Abstract
Background and Aims
The MADS-box transcription factor AGAMOUS (AG) is an important regulator of stamen and fruit identity as well as floral meristem determinacy in a number of core eudicots and monocots. However, its role outside of these groups has not been assessed explicitly. Examining its role in opium poppy, a basal eudicot, could uncover much about the evolution and development of flower and fruit development in the angiosperms.
Methods
AG orthologues were isolated by degenerate RT-PCR and the gene sequence and structure examined; gene expression was characterized using in situ hybridization and the function assessed using virus-induced gene silencing.
Key Results
In opium poppy, a basal eudicot, the AGAMOUS orthologue is alternatively spliced to produce encoded products that vary at the C-terminus, termed PapsAG-1 and PapsAG-2. Both transcripts are expressed at high levels in stamens and carpels. The functional implications of this alternative transcription were examined using virus-induced gene silencing and the results show that PapsAG-1 has roles in stamen and carpel identity, reflecting those found for Arabidopsis AG. In contrast, PapsAG-2, while displaying redundancy in these functions, has a distinctive role in aspects of carpel development reflected in septae, ovule and stigma defects seen in the loss-of-function line generated.
Conclusions
These results describe the first explicit functional analysis of an AG-clade gene in a basal eudicot; illustrate one of the few examples of the functional consequences of alternative splicing in transcription factors and reveal the importance of alternative transcription, as well as gene duplication, as a driving force in evolution.
Keywords: Flower development, MADS-box, Papaver somniferum, alternative transcription, AGAMOUS, gene-silencing
INTRODUCTION
A key means of generating morphological diversity is through duplication of critical regulatory genes and consequent diversification in the deployment of their function (Force et al., 1999; Moore et al., 2005). The permutations for diversity are amplified by the fact that such duplicated genes may interact with myriad other components and programmes in different ways. In Arabidopsis, a number of MADS-box transcription factors have been shown to be required for specifying floral organ identity, development and determinacy. Homologues of these genes have been postulated to regulate analogous processes in other angiosperms, while alterations in the number and function of such genes is thought to have contributed to the morphological diversity of angiosperm floral structures (Irish and Litt, 2005; Kaufmann et al., 2005).
In Arabidopsis, the AGAMOUS (AG) MADS-box gene is required for determining stamen and carpel identity and floral determinacy. However, functional analyses of AG-related gene duplicates have suggested that there is some variation in the roles of such gene products in other species. For instance, the AG orthologue in antirrhinum is FARINELLI (FAR), mutations of which affect the stamens only (Causier et al., 2005). Instead, PLENA (PLE), a paralogue of AG and FAR, is required in antirrhinum to condition stamen and carpel differentiation and determinacy (Bradley et al., 1993; Causier et al., 2005). Phylogenetic analyses suggest that the gene duplication giving rise to the AG and PLE paralogous lineages occurred in the ancestor of the core eudicots (Kramer et al., 2004; Zahn et al., 2006). An independent duplication event in the monocots gave rise to two AG-related genes in maize and rice. These two genes were suggested to have discrete functions in maize (Mena et al., 1996). Genetic analyses in rice have confirmed the subfunctionalization of these duplicates with OsMADS58 controlling carpel development and floral determinacy and OsMADS3 controlling stamen identity (Yamaguchi et al., 2006).
Little is known, though, of the potential roles of AG-related genes in the basal eudicots. In Eschscholzia californica, two AG-related genes have been identified, EScaAG1 and EScaAG2, that have similar expression patterns in stamens and carpels, with EScaAG1 being expressed at much higher levels, suggesting that EScaAG2 may be non-functional (Zahn et al., 2006). In Aquilegia formosa, a single AG-like gene has been identified and shown to be expressed in stamens, stamenodia and carpels (Voelckel et al., 2010). In Thalictrum dioecium, ThdAG1 is expressed in developing stamen and carpel primordia, while ThdAG2 expression is limited to the developing ovules, suggesting that these genes may have distinct roles (Di Stilio et al., 2005). These genes were isolated by extensive degenerate RT-PCR sampling or from cDNA libraries generated from floral tissues. All of these expression analyses are generally consistent with a potential role for basal eudicot AG homologues in specifying reproductive identity. As of yet, though, functional analyses have not been carried out for any AG-related basal eudicot gene.
Opium poppy (Papaver somniferum) is a basal eudicot species in the order Ranunculales and family Papaveraceae. The Ranunculales is well supported as the sister group to all the other eudicots (Angiosperm Phylogeny Group, 2009). The opium poppy is largely recognizable by its distinctive fruit, the capsule, which is rich in opiate-containing laticiferous vessels, and so this fruit has been the subject of considerable physiological and biochemical studies (Kapoor, 1997). In addition, opium poppy is ideally suited to analyses of gene function, as loss of function of individual genes can be achieved through highly efficient virus-induced gene silencing (VIGS; Hileman et al., 2005).
In a search for the MADS-box genes involved in fruit development in opium poppy a PapsAG locus was identified that undergoes alternative splicing to produce two abundantly expressed transcripts, PapsAG-1 and PapsAG-2. These transcripts differ only in the encoded C-terminal domains. The functions conferred by these alternatively spliced forms were explored and each was shown to have some unique function in floral development. These observations have important ramifications for developing new models for the evolution of floral homeotic gene function.
MATERIALS AND METHODS
Isolation of PapsAG genes
RT-PCR reactions using the QVT1 and QVT2 degenerate forward primers with an oligo dT reverse primer (Hileman et al., 2006) were used to isolate MADS-box genes from carpel and flower cDNA of Papaver somniferum (Persian White). Total RNA was extracted using Trizol reagent (Invitrogen, Cleveland, OH, USA) and converted to cDNA using SuperscriptIII (Invitrogen) as per manufacturers instructions. Isolated sequences were cloned into pCR4-TOPO sequencing vector (Invitrogen) and sequenced to identify AG-like clones for further analysis. A total of 23 and 18 sequenced clones for PapsAG-1 and PapsAG-2 were identified and used to generate consensus cDNA sequences.
Sequence and phylogenetic analyses
Translated sequences of AG orthologues were aligned using CLUSTAL_X (Thompson et al., 1997) and alignments refined by hand using BioEdit (Hall, 1999). Sequences for PapsAG-1 and PapsAG-2 were deposited in Genbank with accession numbers GU123602 and GU123603.
Expression analyses using RT-PCR
Total RNA was extracted using the Trizol reagent (Invitrogen) and approx. 300 ng was used in 10-μL cDNA synthesis, reactions using Superscript™III reverse transcriptase (Invitrogen). For cDNA synthesis the poly(T) primer used was 5′-GACTCGAGTCGACATCGA(T)17. Primers for testing expression of PapsAG-1 and PapsAG-2 were: AGbF 5′-TATGACTCTCGGAACTTTCTC-3′ forward primer with AG1R 5′-ACATAGAATAGACTCAGC-3′ and AG2R 5′-GTAATGTAGTCAAATCCAGATG-3′ reverse primers. Actin primers were ACT1: 5′-ATGGATCCTCCAATCCAGAC-3′ and ACT2: TATTGTGTTGGACTCTGGTG-3′. PCR consisted of cycles of 94 °C for 30 s, 53 °C for 45 s, 72 °C for 1 min, preceded by a 5-min denaturation at 94 °C and followed by an extension at 72 °C for 6 min; 30 cycles for PapsAG-1 and PapsAG-2 and 26 cycles for Actin.
Genomic DNA PCR
Genomic DNA was extracted from leaf tissue using established protocols (Aldrich and Cullis, 1993). This was used in PCR reactions with AGbF/AG2R and AGbF/AG1R primer combinations using 30 cycles of 94 °C for 1min, 53 °C for 2 min, 72 °C for 3 min, preceded by a 5-min denaturation at 94 °C and followed by an extension at 72 °C for 6 min. PCR products were cloned into the pCR4-TOPO sequencing vector (Invitrogen) and 20 (10 for each primer set) clones were sequenced, then aligned with the cDNA sequences to map the location of the intron and predict the splicing sites.
Expression analyses using in situ hybridization
Hybridizations were carried out as previously described (Drea et al., 2005, 2007) with minor modifications. Gene-specific regions derived from the C-terminal domain and 3′-UTR of PapsAG-1and PapsAG-2 sequences were used to generate digoxyenin-labelled RNA probes. PCR fragments amplified with AG1sF/AG2sF forward primers and AG1T7R/AG2T7R reverse primers were cleaned using a Qiagen PCR purification kit and used in an in vitro transcription reaction with dig-UTP and T7 RNA polymerase (Roche).
AG1F: 5′-GAAGATAGAAGACATCAAACC-3′AG2F: 5′-ATGATGGCATTCTCTTTCAAG-3′
AG1T7R: 5′-GATCTAATACGACTCACTATAGGGAGTCAACATAGAATAGACTCAGC-3′
AG2T7R: 5′-GATCTAATACGACTCACTATAGGGAGTAATGTAGTCAATCC-AGATG-3′
T7 RNA polymerase sites are underlined. Probe lengths were 250 bp and 209 bp, respectively.
Virus-induced gene silencing
Gene-specific regions of PapsAG-1 and PapsAG-2, as well as concatenated PapsAG-1/PapsAG-2 sequences were introduced into the TRV2 vector (Liu et al., 2002), transformed into Agrobacterium strain GV3101 and used to infiltrate poppy seedlings at the three-to-five leaf stage as previously described (Hileman et al., 2005). Individual resulting plants were assayed for the presence of the viral vector using RT-PCR as well as for any visible phenotype. Three constructs incorporating regions of PapsAG-1 and/or PapAG2 into the TRV2 vector cut with XbaI and BamHI were assembled as follows:
AG1F: 5′-GCTCTAGAATGATGGCATTCTCTTTCAAG-3′
AG2F: 5′-GCTCTAGAGAAGATAGAAGACATCAAACC-3′
AG1R: 5′-CGGGATCCACATAGAATAGACTCAGC-3′
AG2R: 5′-CGGGATCCGTAATGTAGTCAAATCCAGATG-3′
Primers AG1F and AG1R were used to amplify a fragment for the vigsAG1 construct; primers AG2F and AG2R used to amplify a fragment for the vigsAG2 construct.
PapsAG-2 was reamplified with AG2BAM and AG2XHO and ligated into a BamHI- and XhoI-cut TRV2 vector already containing PapsAG-1.
AG2BAM: 5′-CGGGATCCCTCGAATCAGCCAATCAC-3′
AG2XHO: 5′-CCGCTCGAGGTAATGTAGTCAAATCCAG-3′
Constructs vigsAG1 and vigsAG2 contain 255 nt and 239 nt of 3′-UTR sequences specific to PapsAG-1 and PapsAG-2; vigsAG-D contains 313 nt and 297 nt of the PapsAG-1 and PapsAG-2 3′ sequences concatenated in the TRV2 vector. At least 100 young seedlings were infiltrated with each of the three constructs and with an empty vector construct (containing no insert) as previously described (Hileman et al., 2005; Drea et al., 2007) and flowers analysed for phenotypes and gene silencing using semi-quantitative RT-PCR.
Scanning electron microscopy
Plant material was fixed overnight in FAA (3·7 % formaldehyde, 5 % acetic acid, 50 % ethanol) and then transferred to 70 % ethanol. Samples were dehydrated in an ethanol series (80 %, 90 %, 100 % ethanol) before critical-point drying in a Bal-Tec 030 critical-point drier. Samples were coated in gold using a Polaron SC7640 sputter coater and analysed on a Hitachi S3000H scanning electron microscope equipped with digital image capture.
RESULTS
Isolation and analysis of AG orthologues in Papaver somniferum
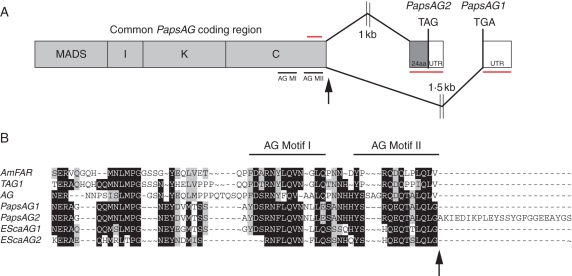
Degenerate RT-PCR with QVT primers (Hileman et al., 2006) was used to isolate AG orthologues from Papaver somniferum flowers and developing capsules. Products were cloned and sequenced and two distinct classes of cDNAs were identified, designated PapsAG-1 and PapsAG-2 (Fig. 1). These cDNAs were identical in sequence to nucleotide position 599 and then sequence diverged completely. In the predicted protein products this corresponded to complete identity as far as the stop codon of PapsAG-1 with PapsAG-2 encoding a further 24 amino acids beyond this point (Fig. 1B).
Fig. 1.
Gene structure of the Papaver somniferum AG gene. (A) Schematic showing how alternative splicing at the 3′ end of the coding sequence generates two transcripts, PapsAG-1 and PapsAG-2 (not drawn to scale). Regions underlined in red are gene-specific fragments used for gene-silencing with VIGS and for in situ mRNA hybridization probes. The region overlined with a red bar indicates the presence of a forward primer outside of the region used in vigs lines for expression analysis. Black arrows indicate the splicing donor site. AG MI, AG Motif I; AG MII, AG Motif II. (B) Alignment of the C-terminal regions of translated PapsAG-1 and PapsAG-2 sequences with AG orthologues from tomato (TAG; Q40168), Antirrhinum majus (AmFAR; CAB42988), Arabidopsis thaliana (AtAG; P17839) and Eschscholzia californica (EScaAG1 and EScaAG2; AAZ53205 and AAZ53206). PapsAG-2 has a 24-amino-acid extension. The conserved AG motifs are indicated.
Alignment of the predicted products of PapsAG-1 and PapsAG-2 with C and D class MADS box proteins from core eudicots, basal eudicots and monocots (Fig. S1 in Supplementary Data, available online) confirmed that PapsAG is an AG orthologue. PapsAG lacks characteristic D-lineage synapomorphies (Dreni et al., 2007) such as Q105 and H113 which are diagnostic for D-lineage MADS box proteins as compared with C-lineage sequences. In addition, both PapsAG-1 and PapsAG-2 encode the AG sequence motifs I and II (Fig. 1B), which are highly conserved in C class proteins (Kramer et al., 2004). Another feature of C-lineage AG-like genes is the presence of an extra intron at the C-terminus corresponding to a splice site at the extreme 3′ end of the genes (Kramer et al., 2004); further investigations were performed to determine if the PapsAG-1 and PapsAG-2 cDNAs were the result of alternative splicing at this site. Using a common forward primer and reverse primers specific for each of the putative 3′-UTRs, the corresponding genomic regions from Papaver somniferum DNA preparations were isolated. Analysis of the resulting sequences revealed that PapsAG-1 follows the splicing pattern typical of AG orthologues and splices to a point approx. 1·5 kb downstream where it encounters the stop codon. PapsAG-2, on the other hand, proceeds to an alternative splice site approx. 1 kb downstream of the donor splice site and incorporates an extra 24-amino-acid coding region in the process (Fig. 1A, B). Examination of the corresponding genomic sequence showed that there were sequence motifs for donor and acceptor splice sites that matched consensus splice sites in plants (Fig. S2 in Supplementary Data; Brown and Simpson, 1998). Furthermore, the two proposed acceptor sites for PapsAG-1 and PapsAG-2 were the only two predicted splice acceptor sites (with confidence levels of 100 % and 94 %, respectively) using the NetPlantGene prediction tool (Hebsgaard et al., 1996).
Alternative splicing affecting AG family members has been reported previously (Kitahara and Matsumoto, 2000; Lightfoot et al., 2008), including a report in crocus where it affects the C-terminal region specifically (Tsaftaris et al., 2005), but any functional consequences have not been assessed in these cases. Protein structure prediction using PredictProtein (Rost et al., 2004) suggests that the 24-amino-acid extension encoded by PapsAG-2 maintains a structural profile similar to that of the unextended version and is most likely involved in protein–protein interactions. In turn, the modifications of the PapsAG-2 C-terminal domain protein sequence would suggest that it might form qualitatively distinct higher-order protein complexes as compared with PapsAG-1.
Expression analysis of PapsAG-1 and PapsAG-2
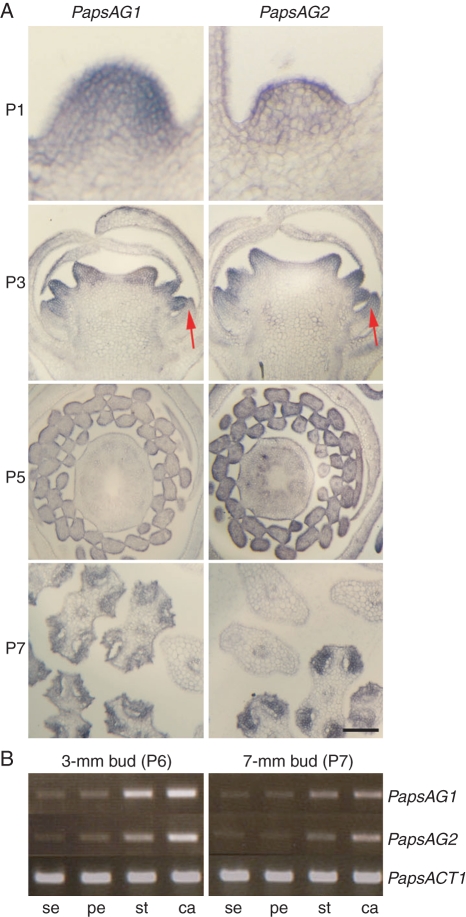
To examine the expression patterns of the two PapsAG transcripts semi-quantitative RT-PCR and in situ hybridizations were employed. Sepals, petals, stamens and carpels were dissected from stage P6 and P7 flowers (stages according to Drea et al., 2007) and, using RT-PCR, both PapsAG-1 and PapsAG-2 showed expression in all whorls (Fig. 2B). Both genes were expressed more strongly in the two innermost whorls, as expected for AG orthologues. A more detailed analysis of expression patterns within the flower was conducted using in situ hybridization on flowers at stages P1, P3, P5 and P7 using probes derived from 3′-UTR sequences and so specific for each transcript. The expression patterns for both transcripts were identical with the possible exception of a somewhat broader expression domain of PapsAG-2 in the petal (Fig. 2A, arrow). Expression of both transcripts was detected very early in the young floral meristem (P1) and through early to mid-stages of flower development was detectable in stamens and carpels (Fig. 2A, P3, P5 and P7). Later in flower development the stamen expression was restricted to the anthers (Fig. 2A, P7) as has been observed for Arabidopsis AG (Ito et al., 2004). The expression in stamens and carpels and PapsAG-2 expression in petals are maintained at stage P5 and both transcripts are detected in septae periphery and in primordial ovule at stages P6 and P7 (Fig. S3 in Supplementary Data). Control experiments using corresponding sense probes and a histone H4 antisense probe were performed (Fig. S4 in Supplementary Data).
Fig. 2.
Expression analyses of PapsAG-1 and PapsAG-2. (A) In situ hybridization of PapsAG-1 and PapsAG-2 transcript-specific probes on young P. somniferum flowers showing similar expression patterns. P1 stage, Young meristem in longitudinal section before visible organ primordia appear; P3 stage, longitudinal section of developing flower bud; P5 stage, cross-section of older flower bud; P7 stage, cross-section through developing anthers with adjacent filament sections. The stages are as in Drea et al. (2007). Scale bar = 200 µm. (B) RT-PCR with PapsAG-1 and PapsAG-2 transcript-specific primers using cDNA from dissected floral organs (sepal, se; petal, pe; stamen, st; carpel, ca) of older bud stages (P6, 3-mm buds; P7, 7-mm buds). Amplification of the P. somniferum ACTIN gene, PapsACT1, was used as a control.
Functional dissection of AG orthologues in Papaver somniferum
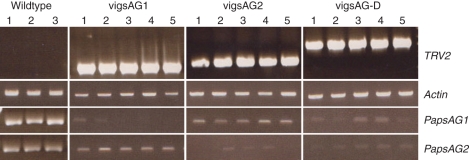
Both PapsAG-1 and PapsAG-2 are expressed at high and comparable levels in the poppy flower, indicating that they are potentially functional and do not represent pseudogenes. Given the considerable sequence similarity and nearly identical expression patterns of the PapsAG-1 and PapsAG-2 transcripts, it was essential to ascertain whether the encoded functions were similar or distinct. To investigate the functions of both transcripts' products, VIGS constructs were generated to silence each transcript individually and both transcripts simultaneously. At least 100 young seedlings were infiltrated with each of the three constructs as previously described (Hileman et al., 2005; Drea et al., 2007) and flowers analysed for phenotypes and gene silencing using semi-quantitative RT-PCR. Table 1 summarizes the phenotype frequencies and type for each of the three constructs. In the cases of vigsAG1 and vigsAG-D at least 50 % of the infiltrated plants produced abnormal flowers, whereas only approx. 12 % of vigsAG2 flowers were identified as having defects. This could be due to the possibility of both the more subtle functional roles of the PapsAG-2 transcript and the increased difficulty in recognizing these more subtle defects in the flowers. Flowers were examined at two main stages of development: when the flower is pendant prior to anthesis just before its upright extension and flower opening, corresponding to stage P8 (Fig. 3), and at anthesis (Fig. 4). RNA was extracted from the inner two whorls of the flowers examined and RT-PCR was performed to test for presence of the TRV2 construct containing the correct sized insert and to test for down-regulation of the PapsAG-1 and PapsAG-2 transcripts (Fig. 6). Control experiments with TRV2 containing no insert (TRV2-E) were also carried out but no defects were observed (Fig. S5 in Supplementary Data).
Table 1.
Summary of phenotypes identified using VIGS to silence PapsAG1 and PapsAG2 genes individually and both genes simultaneously
| Phenotype |
||||
|---|---|---|---|---|
| vigs line | Whorl 3 | Whorl 4 | Number | Total |
| vigsAG1 | Anthers partially transformed | Carpel (stigma and ovules) deformed, gynophores extended | 51 | 51 |
| vigsAG2 | Normal | Stigma and ovule defects | 12 | |
| Normal | Pedicel-like bend in carpel | 4 | 16 | |
| vigsAG-D | Petalloid | Carpel petalloid or sepalloid with recurring flower inside | 37 | |
| Petalloid | Severely sepalloid and hollow; some contain rudimentary ovules | 23 | ||
| Some petalloidy – anthers partially transformed | Carpels deformed | 5 | ||
| Morphologically normal but some partially green | Carpels deformed | 4 | 69 | |
Approximately 100 seedlings for each construct were infiltrated and screened for VIGS-induced phenotypes.
Fig. 3.
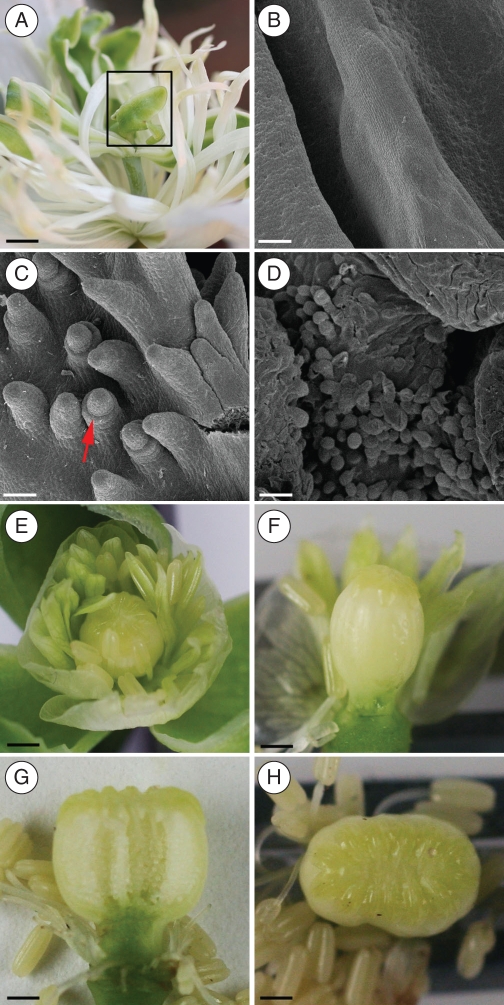
Phenotypes of vigsAG1,vigsAG2 and vigAG-D plants at pendant flower stage: (A) wild-type poppy flower; (B) vigsAG1 flower showing extended gynophore (black arrow), partially transformed anthers (red arrow) and open stigma (blue arrow); (C) vigsAG1 capsule cut in transverse showing sepals developing inside (arrow); (D) vigsAG2 curved capsule (inset shows normal stamens from a wild-type flower); (E, F) vigsAG2 flower showing defective stigma (red arrow); (G, I) vigsAG-D flowers showing petaloid stamens and sepaloid carpel with petals inside (G), petaloid carpel with another flower inside with extra pedicel indicated (red arrow, H) and elongated sepaloid carpel which was completely empty inside (inset, I). Scale bars = 2 mm.
Fig. 4.
Distinct effects of PapsAG-1 and PapsAG-2 gene silencing in capsule development: (A) wild-type stigma; (B) wild-type capsule cut open in transverse orientation showing ovules and septae; (C, E) vigsAG1 deformed stigmas; (D) vigsAG1 capsule cut in longitudinal orientation showing defective ovule development particularly at the distal end; (F) vigsAG1 open flower showing extended gynophores and modified anthers; (G, H) vigsAG2 stigma and open capsule cut in transverse orientation showing the deformed stigma and corresponding ovule defects within the capsule; (I, J) scanning electron micrographs of vigsAG2 stigmas, showing a retracted stigmatic ray (blue arrow) and a stigmatic pore in reverse orientation, i.e. it is directed towards the centre of the stigma (red arrow, I); disorganized stigma with ectopic papillae (blue arrow) and ovule (red arrow, J). Scale bars: (A–E, G, H) = 2 mm; (F) = 5 mm; (I) = 200 µm; (J) = 100 µm.
PapsAG-1 loss of function affects stamen and carpel whorls
In wild-type P. somniferum flowers, the globular ovary is crowned by a radial arrangement of fused stigmatic rays containing papillae-lined pores for the capture of the copious pollen grains and conduction of pollen tubes to the ovules within (Fig. 4A, B). The multicarpellate gynoecium is attached to the pedicel by a thin gynophore, and lacks an obvious style. The gynoecium is paracarpous, in that the margins of the fused carpels form projections called septae rather than meeting at the centre of the ovary (Bernáth, 1998). The surrounding stamens are numerous and hypogynous. In contrast to the unfixed number of stamens and carpels, the perianth consists of two (or sometimes three) sepals and four petals. The flowers progress through a series of well-characterized stages (Drea et al., 2007), and by stage 8, when the flower bud is pendant, all floral organs are distinct (Fig. 3A).
Flowers of infiltrated plants were opened manually and visually examined for any obvious defects at stage 8 and in open flowers. vigsAG1 flowers displayed a consistent phenotype consisting of partially transformed stamens – anthers appeared petalloid, more obviously at the extreme distal end, an open stigma where the rays have not converged centrally, and extended gynophores (Fig. 3B, C). Though the capsule is recognizable in general morphology, inside the developing capsule ectopic petals and/or sepals were observed in many cases (Fig. 3C). The effects on gynophore development were more apparent in mature flowers, in which the gynophore was quite elongated and resembled an extended pedicel (Fig. 4F). Examination of mature flowers also revealed the extent of the defects in ovule development where both aborted and unfertilized ovules were observed, particularly in the distal region of the capsule (Fig. 4D). Stigma defects produced either over-papillated or more naked surfaces (Fig. 4C, E). Overall it appeared that PapsAG-1 affects both the stamen and carpel whorl identity and differentiation. The presence of ectopic outer-whorl tissues within the carpel of some vigsAG1 plants also suggests that the gene plays a role in determinacy.
PapsAG-2 loss of function affects capsule development
The effects of PapsAG-2 VIGS were more difficult to identify presumably due to more subtle effects on flower development. Stage 8 flower buds were overtly normal in appearance and it was only on close examination of the developing capsules that defects in stigma arrangement and overall capsule shape were observed (Fig. 3E, F). Stamens appeared to be normal in all of these plants. In a subset of the defective flowers identified, the entire capsule was curved (Fig. 3E), which was reminiscent of the pedicel curvature observed in pendant opium poppy flowers at stage 8. The gynophores of these flowers, however, appeared to be normal in all cases. In mature flowers the stigma defects were more obvious. The radial arrangement of the stigmatic rays, as observed in wild-type plants (Fig. 4A), was distorted (Fig. 4G) and when the capsule was opened it was seen that there were corresponding defects in ovule development (Fig. 4H) – ovules were very white and enlarged or underdeveloped. The stigmas of vigsAG2 plants were examined more closely using scanning electron microscopy. As was the case for vigsAG1, stigmas were occasionally over-papillate or contained naked surfaces (Fig. 4I, J). Papillae sometimes developed along the centre of the rays where no pores were forming and occasionally an ovule was found developing amongst the papillae (Fig. 4J). In other cases, the pores were reversed in orientation with the turn directed toward the centre of the radial surface, rather than around the stigma boundaries (Fig. 4I). Overall the effect of PapsAG-2 was restricted to the innermost whorl and could be interpreted as being the result of a mild determinacy loss within the carpel.
Both PapsAG transcripts act redundantly to confer floral determinacy and organ identity
Silencing of both PapsAG transcripts simultaneously generated flowers with a dramatic phenotype involving a complete loss of stamen and carpel identity and a high incidence of loss of determinacy (Fig. 3G–I). vigsAG-D flowers consisted of stamens that were considerably or completely transformed into petals (Fig. 4A). Carpels were affected in three main ways: (1) individual carpels could be transformed into a sepal that enclosed developing petals (Fig. 3G), or (2) they could surround an empty interior (Fig. 3I) or (3) they could be transformed into petals (Fig. 3H). The identity of the transformation to petals or sepals was assigned based on visual identification (colour and form) and by scanning electron microscopy on the transformed tissues.
An internal examination of transformed carpels (region boxed in Fig. 5A) using scanning electron microscopy showed a completely smooth adaxial wall (Fig. 5B). Where internal structures were observed within these carpels, they consisted of some rudimentary ovule development on the adaxial surface where the integuments are only barely discernable (Fig. 5C) and/or short papillae at the distal end (Fig. 5D). These observations indicate that both PapsAG-1 and PapsAG-2 have a role in the specification of floral determinacy, as well as shared roles in determining aspects of floral organ identity. Of the vigsAG-D lines, 13 % displayed a weaker phenotype (Table 1) where there was a more partial transformation of stamens, recognizable carpels with some defects and a normal (unextended) gynophore (Fig. 5E–H). In these cases the carpels produced a smaller stigmatic ray area (Fig. 5E, F) or were misshapen (Fig. 5G, H). The existence of such phenotypes suggests that stamen and carpel identity can be uncoupled from the determinacy function.
Fig. 5.
Phenotypes of vigsAG-D plants: (A) strongly transformed vigsAG-D flower with no distinguishable stamens and rudimentary gynoecium (boxed); (B–D) scanning electron micrographs of adaxial transformed carpels (boxed in A) showing the lack of any ovule initiation (B), in some lines there are rudimentary ovules without developing integuments (red arrow, C) and some stigmatic papillae at the distal end (D). (E–H) Phenotypes of weaker vigsAG-D lines showing partially petaloid (E, F) or generally normal stamens (G, H) and recognizable carpels that show a reduced stigmatic ray area (E, F) or resemble fused carpels (G, H). Scale bars: (B–D) = 50 µm; (A, E–H) = 2 mm.
Fig. 6.
RT-PCR of vigs lines. RT-PCR on RNA/cDNA extracted from the two innermost whorls (stamen and carpels) of pendant flowers from vigs lines for five representative plants transformed with each construct to test for the presence of the TRV construct and for reduced expression of the PapsAG transcripts. Three individual wild-type plants with tissue from the same stage (pendant) were included for comparison.
DISCUSSION
PapsAG-1 and PapsAG-2 are derived from an alternative splicing event
Sequence and phylogenetic analyses of MADS box genes have shown the prevalence of ancient and recent gene duplications in establishing the repertoire of these genes in a number of extant angiosperms (Mena et al., 1996; Kramer et al., 2004; Yamaguchi et al., 2006; Zahn et al., 2006). Gene duplication can have various functional consequences such as neofunctionalization, subfunctionalization or pseudogenization (Drea et al., 2006, 2007; Yamaguchi et al., 2006; Dreni et al., 2007) but a single gene can also potentially generate multiple forms through the production of alternative transcripts from the same locus.
Genomic analyses in rice and Arabidopsis suggest that 20 % of genes are alternatively spliced with intron retention being the most common consequence (Wang and Brendel 2006). It is not unusual in genes encoding proteins with modular structure where exon gain or loss can result in the acquisition or relinquishing of discrete functional modules such as the acquisition of target domains in organelle-localized gene products (Long et al., 1996) or potential neofunctionalization through sequence modification in transmembrane regions (Drea et al., 2006). The FCA gene, encoding an RNA-binding protein required for flowering, produces multiple transcript forms (Macknight et al., 2002). These vary in both spatial and temporal expression of the transcripts and in the abundance of the resulting proteins with subsequent effects on the timing of flowering responses. Alternative splicing and RNA processing within the MADS-box genes in general and the C-class genes in particular has been reported (Kitahara and Matsumoto 2000; Cheng et al., 2003; Lee et al., 2005; Lightfoot et al., 2008) but the functional significance of these events has not yet been elucidated. Where alternative splicing of C-class genes was reported, the splicing site was not the extreme C-terminal splice junction (intron 8) except for the report in Crocus (Tsaftaris et al., 2005), though the number and position of introns are generally conserved in orthologues from various species (Kramer et al., 2004).
The two isoforms of the encoded PapsAG products differ only in the length of the C-terminus region. Divergence in the C-terminal domains encoded by various MADS-box genes has been deemed to be an important determinant of whether a gene duplicate is retained (Janssens et al., 2008) and can affect the nature of the interactions with other MADS-box proteins in higher-order complexes (Geuten et al., 2006). Since the analyses performed here were unable to discern any major differences in the expression patterns of the PapsAG-1 and PapsAG-2 transcripts, it is suspected that their distinct functions are likely to be due to differences in their protein–protein interactions or in transcriptional activation potential.
Conservation and diversification in AG function
This study shows that the Papaver somniferum AG orthologue is required to specify stamen and carpel identity as well as floral determinacy, similar to the role of Arabidopsis AG. However, loss of AG function in Arabidopsis results in homeotic conversions of stamens into petals and carpels into sepals (Bowman et al., 1991). This is in contrast to the loss of PapsAG function in vigsAG-D plants, in which homeotic conversions of carpels to petals were observed in addition to other defects. This phenotype is more similar to the complete loss of AG-like gene function in Antirrhinum plena farinelli double mutants, which display homeotic conversions of both stamens and carpels into petaloid tissue (Davies et al., 1999). This has been postulated to be due to misregulation of B-class MADS box genes in antirrhinum through disruption of PLENA and FAR interactions with a third whorl specific factor (Davies et al., 1999). In P. somniferum, such a postulated third whorl factor would presumably differentially interact with the duplicated B class MADS box genes which each possess distinct functions (Drea et al., 2007).
In Arabidopsis, the role of AG in floral determinacy has been associated with a higher level of AG activity as compared with that required for organ identity specification (Mizukami and Ma, 1995). Furthermore, analyses of partial loss-of-function AG alleles in Arabidopsis has provided support for the idea that the C-terminus of the K domain is required for distinct third and fourth whorl functions as well as correlating determinacy with increased gene function (Sieburth et al., 1995). In other species, these two AG roles appear to have been subfunctionalized; for instance, in rice the AG orthologues OsMADS3 and OsMADS58 have distinct roles, with OsMADS3 being required predominantly for stamen identity specification and OsMADS58 being the main player in conferring floral determinacy (Yamaguchi et al., 2006).
This study shows that AG activities in opium poppy are uncoupled through having a single gene encode two alternative transcripts encoding distinct proteins with different lengths of C-terminal domains. PapsAG-1 mediates organ identity and determinacy functions in both the stamen and the carpel whorls, whereas PapsAG-2 function appears to be largely restricted to the carpel. This parsing of functions into two transcripts is distinct from gene duplication and subfunctionalization, and may reflect a lineage specific mechanism to encode distinct protein functions. A similar observation has been made for the multiple splice forms that are unique to arthropods and that have not been observed in vertebrate systems; nonetheless, the overall developmental function of these receptor isoforms is conserved (Schmucker and Chen, 2009). Though the PapsAG transcripts show some functional redundancy, the unique functions encoded by each transcript would account for the retention of both, in a manner similar to that postulated for the retention of duplicated, subfunctionalized genes (Force et al., 1999; Moore and Purugganan, 2003; Moore et al., 2005). As such, the production of alternative transcripts may provide another means to diversify gene function, and may reflect fine tuning of the types of multiprotein complexes that can be formed to mediate different developmental functions.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
P.H. is funded by a BBSRC studentship and S.D. by the University of Leicester and a Royal Society Research Grant.
LITERATURE CITED
- Aldrich J, Cullis C. RAPD analysis in flax: optimization of yield and reproducibility using klen Taq 1 DNA polymerase, chelex 100, and gel purification of genomic DNA. Plant Molecular Biology Reporter. 1993;11:128–141. [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Bernáth J, editor. Amsterdam: Harwood Academic Publishers; 1998. Poppy: the genus Papaver. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Brown JWS, Simpson CG. Splice site selection in plant pre-mRNA splicing. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:77–95. doi: 10.1146/annurev.arplant.49.1.77. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, et al. Evolution in action: following function in duplicated floral homeotic genes. Current Biology. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Developmental Cell. 2003;4:53–66. doi: 10.1016/s1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO Journal. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) – a new model for the study of dioecy. The Plant Journal. 2005;41:755–766. doi: 10.1111/j.1365-313X.2005.02336.x. [DOI] [PubMed] [Google Scholar]
- Drea S, Corsar J, Crawford B, Shaw P, Dolan L, Doonan JH. A streamlined method for systematic, high resolution in situ analysis of mRNA distribution in plants. Plant Methods. 2005;1:8. doi: 10.1186/1746-4811-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea SC, Lao NT, Wolfe KH, Kavanagh TA. Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. The Plant Journal. 2006;46:723–735. doi: 10.1111/j.1365-313X.2006.02741.x. [DOI] [PubMed] [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF. Functional analyses of genetic pathways controlling petal specification in poppy. Development. 2007;134:4157–4166. doi: 10.1242/dev.013136. [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, et al. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. The Plant Journal. 2007;52:690–699. doi: 10.1111/j.1365-313X.2007.03272.x. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K, Becker A, Kaufmann K, et al. Petaloidy and petal identity MADS-box genes in the balsaminoid genera Impatiens and Marcgravia. The Plant Journal. 2006;47:501–518. doi: 10.1111/j.1365-313X.2006.02800.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Research. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy) The Plant Journal. 2005;44:334–341. doi: 10.1111/j.1365-313X.2005.02520.x. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Molecular Biology and Evolution. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Current Opinion in Genetics and Development. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, et al. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- Janssens SB, Viaene T, Huysmans S, Smets EF, Geuten KP. Selection on length mutations after frameshift can explain the origin and retention of the AP3/DEF-like paralogues in Impatiens. Journal of Molecular Evolution. 2008;66:424–435. doi: 10.1007/s00239-008-9085-5. [DOI] [PubMed] [Google Scholar]
- Kapoor LD. Opium poppy: botany, chemistry and pharmacology. Philadelphia, PA: The Haworth Press; 1997. [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kitahara K, Matsumoto S. Rose MADS-box genes ‘MASAKO C1 and D1’ homologous to class C floral identity genes. Plant Science. 2000;151:121–134. doi: 10.1016/s0168-9452(99)00206-x. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cho YS, Yoon HS, et al. Conservation and divergence of FCA function between Arabidopsis and rice. Plant Molecular Biology. 2005;58:823–838. doi: 10.1007/s11103-005-8105-8. [DOI] [PubMed] [Google Scholar]
- Lightfoot DJ, Malone KM, Timmis JN, Orford SJ. Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Molecular Genetics and Genomics. 2008;279:75–85. doi: 10.1007/s00438-007-0297-y. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. The Plant Journal. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Long M, de Souza SJ, Rosenberg C, Gilbert W. Exon shuffling and the origin of the mitochondrial targeting function in plant cytochrome c1 precursor. Proceedings of the National Academy of Sciences of the USA. 1996;93:7727–7731. doi: 10.1073/pnas.93.15.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. The Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Molecular Biology. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The early stages of duplicate gene evolution. Proceedings of the National Academy of Sciences of the USA. 2003;100:15682–15687. doi: 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Grant SR, Purugganan MD. Molecular population genetics of redundant floral-regulatory genes in Arabidopsis thaliana. Molecular Biology and Evolution. 2005;22:91–103. doi: 10.1093/molbev/msh261. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Research. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes and Development. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. The Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaftaris AS, Pasentsis K, Polidoros AN. Isolation of a differentially spliced C-type flower specific AG-like MADS-box gene from Crocus sativus and characterization of its expression. Biologia Plantarum. 2005;49:499–504. [Google Scholar]
- Voelckel C, Borevitz JO, Kramer EM, Hodges SA. Within and between whorls: comparative transcriptional profiling of Aquilegia and Arabidopsis. PLoS One. 2010;5:e9735. doi: 10.1371/journal.pone.0009735. doi:10.1371/journal.pone.0009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proceedings of the National Academy of Sciences of the USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. The Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack JH, Arrington JM, et al. Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evolution and Development. 2006;8:30–45. doi: 10.1111/j.1525-142X.2006.05073.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.