·7 %). CmLFY expressed at relatively high levels in inflorescences (45 %).
Fig. 4.
Expression of floral identity genes in selected vegetative and reproductive tissues of C. maximus. Relative mRNA levels were determined using qRT-PCR and normalized using housekeeping genes 18S, GAPDH and β-actin as internal controls. Values are mean ± s.e. of four replicates. Inflo, inflorescences 2–3 mm in length; Flo Bud 1, early-stage flower buds 2–3 mm in length; Flo Bud 2, mid-stage flower buds 18–20 mm in length; Seed pods, 2–3 weeks after flowering.
No CmAP1 activity was detected in adult leaves, vegetative shoot tips or seed pods. The highest level of expression was in early-stage flower buds (100 %) and in mid-stage flower buds (82 %), but a relatively low level (9 %) was detected in inflorescences.
CmPI expression was not detected in leaves, vegetative shoot tips or seed pods. Inflorescences and early-stage flower buds had low levels of expression (19 and 1·5 %, respectively) relative to high expression in mid-stage flower buds (100 %).
The highest CmAG expression was detected in mid-stage flower buds, whereas expression was low in inflorescences (2·4 %) and early-stage flower buds (35 %). No CmAG expression was detected in adult leaves and vegetative shoot tips but significant (35 %) CmAG expression was detected in seed pods.
Organ-specific expression of floral organ identity genes
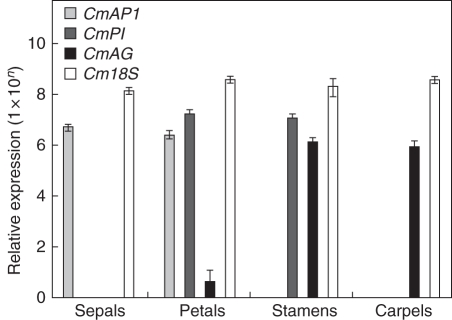
Sepals, petals, stamens and carpels were excised from flower buds in early spring (September) when the flower buds were beginning to show the tips of petals (Fig. 2O, Stage 2). Expression of CmAP1, CmPI and CmAG was assessed using Cm18S as an internal control (Fig. 5). CmAP1 expression was found only in sepals and petals, CmPI expression was detected in petals and stamens, but not in sepals or carpels, and CmAG expression occurred at high levels in stamens and carpels but was also detected in petals at a low level (albeit 1000-fold lower than in the other organs).
Fig. 5.
Expression of floral organ identity genes and the 18S orthologue in different floral organs of C. maximus. Ten flower buds at late developmental stage (18–20 mm in length showing petal tip) were individually dissected to obtain a mixed sample of each floral organ. Relative mRNA levels were determined using qRT-PCR, calculated and presented on a logarithmic scale across all the tested genes. Values are mean ± s.e. of three replicates.
Expression profiles of floral organ identity genes during a growth cycle
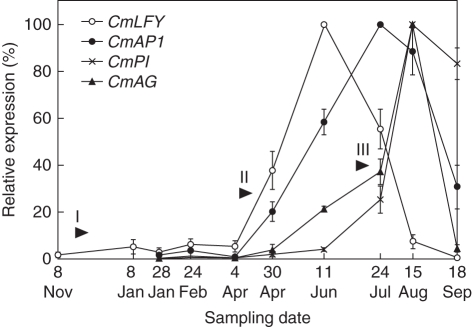
Given that the floral identity genes showed differential expression during different stages of flower development, and given that there is a continuous cycle of inflorescence formation and abortion up until those formed in May which develop through to flowering, a detailed expression profile of the genes was obtained across a developmental cycle (Fig. 6). In this cycle, shoot tips were harvested from November, inflorescences were harvested from January to April, and from June to September samples were of flower buds.
Fig. 6.
Expression profiles of floral identity genes in C. maximus during a complete growth cycle. Relative mRNA levels were determined using qRT-PCR and normalized using housekeeping genes 18S, GAPDH and β-actin as internal controls. Values are mean ± s.e. of four replicates. Arrows show the starting points of: I, floral meristem initiation; II, floral organ initiation; III, floral organ differentiation and development. Sample descriptions: 8 Nov to 8 Jan, shoot tips with/without inflorescence primordia; 28 Jan to 30 Apr, inflorescences (15–20 mm) with floral primordia; 11 Jun, inflorescences (25–35 mm) with flower bud (approx. 1 mm) and all floral organs initiated; 24 Jul, flower buds (2–3 mm) with well-differentiated floral organs and elongated anthers and pistils; 15 Aug, flower buds (10–12 mm) showing petal tip, with well-formed pollen and ovules; 18 Sep, mature flower buds before opening.
In shoot tips, CmLFY expression remained at a very low level from August to April (2–5 %) compared with its highest expression in June. In inflorescences and floral buds, expression of the four GOIs was low from January to early April. Increasing levels of these genes were detected from April to September but this occurred in a sequential manner: CmLFY peaked in June, CmAP1 in July, and CmPI and CmAG in September. CmPI expression remained high (>80 %) during flowering in September whereas that of the other genes had declined by that time.
DISCUSSION
Floral development in Clianthus follows an unusual pattern with significant resource being allocated to inflorescence development throughout much of the year, only for the bulk of those inflorescences to be aborted. Only those inflorescences formed during a few weeks in autumn proceed to maturity. Light and scanning electron microscopy of Clianthus established that organogenesis had not begun in the inflorescences destined to be aborted. The pattern of organogenesis, once begun, essentially followed the order detailed for the model plants of sepals, petals, stamens and carpel. However, the inner stamen whorl was not established until after the carpel was initiated and the carpel primodium had enlarged. The carpel was also much more advanced than that of the floral organs in whorls 2 and 3, and its height was superior to that of the other organs until the formation of the gametocytes. In addition, there was a significantly delayed expansion of the petals.
Genes controlling the specification of floral organ identity and development have been isolated and characterized in a variety of plant species. Intensive studies and analysis of homeotic mutants in Arabidopsis and Antirrhinum led to the ABC model of floral organ identity specification (Bowman et al., 1991; Coen and Meyerowitz, 1991; Weigel et al., 1992). Analyses have largely substantiated the applicability of this model, but simultaneous and quantitative expression of A, B, and C orthologues has rarely been carried out in plants across an entire life cycle (e.g. Song et al., 2008b in Sophora). This is of particular interest in species that show unusual floral development, such as the mass abortion of inflorescences exhibited by Clianthus, and different patterns of organogenesis, as in the papilionoid legumes (Tucker, 2003).
To further establish the genetic control of inflorescence and floral development, the Clianthus equivalents of LFY, AP1, PI and AG were isolated. The combined information from the BLAST searches against the GenBank database, the multiple sequence alignments, the phylogenetic trees aligning the sequences into the expected taxonomic relationships and the gene structures all suggest that we have isolated from Clianthus the gene orthologues of LFY/FLO, AP1/SQUA, PI/GLO and AG/FARINELLI or SHP/PLE from Arabidopsis and Antirrhinum, respectively. qRT-PCR then enabled us to monitor simultaneously the expression of these genes during inflorescence development and abortion, and flower development. The high sequence similarity lends support to the hypothesis that the ABC model would be broadly applicable to Clianthus. Optimization of the qRT-PCR approach used has been described previously (Song et al., 2008b).
Expression of CmAP1, CmPI and CmAG agreed with that predicted by the ABC model: as expected, the Class A gene CmAP1 was expressed in the sepals and petals; the Class B gene CmPI was expressed in petals and stamens; and the Class C gene CmAG was expressed in stamens and carpels. The additional expression of CmAG in seed pods was not unexpected as this was also found in S. tetraptera (Song et al., 2008b).
In those C. maximus inflorescences that did not abort but proceeded through to flowering, changes in the levels of the four GOIs did not track in the order of the model plant Arabidopsis (i.e. A then AB, BC and then C), but reflected the developmental sequence as shown by scanning electron microscopy. Sepals appeared first as did the increase in the Class A gene, CmAP1. However, the Class C gene, CmAG, then increased reflecting the strong development of the carpel. Subsequently, CmPI increased and both genes peaked simultaneously, along with CmAP1 expression, possibly reflecting a requirement for expression of all three homeotic genes throughout organ development. S. tetraptera, which also exhibits an unusual sequence of floral organ development, showed a similar altered pattern of gene expression (Song et al., 2008b).
While the alignment of the floral identity genes is as predicted, the abortion of inflorescences is an unusual phenomenon. In a number of woody species with prolonged patterns of floral development, and in which expression of LFY and/or AP1 has been monitored, a bimodal pattern of expression has been observed with expression of LFY and/or AP1 elevated at the initial stages of floral development followed by a decrease, usually through a winter dormancy period, after which expression of these genes again increased. Such a bimodal pattern of expression was first shown, by northern blot analysis, for both LFY- and AP1-equivalents, in Actinidia deliciosa (Walton et al., 2001). A similar bimodal pattern was shown for the LFY-equivalent of Vitis (Carmona et al., 2002). In M. excelsa flowers, which develop in one season, the temporal expression patterns showed both LFY- and AP1-equivalents to be elevated during early floral initiation in autumn, down-regulated during winter and up-regulated during floral organ differentiation in spring. In contrast to the above winter-dormant species, S. tetraptera exhibits a summer–autumn dormancy during floral development but here, again, a definitive bimodal pattern of expression occurs: peaks of expression of StLFY and StAP1 coincided with floral organ initiation and subsequently with floral organ differentiation, as shown by Song et al. (2008b) using qRT-PCR.
In C. maximus, at the transition from the vegetative to the reproductive phase with the formation of the inflorescence, we might have expected enhanced activity of CmLFY and/or CmAP1 as occurred in the woody species mentioned above. However, throughout the months when inflorescences were being formed and aborted, little activity of CmLFY was detected and no expression of CmAP1. Both LFY and AP1 are considered necessary for floral meristem initiation (Liu et al., 2009), and Kaufmann et al. (2010) suggest that AP1 acts to down-regulate floral repressors during the earliest stages of flower development. It is clear from our work that expression of both CmLFY and CmAP1 is essentially lacking in Clianthus plants until late autumn (May). The sequence of events that occurs at this time that lead to the up-regulation of CmLFY and CmAP1 is yet to be determined.
In conclusion, predictions of gene expression based on the ABC model were upheld despite the unusual mass abortion of inflorescences and the non-standard pattern of organ formation. We suggest that the lack of expression of LFY and AP1 in inflorescences may have been the cause of inflorescence abortion.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by the New Zealand Foundation for Research, Science & Technology Public Good Science Fund under the Native Ornamental Plants Programme through subcontract C02X0015 to Crop & Food Research and a Massey PhD scholarship (J.S.). We acknowledge Mr Neil Andrew for his technical assistance (SEM).
LITERATURE CITED
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Cubas P, Martinez-Zapater JM. VFL, the grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiology. 2002;130:68–77. doi: 10.1104/pp.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3·0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Henriod RE, Jameson PE, Clemens J. Effects of photoperiod, temperature and bud size on flowering in Metrosideros excelsa (Myrtaceae) Journal of Horticultural Science and Biotechnology. 2000;75:55–61. [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- de Lange PJ, Norton DA, Heenan PB, et al. Threatened and uncommon plants of New Zealand. New Zealand Journal of Botany. 2004;42:45–76. [Google Scholar]
- Liu C, Thong Z, Yu H. Coming into bloom: the specification of floral meristems. Development. 2009;136:3379–3391. doi: 10.1242/dev.033076. [DOI] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nature Reviews Genetics. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Murdoch J, Gardiner SE, Young A, Jameson PE, Clemens J. Molecular markers and a sequence deletion in intron 2 of the putative partial homologue of LEAFY reveal geographical structure to genetic diversity in the acutely threatened legume genus Clianthus. Biological Conservation. 2008a;141:2041–2053. [Google Scholar]
- Song J, Clemens J, Jameson PE. Quantitative expression analysis of the ABC genes in Sophora tetraptera, a woody legume with an unusual sequence of floral organ development. Journal of Experimental Botany. 2008b;59:247–259. doi: 10.1093/jxb/erm305. [DOI] [PubMed] [Google Scholar]
- Sreekantan L, McKenzie MJ, Jameson PE, Clemens J. Cycles of floral and vegetative development in Metrosideros excelsa (Myrtaceae) International Journal of Plant Sciences. 2001;162:719–727. [Google Scholar]
- Sreekantan L, Clemens J, McKenzie MJ, Lenton JR, Croker SJ, Jameson PE. Flowering genes in Metrosideros fit a broad herbaceous model encompassing Arabidopsis and Antirrhinum. Physiologia Plantarum. 2004;121:163–173. doi: 10.1111/j.0031-9317.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SC. Floral development in legumes. Plant Physiology. 2003;131:911–926. doi: 10.1104/pp.102.017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Podivinsky E, Wu RM. Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiologia Plantarum. 2001;111:396–404. doi: 10.1034/j.1399-3054.2001.1110318.x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.