Abstract
A hidden Markov model-based gene-finding system called Genie was applied to the genomic Adh region in Drosophila melanogaster as a part of the Genome Annotation Assessment Project (GASP). Predictions from three versions of the Genie gene-finding system were submitted, one based on statistical properties of coding genes, a second included EST alignment information, and a third that integrated protein sequence homology information. All three programs were trained on the provided Drosophila training data. In addition, promoter assignments from an integrated neural network were submitted. The gene assignments overlapped >90% of the 222 annotated genes and 26 possibly novel genes were predicted, of which some might be overpredictions. The system correctly identified the exon boundaries of 70% of the exons in cDNA-confirmed genes and 77% of the exons with the addition of EST sequence alignments. The best of the three Genie submissions predicted 19 of the annotated 43 gene structures entirely correct (44%). In the promoter category, only 30% of the transcription start sites could be detected, but by integrating this program as a sensor into Genie the false-positive rate could be dropped to 1/16,786 (0.006%). The results of the experiment on the long contiguous genomic sequence revealed some problems concerning gene assembly in Genie. The results were used to improve the system. We show that Genie is a robust hidden Markov model system that allows for a generalized integration of information from different sources such as signal sensors (splice sites, start codon, etc.), content sensors (exons, introns, intergenic) and alignments of mRNA, EST, and peptide sequences. The assessment showed that Genie could effectively be used for the annotation of complete genomes from higher organisms.
INTRODUCTION
The Genome Annotation Assessment Project (GASP) was organized by the Berkeley Drosophila Genome Project to determine the accuracy of current computational gene annotation methods when applied to the Drosophila melanogaster genome sequence. Gene annotation were submitted by 12 groups for the well-annotated Adh region of this genome (Reese et al. 2000). The predictions we submitted were computed using the Genie suite of software tools for gene finding. The Genie system is a generalized hidden Markov model (GHMM) that incorporates signal and content sensors as described in a recent review (Haussler 1998). Signal sensors model statistical information from functional sites in genomic DNA such as splice sites, start and stop codons, branch points, and promoters. In contrast, content sensors model global statistical properties of genes. The most studied model is the sensor to predict coding regions, referred to as coding exons or simply exons. In Genie, these content sensors are mostly based on the coding usage and coding preferences (summarized in Fickett and Tung 1992) as well as a length distribution for these content sensors. The applied Genie system is a newly trained version of the original work first described by Kulp et al. (1996). This initial version was trained and optimized for human genes. The work was a first implementation and optimization of earlier theoretical work by Stormo and Haussler (1994). Improvements on the splice site models as well as a description of the training for Drosophila melanogaster and initial results for this organism were reported in Reese et al. (1997). The team further developed the system to integrate so-called homology information into a statistical gene-finding framework (Kulp et al. 1997).
For the GASP experiment, three annotation files were submitted. The first, named Genie, was generated using statistical information from the cleaned gene collection as described in the next section. The second submission, named GenieEST, used the same signal sensors as Genie but extended the content sensors by incorporating EST information for the determination of the splice boundaries. The third submission, named GenieESTHOM, used, in addition to all the models from GenieEST, protein homology information from BLASTX runs (Altschul and Gish 1996) against the nonredundant protein Genbank database (nr). This run resulted in DNA–protein alignments to related protein sequences in Drosophila melanogaster, as well as to related protein sequences in other organisms.
METHODS
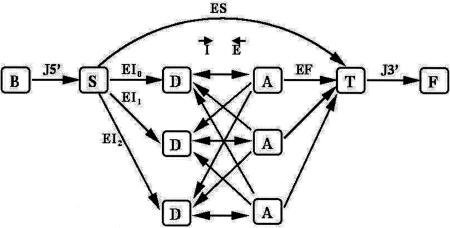
A GHMM is a probabilistic state machine representing gene structure, that is, a generative model that outputs random sequences along with a probability associated with each sequence. Figure 1 shows a schematic representation of the underlying model. Arcs in the graph are content sensors, that is, variable length features such as exons and introns, and the nodes in the graph are signal sensors corresponding to transitions between contents. The independent probabilities in the model are transition probabilities at nodes, length distributions on arcs, and likelihood models for each content sensor. The software that implements this model is very modular, and allows for easy integration of new nodes, arcs, and sensors.
Figure 1.
A GHMM including frame constraints. (B) The beginning state; (J5′) the 5′ UTR content sensor; (S) the start codon signal sensor; (EI) the initial exon content sensor; (D) the 5′ splice site sensor; (A) the 3′ splice site sensor; (E) the internal exon content sensor; (I) the intron content sensor; (EF) the final exon content sensor; (T) the start codon signal sensor; (F) the end state. (ES) The single exon gene content sensor. For multiple genes in genomic regions such as Adh, an additional arc loops from F to B and models the intergenic region including the promoter sensor.
Given a candidate sequence, the probability of the sequence given the model can be efficiently derived with a depth-first search. Further, by use of the same algorithm, the maximum probability path through the graph can be determined, which reveals the state sequence corresponding to the most likely gene structure. The details of the system are described elsewhere (D. Kulp, in prep.).
For the application to the Drosophila genomic sequence, Genie was trained on a dataset provided by the organizers of the GASP experiment (http://www.fruitfly.org/GASP/data/data.html), consisting of genomic DNA entries containing full coding region annotations from GenBank. The complete dataset consisted of 275 multiple exon and 141 single exon Drosophila genes. In addition to this well-annotated gene structure data set, all available coding sequences from mRNA sequence entries in GenBank for Drosophila melanogaster were used for training the codon usage/codon preference Markov models. For GenieESTHOM the genomic Adh sequence was run against the nonredundant (nr) GenBank protein database.
EST/cDNA alignments were used to predict intron splice pairs in GenieEST. By use of BLASTN (Altschul and Gish 1996), pairs of hits to the same subject sequence were extracted. When such pairs were approximately contiguous in the subject sequence and aligned near GT/AG splice boundaries, then an intron was predicted. The content sensor models for splice sites and introns were modified such that the probability was artificially raised for these so-called EST introns, effectively constraining the system to ensure that the introns were correctly annotated according to the EST/cDNA evidence.
As first described in Kulp et al. (1997), protein sequence homology is included as part of the content sensor for protein-coding regions in GenieESTHOM. By use of BLASTX, candidate homologs are identified and assigned a likelihood probability similar to the BLAST S score. The likelihood of a coding region that includes a protein database hit may be higher than by statistical analysis alone depending on the degree of similarity.
Part of the gene structure GHMM includes the core promoter region. The content sensor for this region is a time-delay neural network (Reese 2000). The low specificity of independent promoter prediction is compensated in this approach by integrating promoter prediction into the complete gene prediction. Thus, in effect, possible promoter sites are only considered if they occur upstream of a probable coding region.
We have submitted annotations for the gene-finding category as well as the promoter prediction category. In the following sections, both classes will be discussed separately.
Gene Finding
We submitted a total of 241, 246, and 258 gene predictions from Genie, GenieEST, and GenieESTHOM, respectively. In general, all three programs scored well in the gene-finding category (see Table 3 in Reese et al. 2000). We divide the summary of the results into the proposed three categories: Base level, Exon level, and Gene level, and discuss the performance for all three submissions.
Base Level Results
All three programs achieve >95% sensitivity (Reese et al. 2000). The extra information from ESTs and homology improve the sensitivity of the statistical Genie outcome by 1%. Most of the bases belonging to coding exons seem to be predicted by Genie, which makes the tool robust and sensitive for a first scan of genomes to identify most of the proteome of an organism.
In specificity, one can see a drop in performance for the Genie annotations that use homology information (GenieESTHOM) to 83% from 92% for Genie and 91% for GenieEST, respectively. This is surprising and means that Genie uses misleading protein homology information to predict coding regions that are noncoding. We believe this is due to some weaker homology hits that are recognizing protein-like elements in the DNA. These hits could be due to pseudogenes or just simply to elements that are protein like and were originally derived from real protein sequences either through outside integration by transposons, viruses, or simply by evolutionary gene duplication and subsequent degeneration through mutations. For 13 of the overpredicted genes, we know that they overlap transposable elements and, therefore, all 13 are counted as false positives (see Table 6, below for details on the overlapped transposons).
Table 6.
Transposable Elements Predicted by Genie
| Strand | Begin | End | Genie | Genie EST | Genie ESTHOM | Other gene finder hits | Homology hits | Transposon name |
|---|---|---|---|---|---|---|---|---|
| F | 55,422 | 58,941 | — | — | X | 4 | 2 | Fw |
| R | 93,549 | 94,119 | X | X | X | 3 | 1 | G |
| R | 255,612 | 256,662 | — | — | X | 1 | 1 | Doc |
| R | 959,378 | 962,797 | — | — | X | 2 | 1 | Doc |
| R | 1,136,806 | 1,145,466 | — | X | X | 5 | 0 | Roo |
| R | 1,293,597 | 1,298,741 | — | X | X | 5 | 1 | Copia |
| F | 1,474,114 | 1,481,634 | X (2 genes) | — | — | 3 | 2 | Yoyo |
| F | 1,935,760 | 1,943,170 | X | X | X | 3 | 2 | Blood |
| F | 2,076,116 | 2,083,110 | — | — | X | 3 | 2 | 297 |
| F | 2,174,330 | 2,176,188 | — | — | X | 1 | 1 | Copia-like |
| F | 2,177,045 | 2,178,655 | — | — | X | 3 | 2 | Copia-like |
| F | 2,590,477 | 2,595,625 | — | — | X | 5 | 2 | Copia |
| F | 2,603,050 | 2,610,046 | — | — | X | 2 | 1 | 297 |
The transposable elements that have an overlapped prediction by Genie are listed. The Begin and End coordinates are the transposable element coordinates from Ashburner et al. (1999).
Exon Level Results
Predicting splice sites, translation initiation, and termination is difficult to accomplish within a purely computational framework because these sites can be very divergent and might be regulated through the over- or under-representation of nucleotides in the respective consensus sequences. Prediction is further confounded by external enhancer or repressor-binding sites that are not well understood. The low rate of missed exons of 8.1%, 4.8%, and 3.2% for Genie, GenieEST, and GenieESTHOM, respectively, and the high sensitivity scores of >70% suggest that Genie finds almost all of the exons, but has more trouble predicting the precise boundaries correctly. GenieEST demonstrates significant improvement (sensitivity of 77% compared with 70%) in splice site identification, which is to be expected from the EST alignments.Sensitivity improves to 79% in GenieESTHOM. This tendency of improved scores for GenieEST and GenieESTHOM reverses itself on the specificity scores and wrong exon scores. Here, the best scores are from the pure statistical Genie program. This fact might reflect the data quality in the std3 reference set of presumed correct gene structures, in which quite a number of the genes are based on pure GENSCAN (Burge and Karlin 1997) predictions, a program similar in structure and concept to the statistical Genie program.
Gene Level Results
All three versions of Genie have problems assembling complete genes absolutely correctly. It is clear that this is a very hard problem, and so we find a sensitivity of 44% for GenieEST and GenieESTHOM to be very promising. We suspect that this is due to a well-balanced integration of statistical sensors combined with the strength of sequence similarity methods. Specificity is almost equal for Genie and GenieEST, but drops for GenieESTHOM, due to misleading hits to low-scoring protein-like elements. The relatively low number of wrong genes (10.7%) for the pure statistical Genie implies that users can have confidence that predicted genes do correspond, at least in part, to true protein-coding regions. Nothing in the training of Genie or in the application constrained Genie from predicting the transposases and the reverse transcriptases in the transposable elements as genes, and, of course, there might also be new genes that Genie recognizes that are not yet in the biological annotation from std3 (see Table 1 for details).
Table 1.
Possible Novel Genes
| Strand | Begin | End | Genie | Genie EST | Genie ESTHOM | Other gene-finder hits | Homology hits | Comments |
|---|---|---|---|---|---|---|---|---|
| F | 21,599 | 21,988 | X | — | — | 5 | 0 | |
| F | 131,015 | 131,248 | X | X | X | 5 | 0 | |
| F | 267,633 | 268,061 | X | X | X | 1 | 0 | |
| R | 306,476 | 306,985 | X | X | X | 1 | 0 | |
| R | 328,048 | 328,733 | X | X | X | 4 | 0 | |
| F | 329,808 | 331,184 | X | X | X | 6 | 0 | |
| F | 403,468 | 405,391 | X | X | X | 5 | 2 | |
| F | 408,759 | 412,000 | X | X | X | 6 | 2 | |
| R | 426,746 | 427,525 | X | X | X | 6 | 0 | |
| R | 603,442 | 604,456 | — | X | X | 5 | 0 | |
| F | 754,773 | 754,919 | X | X | X | 2 | 0 | |
| R | 846,339 | 845,892 | — | X | X | 3 | 0 | |
| R | 870,684 | 870,866 | X | X | X | 4 | 0 | |
| R | 910,572 | 911,055 | X | X | X | 5 | 0 | |
| F | 1,115,807 | 1,116,493 | X | X | X | 2 | 0 | |
| F | 1,117,474 | 1,117,608 | X | X | X | 3 | 0 | |
| R | 1,263,535 | 1,264,137 | — | X | X | 3 | 0 | |
| R | 1,365,077 | 1,365,732 | X | X | X | 4 | 0 | |
| R | 1,850,650 | 1,851,240 | X | X | X | 4 | 0 | |
| F | 2,453,955 | 2,454,498 | — | X | X | 4 | 0 | |
| F | 2,580,916 | 2,581,059 | X | X | X | 1 | 0 | possible FP |
| R | 2,584,165 | 2,584,914 | — | X | X | 3 | 0 | |
| R | 2,741,387 | 2,742,230 | X | X | X | 4 | 0 | |
| R | 2,762,639 | 2,774,287 | X | X | X | 2 | 0 | |
| F | 2,779,268 | 2,779,566 | X | X | X | 2 | 0 | |
| F | 2,843,324 | 2,843,386 | X | X | X | 1 | 0 | very short, possible FP |
Twenty-six Genie gene predictions that have no overlaps to any gene structure in std3. (Strand) The strand on which the predicted genes are located, consistent with the annotation in Ashburner et al. (1999) “Genes on the top [forward strand] of each map are transcribed from distal to proximal (with respect to the telomere of chromosome are 2L); those on the bottom [reverse strand] are transcribed from proximal to distal.” Begin and End gene coordinates note the first and last base of the predicted coding gene region by Genie. Genie, GenieEST, and GenieESTHOM label the Genie program variant. (Other gene finder hits) The count of how often this newly predicted gene is also overlapped by one or more of the other six gene-finding programs. (Homology hits) The count of how often a newly predicted gene overlaps any homology hits.
The statistic of split genes and joined genes describes the behavior of a program to assemble and separate genes from each other. The split gene numbers range from 1.17 to 1.15 for the three Genie programs, which indicates quite a high number of genes that are split into one or more genes. A total of 15%–17% of all genes are split into one or more separate gene predictions. The joint gene numbers are much lower (1.08–1.09), indicating the tendency of Genie to prefer to break up genes instead of joining them. Compared with other gene finders, both numbers are high, suggesting that other programs have better solutions for this problem.
Promoter Prediction
A total of 234 and 237 transcription start-site predictions were submitted for GeniePROM and GenieESTPROM, respectively. The success rate of the promoter assignment of ∼30% (27.1% for GeniePROM and 32.6% for GenieESTPROM) is in the same order as other programs, but indicates that promoter recognition is very difficult due to the complex initiation process. Because Genie's promoter assignments are in the context of gene identification and, as such, modeled in the complete generalized HMM to occur upstream of the start codon, the false-positive rate is low. For the evaluation of 853,180 negative bases, the rate is 1/14,710 for GeniePROM and 1/16,729 for GenieESTPROM, respectively (see Table 4 in Reese et al. 2000). It is interesting to recognize that the EST integration improves promoter identification, which might be due to an extension of the 5′ region of gene using information from a 5′ EST sequence. Because of the integration into a gene-finding system, the numbers should be compared with the similar MAGPIE system, and it can be seen that whereas GenieESTPROM misses two more promoters (30 vs. 33), the false-positive rate for it is lower (1/16,729 vs. 1/14,968).
Selected Gene Annotations
In this section, we discuss selected predictions or nonpredictions from Genie, GenieEST, GenieESTHOM, and GenieESTPROM compared with the standard sets std1 and std3, as well as the behavior of Genie compared with other gene-finding systems on the basis of the selected examples from Reese et al. (2000).
As indicated in Figure 1 and the corresponding legend, the three Genie submissions are grouped together in all the figures from Reese et al. (2000). They are the group of gene finders that are the farthest apart from the genomic sequence axis closest to the protein homology annotations.
In the “busy” region [in Reese et al. (2000); Fig. 2A], all three Genie submissions predict the first four (DS02740.4, DS02740.5, I(2)35Fb, DS02740.8) and the last of the forward strand genes (fzy) correctly. The fifth gene (DS02740.10), between 2,752,000 and 2,755,000 is only predicted by GenieEST and GenieESTHOM. This indicates that coding potential is not strong enough to distinguish protein coding from intergenic, and the additional information from EST sequences is necessary to identify the coding regions for this gene. Although GenieEST and GenieESTHOM predict the first three exons correctly, both miss the 3′ splice site for the fourth exon and then select a 3′ splice site in a different frame so that a stop codon is introduced in the middle of the real fourth exon. Thus, both programs miss the last four exons. It is possible that the coding potential for these remaining four exons is low, which is suggested by the fact that nothing is predicted with the statistical Genie. The fifth gene (Sed5) in this region on the forward strand is very interesting. Although two of the seven gene-finding programs follow the suggested annotation in std3, four others agree with a longer first coding exon annotation. All three Genie programs predict a longer initial coding exon. This is very interesting because of the difficulty in determining the exact start of translation of a gene. Most of the biologists predict the first ATG in a 5′ EST sequence, followed by a long ORF as the real start codon, but this is not a strict rule and might be wrong in some cases.
On the reverse strand, the complete Genie suite predicts a two-exon gene at 2,741,000–2,742,500, where no gene exists in the std3 reference set. Because this prediction agrees with four other gene-finding programs and does not overlap any of the transposon annotations from Ashburner et al. (1999), this might be a real gene missed in the std3 annotation. This gene also does not show any protein homology and might therefore be a novel gene (see Table 1 for details). Further EST screening and subsequent full-length sequencing studies may confirm this hypothesis. The next gene (heix) is correctly predicted by agreement of all three Genie programs, but the third gene (DS02740.9) on the reverse strand is not. The statistical Genie misses the first two exons and introduces a wrong start codon. EST sequence information extends the GenieEST and GenieESTHOM predictions, correctly identifying the final two and the second exon. However, both programs miss the initial exon, which is only 3bp long; the Genie model has a minimum length limitation of 6bp. The fourth and fifth genes (anon-35Fa and cni, respectively) gene on the reverse strand, both short genes with four and five exons, respectively, are predicted completely correctly. The last and longest gene (cact or cactus) in this region spans almost 12 kb from 2,762,639 to 2,774,287. The interesting fact about this gene is that it has a very long intron between the third and fourth exon spanning 8 Kb. Whereas most of the other gene-finding programs predict this intron correctly, all three Genie programs miss this intron and split this gene into two separate genes. This is a typical behavior of Genie and is addressed in the next version of the program.
Genie's overall low false-positive rate is demonstrated in the gene-poor region, [Fig. 2B in Reese et al. (2000)]. In this gene desert Genie predicts only two genes both on the reverse strand. The first is a single exon gene (DS01759.1), which is correctly predicted by all three Genie programs. GenieEST and GenieESTHOM both agree on an additional gene after this first single exon gene. Whereas other gene-finding programs predict single exon genes or genes containing two exons here, GenieEST and GenieESTHOM predict a gene with four exons. Although the exact structure of a possible gene in this region can only be wild speculation, it seems probable that there is a novel gene in this region.
All Genie programs predict the genes Adh and Adhr [Fig. 3A in Reese et al. (2000)] correctly. This is not surprising for GenieEST and GenieESTHOM because there are many ESTs available for both genes. But even without EST evidence, Genie predicted these duplicated genes correctly. As described in Ashburner et al. (1999), both genes are active but under regulation of only one and the same promoter. The integrated promoter prediction (GenieESTPROM) indicates a possible TSS at 1,111,271 for the Adhr gene with a reasonable score. It would be interesting to verify this prediction by biological experiments.
Analysis of the gene outspread (osp), (Fig. 3B), reveals a structural error in the gene model of Genie. The osp gene, the first gene on the reverse strand, contains many very long introns and contains, in one of these introns, the Adh/Adhr gene duplication on the opposite strand. In another intron, osp contains one gene (DS09219.1) on the same strand and another, equal to Adh/Adhr, on the opposite strand (DS07721.1). The current Genie model is built so that it does not allow gene(s) within or overlapping other genes either on the same or on the reverse strand. Therefore, the suite of Genie annotations break up the osp gene. The seven 3′ exons are predicted correctly, but Genie introduces an erroneous first exon to complete this gene prediction. The exon at 1,104,419–1,104,995 overlaps with a Genie prediction of a single exon gene from 1,104,411 to 1,104,965. The correct prediction of most protein-coding bases in osp, despite the program's inability to identify the full gene structure in this complex situation, demonstrates its graceful degradation on odd gene structures and may explain its high base-level sensitivity relative to the number of totally correct gene predictions. Although the remaining seven 5′ exons from osp are missed, the GenieEST and GenieESTHOM versions introduce a wrong three-exon gene in the middle of an intron. These EST-based Genie versions are forced to predict this gene through a mistaken EST sequence hit and alignment, which belongs to the overlapping DS09219.1 gene transcript (see Table 9, below, for details).
Table 9.
Erroneous EST UTR Predictions
| Strand | Begin | End |
|---|---|---|
| R | 40,843 | 43,076 |
| R | 346,994 | 356,311 |
| R | 393,573 | 398,794 |
| F | 507,364 | 512,758 |
| F | 849,268 | 851,919 |
| R | 1,372,338 | 1,373,546 |
| R | 1,756,026 | 1,761,674 |
| F | 2,491,469 | 2,497,464 |
| R | 2,698,932 | 2,706,347 |
| R | 2,709,485 | 2,711,209 |
Coding gene predictions by GenieEST and GenieESTHOM that are either complete overpredictions or partially wrong by extending the coding regions into the 5′/3′ UTR due to a wrong underlying gene model structure for noncoding ESTs (see text for details). The Begin and End coordinates are the GenieEST and GenieESTHOM predictions.
Additional evidence for the general Genie behavior of splitting genes comes from the most complex gene in the Adh region, the Ca-α 1D gene [Fig. 3C in Reese et al. (2000)]. This long gene with >30 exons is split by Genie into three separate genes. Most of the long exons are covered by Genie predictions, whereas some of the short exons are missed entirely.
In Figure 3D in Reese et al. (2000), the idgf cluster of three genes of the same family (idgf1, idgf2, and idgf3) shows the benefit of using EST information and the additional benefit of using homology information. The first intron is missed by the statistical Genie but recovered through the additional EST alignment information that spans this intron in GenieEST. Idgf3 is correctly predicted, but only in GenieESTHOM is ldgf1 predicted correctly from start to stop. In this case, the protein homology information extends the initial exon to a different start codon farther upstream.
Additional Selected Observations of the Genie Annotation
In Table 1 we list 26 potential novel genes that are predicted by at least one of the Genie programs and, in addition, have evidence through an overlap from at least one other gene-finding or homology program. The number seems to be very high (>11.7%), but because the process of annotating genes in genomic DNA is so hard, and because not all programs were available at the time of the annotation (Ashburner et al. 1999), we believe that at least the majority of these predictions are real genes. All predictions that overlap with an annotated transposable element were removed from this list.
Table 2 lists the 19 genes from the reference std3 set for which no overlap of a Genie prediction exists. Thus, <10% of the annotated genes in std3 are missed by Genie. The individual submission scores are as low as 4.6%. We note that nine of these std3 annotations are based solely on predictions from the human version of GENSCAN (Burge and Karlin 1997) and/or gene finder (P. Green, unpubl.) predictions, the two gene-finding programs used for the annotations in Ashburner et al. (1999). For an additional six of these genes, Ashburner et al. (1999) augmented their evidence with BLAST hits with low P values. For Mst35Bb, there exists a very reliable cDNA alignment and it is certainly a real missed gene by Genie. The three remaining genes (DS07721.1, DS003192.3, DS003192.4) are all based on cDNA alignments. None of these genes is predicted by any gene-finding program, and for only one, DS003192.4, there are two homology annotations but on the opposite sequence strand. Therefore, we believe that these alignments are very questionable and might be the result of typical cDNA-cloning artifacts as mentioned briefly in Reese et al. (2000). To summarize the analysis of the 19 overpredicted genes, it is possible that the missed prediction rate of Genie is below the noted 4.6% and that very few real genes are missed.
Table 2.
Genes Entirely Missed by Genie
| Strand | Begin | End | Other gene-finder hits | Homology hits | Gene names (Ashburner et al. 1999) | Evidence (Ashburner et al. 1999) | Comments |
|---|---|---|---|---|---|---|---|
| R | 230,985 | 240,152 | 3 | 0 | DS08249.5 | gene prediction only | — |
| F | 498,520 | 507,581 | 2 | 0 | DS01514.3 | gene prediction only | — |
| R | 523,395 | 525,283 | 3 | 0 | DS05899.7 | gene prediction plus low P value BLAST hit | — |
| R | 533,592 | 536,913 | 3 | 0 | DS05899.6 | gene prediction only | — |
| F | 1,152,128 | 1,152,385 | 0 | 0 | DS07721.1 | cDNA (?) | very suspicious annotation |
| R | 1,285,030 | 1,286,199 | 3 | 1 | DS06874.6 | gene prediction plus low P value BLAST hit | — |
| F | 1,300,469 | 1,315,922 | 3 | 0 | DS06874.7 | gene prediction only | — |
| R | 1,368,793 | 1,369,282 | 5 | 0 | Mst35Bb | cDNA | — |
| F | 1,484,701 | 1,489,834 | 2 | 0 | DS00929.16 | gene prediction only | suspicious annotation |
| R | 1,520,808 | 1,521,371 | 1 | 1 | DS00929.7 | gene prediction plus low P value BLAST hit | overlaps gene on opposite strand |
| F | 1,628,242 | 1,628,412 | 0 | 0 | DS003192.3 | cDNA (?) | very suspicious annotation |
| R | 1,663,026 | 1,663,163 | 0 | (2 opposite strand) | DS003192.4 | cDNA (?) | very suspicious annotation |
| R | 1,782,412 | 1,786,409 | 2 | 0 | Ms(2)35Ci | gene prediction only | — |
| R | 1,875,987 | 1,895,879 | 5 | 0 | DS03023.4 | gene prediction only | gene on opposite strand |
| R | 2,109,315 | 2,113,209 | 4 | 0 | BACR44L22.5 | gene prediction only | — |
| F | 2,158,476 | 2,159,460 | 3 | 2 | DS07108.5 | gene prediction plus low P value BLAST hit | — |
| F | 2,236,081 | 2,241,876 | 3 | 0 | DS02252.3 | gene prediction plus low P value BLAST hit | 5000-bp single-exon gene |
| R | 2,286,435 | 2,287,433 | 3 | 0 | DS02252.4 | gene prediction plus low P value BLAST hit | — |
| F | 2,398,367 | 2,410,394 | 5 | 0 | DS07486.5 | gene prediction only | — |
The gene names from Ashburner et al. (1999) are listed. In addition, the evidence for that gene annotation from that paper is given. The Begin and End gene coordinates are from the std3 annotations. In addition, we list the number of overlaps by other gene finders and the two homology programs.
One of the biggest problems with the Genie programs in the Adh annotation are joined and split genes. Tables 3 and 4 show that Genie is parameterized to favor splitting genes versus joining genes. Only 9 Genie annotations span two or more std3 genes (joined genes), whereas 19 std3 genes are split into separate Genie-predicted genes (split genes). The problem of joining genes is due to the difficulty of identifying the ends and starts of genes that unfortunately do not encompass strong statistical signals. Careful analysis of the split genes, on the other hand, showed that the length distribution of introns, a geometric distribution that favors short introns, is the reason for so many split genes. Another behavior related to the same problem of the length distributions of introns is the general tendency of Genie to introduce erroneous exons within otherwise long introns. Table 5 lists 11 typical examples.
Table 3.
Joined Genes
| Strand | Begin | End | Genie | Genie EST | Genie ESTHOM | No. of joined genes | Names of joint genes |
|---|---|---|---|---|---|---|---|
| R | 336,668 | 339,013 | X | X | X | 2 | DS00941.11, DS00941.12 |
| F | 341,713 | 343,984 | X | X | X | 2 | DS00941.14, DS00941.15 |
| F | 454,701 | 458,802 | X | X | X | 2 | DS00180.5, DS00180.12 |
| R | 458,837 | 463,657 | X | X | X | 2 | DS00180.7, DS00180.8 |
| F | 471,109 | 476,389 | X | X | — | 2 | DS00180.11, DS00180.14 |
| R | 839,712 | 843,808 | X | X | X | 3 | DS01068.10, DS01068.4, DS01068.5 |
| F | 1,599,218 | 1,607,306 | X | X | X | 2 | DS04929.3, stc |
| R | 2,102,169 | 2,104,442 | — | — | X | 2 | BACR44L22.8, BACcr44L22.2 |
| R | 2,786,019 | 2,792,601 | X | X | X | 3 | DS02740.18, DS02740.19, DS09218.1 |
All predictions in which Genie joins one or more genes from std3 are listed. The Begin and End gene coordinates are from the Genie predictions. The last two columns list the number of genes joined and their respective names.
Table 4.
Split Genes
| Strand | Begin | End | No. of split genes | Comments |
|---|---|---|---|---|
| F | 45,358 | 130,409 | 5 | gene on opposite strand |
| F | 373,286 | 391,500 | 3 | |
| F | 445,189 | 456,317 | 3 | |
| R | 477,171 | 487,236 | 2 | |
| F | 568,986 | 575,533 | 2 | |
| R | 679,874 | 691,416 | 2 | |
| F | 757,457 | 821,487 | 4 | one gene on same strand |
| R | 1,094,414 | 1,182,415 | 2 | two genes on opposite strand |
| R | 1,398,183 | 1,413,067 | 2 | |
| F | 1,506,022 | 1,521,842 | 2 | gene on opposite strand |
| F | 1,558,915 | 1,561,694 | 2 | |
| F | 1,565,296 | 1,585,380 | 3 | |
| R | 1,653,146 | 1,667,970 | 2 | |
| R | 1,718,580 | 1,737,780 | 2 | |
| R | 1,747,063 | 1,752,780 | 2 | |
| R | 2,220,563 | 2,224,367 | 2 | |
| F | 2,463,394 | 2,488,789 | 2 | |
| F | 2,619,967 | 2,639,006 | 3 | |
| R | 2,714,362 | 2,736,449 | 2 |
All std3 gene annotations are split into two or more genes by all three Genie programs. The Begin and End gene coordinates are from the std3 annotations. Comments are given for the reason of the splitting when genes within genes occur.
Table 5.
Missed Long Intron(s)
| Strand | Begin | End | Genie | Genie EST | Genie ESTHOM |
|---|---|---|---|---|---|
| R | 268,751 | 273,483 | X | X | X |
| R | 654,984 | 667,105 | X | X | X |
| F | 828,047 | 833,672 | X | X | X |
| R | 880,856 | 901,495 | X | X | X |
| R | 1,051,748 | 1,057,314 | — | — | X |
| R | 1,271,377 | 1,276,359 | X | X | X |
| R | 1,421,921 | 1,432,223 | X | X | X |
| F | 1,974,488 | 1,983,855 | X | X | X |
| F | 2,040,123 | 2,057,901 | X | X | X |
| F | 2,505,534 | 2,530,156 | X | X | X |
| F | 2,683,427 | 2,694,719 | X | X | X |
Genes that have long introns that are missed by any Genie program are listed and it is indicated which program misses them. The Begin and End coordinates are from the std3 annotations.
A simple, but unfortunate oversight is the poor treatment of transposable elements in the Adh region. Ashburner et al. (1999) found 17 transposable elements, which consist of repetitive elements, but also protein coding-like regions including long ORFs, predominantly for the transposase and the reverse transcriptase proteins. As expected, Genie cannot distinguish these transposon genes from protein-coding genes and, therefore, predicts 13 of the existing 17 genes as protein-coding genes (see Table 6 for a list of predicted transposons). In particular, GenieESTHOM predicts many of the transposable elements to be coding genes, because transposable elements contain protein sequences that result in strong protein alignments. Whereas the statistical Genie version only overlaps 3 of the 17 transposable elements, GenieESTHOM predictions overlap 13. These transposon hits contribute to an increased false-positive rate, wrong exon and wrong gene scores, and lower overall specificity (see Table 3 in Reese et al. 2000).
In Table 7, we report five gene annotations on the basis of Genie predictions that strongly indicate either a different gene structure than reported in std3 or a potentially new alternative splicing form for the listed genes. The underlying evidence along with the Genie predictions come from other gene-finding predictions as well as from EST sequence alignments.
Table 7.
Alternative Splicing Forms Predicted
| Strand | Annotations | Begin | End | Genie | Genie EST | Genie ESTHOM | Other gene finder hits | Gene name | Comments |
|---|---|---|---|---|---|---|---|---|---|
| F | std3 | 159,578 | 163,527 | X | 4 | DS01368.1 | EST alignment verifies last additional intron | ||
| Genie | 159,578 | 164,417 | X | X | X | 5 | |||
| R | std3 | 325,240 | 326,379 | MtPolB | additional first exon | ||||
| Genie | 325,240 | 326,822 | X | X | X | 3 | |||
| R | std3 | 1,334,780 | 1,338,785 | X | 5 | DS03431.1 | missed third exon, (EST verified) | ||
| Genie | 1,334,780 | 1,338,785 | X | X | 1 | ||||
| R | std3 | 1,371,813 | 1,372,351 | 2 | Mst35Bb | longer first exon and shorter last exon | |||
| Genie | 1,371,868 | 1,372,213 | X | X | 3 | ||||
| F | std3 | 1,493,680 | 1,496,198 | 1 | DS00929.1 | wrong first exon and EST intron in second exon | |||
| Genie | 1,495,484 | 1,496,198 | X | X | X | 4 |
Genes in std3 that might have an alternative gene structure as predicted by Genie and with similar predictions from other gene finders are listed.
Through evidence from high-scoring Genie predictions, EST alignments and the other annotation teams eight gene entries in the std3 reference set seem to be very suspicious. In Table 8, predictions from other programs are only listed if they support the suggested corrected gene structure annotated by Genie. Careful cDNA alignment and additional full-length cDNA sequencing should shed light into these cases.
Table 8.
Possible Wrong Annotations in std3
| Strand | Begin | End | Genie | Genie EST | Genie ESTHOM | Other gene finder hits | Homology hits | Gene name | Evidence (Ashburner et al. 1999) | Evidence |
|---|---|---|---|---|---|---|---|---|---|---|
| R | 213,507 | 217,188 | X | X | X | 7 | 1 | DS08249.3 | gene prediction only | last exon questionable |
| F | 281,649 | 284,052 | X | X | X | 6 | 2 | D00797.5 | cDNA (partial?) | missing 10 leading exons |
| R | 941,115 | 944,598 | X | X | X | 4 | — | DS08340.1 | gene prediction only | four extra 3′ exons |
| F | 1,205,439 | 1,213,325 | X | X | X | 5 | — | DS07721.3 | gene prediction only | first exon and last 3 exons questionable |
| R | 1,371,813 | 1,372,351 | — | X | X | 3 | — | TFIIS | known gene | longer first and shorter last exon |
| R | 1,549,142 | 1,549,933 | X | X | X | 6 | — | DS07295.4 | gene prediction only | initial exon and first EST verified intron missed |
| F | 1,721,863 | 1,728,736 | X | — | — | 1 | — | DS07295.4 | gene prediction only | at least first seven exons very questionable |
| R | 1,913,374 | 1,914,948 | X | X | X | 6 | 1 | wor | gene prediction plus BLAST homology hits | additional first EST-verified exon |
Genes from the std3 annotations are listed for which multiple evidence from Genie and other programs exists implying wrong annotations. The Begin and End coordinates are from the std3 annotations. The evidence for the annotation in std3 is given as noted in Ashburner et al. (1999) In addition, comments for rejecting the annotated gene structure by Genie are listed.
DISCUSSION
What Went Right?
The results of GASP show that high specificity on the nucleotide base level can be achieved by using any of the three Genie programs. Splice sites and start and stop codons can be predicted with high confidence. If cDNA sequences exist, Genie can integrate them into the gene structure prediction and refine its splice site predictions.
All three Genie systems, including the EST/cDNA and protein sequence alignments, are fully integrated and automated. While computing the predictions in the 2.9-Mb region, the sequence was divided into 500-kb pieces with 10-kb overlaps. Overlapping predictions in the 10-kb overlaps were then manually resolved. This last manual step is automated in newer versions of the software.
In general, Genie is optimized to have a low false-positive rate for predicted genes. Whereas Ashburner at el. (1999) annotated 222 genes, the Genie programs found 241–258 genes, in which most of the overpredictions are believed to be true, but so far, undetected genes.
Although only ∼30% of the known transcription start sites could be predicted by the integrated neural network-based promoter prediction method—no better or worse than other methods—the false-positive rate, a typical problem in promoter prediction, is reduced. It seems that the integrated transcription start site prediction in a gene-finding system such as Genie might be the only way of scanning for promoters in a complete genome.
What Went Wrong?
Although the base-level predictions and exon-level predictions were very good, the results for gene assemblies partially expressed in joined and split genes are not. The major culprit is a poor model of long introns, which sometimes results in split genes. We have subsequently introduced two intron content sensors into the GenieGHMM—one for the usual shorter introns and one for the rare long introns. To help avoid split genes, we have also modified the EST integration to exploit pairing of 5′ and 3′ reads from the same clones. This information indicates gene boundaries, even when the complete gene lacks EST coverage.
Another oversight that resulted in many false-positive predictions in each performance category was the nontreatment of transposons. Many coding regions in transposable elements were mistaken as genes, especially when using protein homology. A simple prescreening method for transposable elements could have masked out these regions and eliminated them from being predicted by Genie.
A structural mistake in the Genie gene model for GenieEST and GenieESTHOM resulted in erroneous predictions when EST evidence identified introns between noncoding exons (Table 9). At the time of the assessment experiment, Genie's exon and intron models were exclusively based on coding region of genes. The revelation of this weakness has led us to change the underlying gene model, adding the notion of an intron in an UTR region.
All of the above errors were corrected in subsequent versions. Thus, the GASP experiment, the first of its kind to assess gene-finding technologies on a large contiguous genomic sequence region, was extremely useful and helped us directly to improve our system.
However, the challenge of allowing genes within genes is more difficult. The lack of examples of eukaryotic overlapping genes has been a significant impediment. It will be interesting to see how EST alignment information might be helpful for predicting genes within genes. In addition, it should be noted that alternative splicing was not addressed in this GASP experiment. We believe ESTs, clustered by shared splice form or divided by tissue type, will play a key role in addressing this problem.
CONCLUSIONS
Over the years, Genie has become a robust gene-finding system. It allows for automatic training for new organisms, is highly modular, allowing for the integration of new external sensor models, runs fully automatically, even for entire genomes, and the running time is reasonable when applied to complete genomes such as the human genome. The statistical framework allows for a probabilistic assessment of individual predicted features and complete gene predictions. The concept of a generalized or semi-hidden Markov model is very powerful, as can be seen in the high performance scores of all systems on the basis of GHMMs in this experiment.
Acknowledgments
We thank the whole team at Neomorphic Inc. for supporting this work through software development efforts, providing the necessary computing resources and the fruitful research-oriented environment. M.G.R. thanks Gerry M. Rubin for his continuing scientific support and advice for developing this system as a part of his doctoral thesis. M.G.R. was supported by NIH grant HG00750. D.H. gratefully acknowledges the support of DOE Grant DE-FG03-95ER62112.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mgreese@lbl.gov; FAX (510) 486-6798.
REFERENCES
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of drosophila melanogaster. The adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Fickett JW, Tung CS. Assessment of protein coding measures. Nucleic Acids Res. 1992;20:6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler, D. 1998. Computational genefinding. Trends Biochem. Sci. Suppl. Guide Bioinformatics 12–15.
- Kulp D, Haussler D, Reese MG, Eeckman FH. A generalized hidden Markov model for the recognition of human genes in DNA. Intell Systems Mol Biol. 1996;4:134–142. [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. Integrating database homology in a probabilistic gene structure model. Pac Symp Biocomput. 1997;2:232–244. [PubMed] [Google Scholar]
- Reese MG. “Genome Annotation in Drosophila melanogaster.” Ph.D. thesis. Hohenheim, Germany: University of Hohenheim; 2000. [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Reese, M.G., G. Hartzell, N.L. Harris, U. Ohler, and S.E. Lewis. 2000. Genome annotation assessment in Drosophila melanogaster. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Stormo GD, Haussler D. Optimally parsing a sequence into different classes based on multiple types of evidence. Intell Systems Mol Biol. 1994;2:369–375. [PubMed] [Google Scholar]