Abstract
Activation of T-helper cells is dependent upon the appropriate presentation of antigen-derived peptides on MHC class II molecules expressed on antigen presenting cells. In the current study we explored the repertoire of peptides presented on MHC class II molecules on human monocyte derived dendritic cells (moDCs) from four HLA-typed healthy donors. MHC class II-bound peptides could be routinely recovered from small cultures containing 5 × 106 cells. A fraction of the identified peptides were derived from proteins localized in the plasma membrane, endosomes, and lysosomes, but the majority of peptides that were presented on MHC class II originate from other organelles. Subsequently, we studied the antigen-specific peptide repertoire after endocytosis of a soluble antigen. Blood coagulation factor VIII (FVIII) was chosen as the antigen since our current knowledge on MHC class II presented peptides derived from this immunogenic therapeutic protein is limited. Analysis of the total repertoire of MHC class II-associated peptides revealed that per individual sample 20–50 FVIII-derived peptides were presented on FVIII-pulsed moDCs. Repertoires of FVIII-derived peptides eluted from moDCs derived from a panel of four HLA typed donors revealed that some MHC class II-presented FVIII peptides were presented by multiple donors, whereas the presentation of other FVIII peptides was donor-specific. In total 32 different core peptides were presented on FVIII-pulsed moDCs from four HLA-typed donors. Together our findings provide an unbiased approach to identify peptides that are presented by MHC class II on antigen-loaded moDCs from individual donors.
Antigen presenting cells (APCs)1 continuously process endogenous and exogenous antigens into small peptides that are loaded on MHC class I or MHC class II molecules for presentation to T lymphocytes (1). Classically, endogenous antigens are presented on MHC class I molecules for presentation to CD8+ T cells whereas peptides derived from exogenous, internalized antigens are loaded on MHC class II molecules and activate CD4 positive T cells. Over the last decade this concept has been successfully challenged. Firm proof has been obtained for the presentation of exogenous antigens on MHC class I molecules for cross-priming of CD8+ T cells (2). Similarly, inspection of the repertoire of naturally occurring peptides presented on MHC class II molecules revealed that the majority of the presented peptides are in fact derived from endogenous proteins (3, 4). Not surprisingly, a large proportion of naturally presented peptides are derived from proteins that reside in endosomes or lysosomes (3, 4). Recent studies suggest that resident proteins of nonendocytic compartments, such as mitochondria or the nucleus, can also be presented on MHC class II molecules by sampling of intracellular compartments through autophagy (5–8). Current efforts to probe the repertoire of antigen-derived naturally presented peptides are limited by the number of cells needed to obtain substantial amounts of MHC class II bound peptide. Until now the repertoire of naturally presented peptides has been mainly explored using panels of well-characterized immortalized B cells. Typically, around 5 × 109 cells are used for sample preparation (9–11). More recently, MHC class II-presented peptides have been successfully isolated from tissue specimens of patients with multiple sclerosis (12). An elegant study by Wahlstrom and coworkers used human bronchial lavage cells from a pool of patients with sarcoidosis to obtain information on antigenic peptides involved in the pathogenesis of this disease (13). Further advances in MHC peptide identification and quantification by mass spectrometry have already led to the identification of large numbers of MHC class I peptides from more limiting amounts of cells (14) and has allowed for functional analysis regarding the role of the immunoproteasome in the generation of MHC class I peptides (15).
The aim of this study was to investigate whether a significant amount of MHC class II-presented peptides can be eluted from small cultures of human monocyte-derived dendritic cells (moDCs). MoDCs are professional APCs that express high levels of MHC class II that becomes surface-exposed following their maturation (16). Our results indicate that several hundred MHC class II-bound peptides can routinely be eluted from samples containing as few as 5 × 106 moDCs. This allows for the analysis of MHC class II presented peptides from a 50-ml blood draw from individual donors. We subsequently investigated whether pulsing of moDCs with an antigen resulted in the presentation of antigen-derived peptides on MHC class II. Blood coagulation factor VIII (FVIII) was used as a model antigen for this study. Therapeutic administration of FVIII is used to correct the bleeding tendency of hemophilia A patients who lack functional FVIII (17). Up to 25% of patients with hemophilia A develop high affinity antibodies in response to infusion of FVIII, which are also referred to as “inhibitors” (18). FVIII inhibitors are mostly high-affinity IgG antibodies, which are the result of FVIII-specific T-cell activation by professional APCs, followed by T-cell dependent antibody class-switching and affinity maturation (19). At present our knowledge on the repertoire of naturally presented FVIII derived peptides is limited. Here we show that per individual donor between 20 and 50 partially overlapping FVIII-specific peptides could be recovered from moDCs, corresponding to 8–17 different potential CD4+ T-cell epitopes. These findings suggest that small numbers of moDCs can be used to probe the repertoire of presented peptides of potentially immunogenic antigens such as FVIII.
EXPERIMENTAL PROCEDURES
Subjects
Blood was drawn from HLA-typed healthy volunteers in accordance with Dutch regulations and following approval from Sanquin Ethical Advisory Board in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells were isolated from freshly drawn, EDTA anticoagulated blood by separation over a Ficoll-Paque PLUS gradient (d = 1.077, GE Healthcare, Uppsala, Sweden).
Reagents
In this study, the following reagents were used: Recombinant human FVIII (Advate) was kindly provided by Dr. B.M. Reipert (Baxter Healthcare Corporation, Vienna, Austria), CD14 microbeads (MACS, Miltenyi Biotech Bergisch Gladbach, Germany), anti-CD80-FITC, anti-CD83-APC, anti-CD86-APC (BD Biosciences, San Jose, CA) and anti-CD14-PE (Sanquin Reagents, Amsterdam, the Netherlands). Cellgro DC serum-free medium, IL-4, and GM-CSF were obtained from CellGenix (Freiburg, Germany). LPS was obtained from Sigma-Aldrich (St. Louis, MO). Hybridoma L243 (anti-HLA-DR) was obtained from ATCC (Wesel, Germany).
MoDC Preparation and Factor VIII Endocytosis
Monocytes were isolated from the peripheral blood mononuclear cell fraction by positive selection using CD14 microbeads and a magnetic cell separator (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of the monocytes isolated was determined by flow cytometry analysis (20). Monocytes were cultured at a concentration of 0.66 × 106 cells/ml in a 6-well plate (Nunc, Roskilde Denmark) in Cellgro medium supplemented with GM-CSF (1000 IU/ml) and IL-4 (800 IU/ml) for 5 days (20). After 5 days of culture, the immature moDCs were washed and replated in Cellgro medium supplemented with GM-CSF and IL-4 at a concentration of 2.5 × 106 cells/ml in a final volume of 2 ml. Cell were incubated with 100 nm FVIII for 5 h before induction of maturation. After 5 h, the immature moDCs were maturated using 1 μg/ml LPS for 24 h in the presence of 1% human serum. The adherent maturated moDCs were detached by 5 min incubation with phosphate buffered saline (PBS) containing 0.25% trisodiumcitrate and washed before analysis.
Flow Cytometric Analysis of Cell-surface Phenotype
For determining the phenotype of moDCs, immature moDCs or mature moDCs were washed with PBS containing 0.5% bovine serum albumin (PBS/0.5% BSA) and incubated with 50 μl of 1 μg/ml mAb or appropriate isotype controls diluted in PBS/0.5% BSA with 3 mg/ml human gamma globulin for 30 min at 4 °C. Cells were washed twice and resuspended in PBS/0.5% BSA. 4′,6-diamidino-2-phenylindole was added to the cells before analysis to assess cell viability and exclude dead cells from analysis. Cells were analyzed on an LSRII flow cytometer (Beckton Dickinson, San Jose, CA) and analyzed with Flowjo software version 7.5.5 (Tree Star, Inc, Ashland, OR).
Purification of HLA-DR Presented Peptides on moDCs
HLA-DR molecules were purified from FVIII-treated, maturated moDCs or PBS treated control moDCs essentially as described previously (9). Briefly, moDC pellets were resuspended in 50 mm Tris pH 7.0 containing 4% Igepal CA-630 (Sigma). The membrane fraction was solubilized by end-over-end incubation at 4 °C for 30 min. HLA-DR was purified from the detergent-soluble fraction by immuno-affinity chromatography using antibody L243-coupled to CNBr Sepharose 4B (Amersham Biosciences, Buckinghamshire, UK) in the presence of protease inhibitors (Complete Protease Inhibitor Mixture Tablet, 1 tablet per 50 ml buffer, Roche Diagnostics GmbH, Mannheim, Germany) overnight at 4 °C. After washing the Sepharose 5 times with 50 mm Tris-HCl pH 7.0, peptides were eluted from HLA-DR by incubation with 10% acetic acid for 15 min at 70 °C. Eluted peptides were purified from the acetic acid eluate using a C18 ziptip (Millipore, Billerica, MA). In parallel experiments, cell lysates were incubated with isotype control antibody-coupled Sepharose (clone CLB-T4/1, mouse IgG2a, Sanquin Reagents, Amsterdam, The Netherlands).
Analysis of Peptides by Mass Spectrometry
Eluted peptides were separated using a reversed-phase C18 column (50 μm × 20 cm, 5 μm particles) (Nanoseparations, Nieuwkoop, The Netherlands), at a flowrate of 100 nl/min with a 1 h gradient from 0% to 35% (v/v) acetonitrile with 0.1% HAc. Eluted peptides were sprayed directly into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific Inc, Bremen, Germany) using a nanoelectrospray source with a spray voltage of 1.9 kV. The LTQ was operated in a data-dependent mode by performing collision induced dissociation in the ion-trap (35% normalized collision energy) for the five most intensive precursor ions selected from each full scan in the Orbitrap (300–2000 m/z, resolving power 30.000). An isolation width of 2 Da was used for the selected ions (charge ≥2) and an activation time of 30 ms. Dynamic exclusion was activated for the MS/MS scan with a repeat count of 1 and exclusion duration of 30 s. To obtain a high mass accuracy, the LTQ Orbitrap was calibrated on a monthly basis using a calibration solution consisting of caffeine, MRFA, and Ultramark 1621 as recommended by the manufacturer.
Peptide Identification
Peptides were identified using a Sequest search algorithm against UniprotKB nonredundant protein database 25.H_sapiens.fasta (53,784 nonredundant entries actually searched), using Proteome Discoverer release version 1.1 software (Thermo Scientific, Bremen, Germany) (21). Identification of peptides was performed using the following filter settings. During the Sequest search, we allowed a mass deviation of 20 ppm. In general mass deviations were below 3 ppm. Fragment mass tolerance was 0.8 Da. All peptides with a charge state of 2 have a minimal XCorr score of 2.0. For peptides with charge state 3, the minimal XCorr score is 2.25. For charge state 4, 2.5; charge state 5, 2.75; charge state 6, 3.0; charge state 7, 3.2, and charge state >7, 3.4. All peptides not complying to these criteria were excluded. Annotated spectra of individual peptides described in this manuscript are provided in the supplementary information on pages 5–126.
Validation of MHC Class II Bound Peptides by Differential Expression Analysis
MoDCs were prepared from donor A as described previously. Duplicate endocytosis experiments were conducted on 2 × 106 cells, instead of 5 × 106 cells. Cells were incubated with either 100 nm FVIII or with PBS as a control. Immune precipitations were performed as described previously, using either L234-coupled Sepharose (anti-MHC class II) or Sepharose coupled with an isotype control antibody. Each sample was analyzed separately by mass spectrometry. SIEVE™ release version 1.2.0 software (Thermo Scientific, Bremen, Germany) was used to compare the duplicate experiments and subsequently analyze differences in peptide abundance between the different conditions. All frames were analyzed with retention time between 20 and 50 min and m/z between 300 and 2000. Peak intensity threshold was set at 100,000. For peptide identification, the same sequest criteria were used as described previously.
RESULTS
Quantitative Analysis of MHC Class II-Bound Peptides
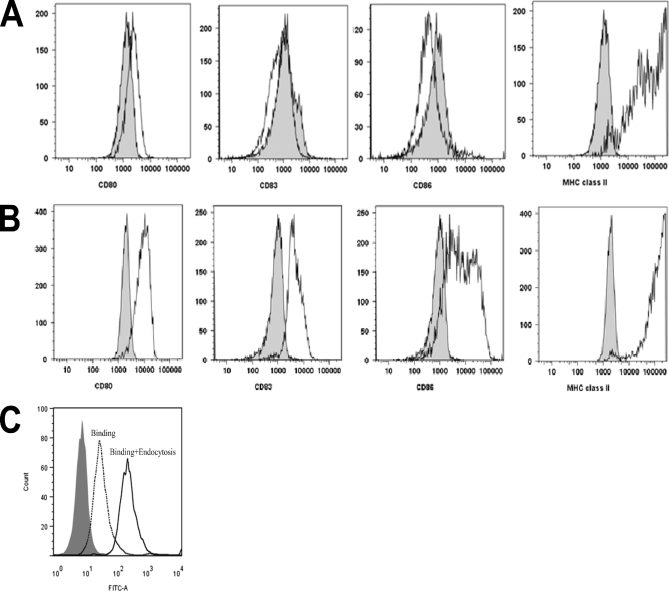
To study the peptide repertoire presented on MHC class II, we used moDCs from HLA-typed healthy donors. Immature moDCs were analyzed for the presence of CD14, CD80, CD83, CD86, and MHC class II on the surface (Fig. 1A). Immature moDCs were defined as CD14 negative and were also checked for the absence of maturation markers CD80, CD83, and CD86. MHC class II was present on immature moDCs. Cells that were maturated with LPS were positive for maturation markers and showed an increased surface expression of MHC class II (Fig. 1B). FVIII is endocytosed by moDCs, leading to activation of FVIII-specific CD4+ T cells (22). We confirm that FVIII is efficiently taken up by immature moDCs (Fig. 1C).
Fig. 1.
Cell surface markers of immature and mature moDCs. Cells were analyzed at the immature (A) and mature (B) state for the presence of cell surface markers CD80, CD83, CD86, and MHC class II. Gray histograms indicate isotype controls. A representative culture is depicted. C, Endocytosis and presentation of FVIII by moDCs. Histogram shows the fluorescence intensity for a FITC-labeled anti FVIII antibody (CLB-CAg-117). Binding of CLB-CAg-117-FITC to untreated cells is shown in gray. Cells stained in the absence of saponin show the membrane binding of FVIII (dotted curve). Increase of fluorescence in cells stained in the presence of saponin indicates that endocytosis of FVIII has taken place. A representative graph of one of the four donors is depicted.
For the analysis of MHC class II-bound peptides, HLA-DR molecules were purified from cell lysates of mature moDCs using anti-HLA-DR antibody L243-Sepharose. HLA-DR-bound peptides were subsequently obtained as described in the Experimental procedures. The sequence of the eluted HLA-DR ligands was determined by liquid chromatography (LC)-MS.
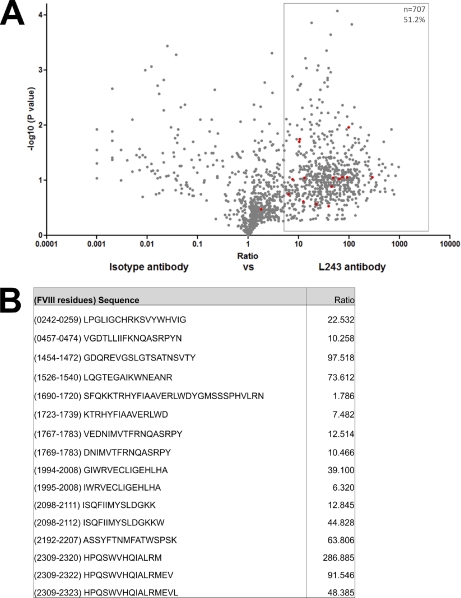
To determine to what extent our method generates specific MHC class II-bound peptides, immunoprecipitations were performed on FVIII-treated moDC samples from donor A (DRB1*0101 and DRB1*1301), using L243-Sepharose, which were compared with control immunoprecipitations, using an isotype control antibody coupled to Sepharose. The relative abundance of peptides between these experiments was compared using SIEVE 1.2. differential analysis software. Fig. 2A clearly shows that the majority of peptides have a ratio isotype:L243 higher than 1, meaning that these peptides are more abundantly present when samples are immunoprecipitated with L243-Sepharose as compared with the samples in which isotype-Sepharose was used. Peptides were regarded as specific MHC class II ligands when the ratio was above 5. Using this stringent threshold, many peptides are regarded as nonspecific. All FVIII peptides, however, with the exception of one peptide, are specific MHC class II ligands (Figs. 2A and 2B). Absolute identification of peptides present in samples immunoprecipitated with the control-Sepharose does, however, identify additional FVIII peptides. These peptides are all derived from the same region in FVIII. This set of nonspecific Sepharose-binding peptides is reproducible for all donors tested (data not shown). The fact that even these peptides may be enriched in samples immunoprecipitated with L243 could mean that, in addition to their background binding to control-Sepharose, they also contain an MHC class II-binding motif. This is also shown in Fig. 2A, where a FVIII peptide, containing the same sequence as the one identified as nonspecific, has a ratio of 7.482. Peptides with this sequence were excluded from all subsequent analyses. To further illustrate that peptides in Fig. 2A with a ratio larger than 5 are in fact true MHC class II-associated peptides, the Propred prediction algorithm was used to compare the predicted binding properties of all peptides with a ratio larger than 5 with peptides that have a ratio less than 5 (23). supplemental Fig. 1 shows that peptides with a ratio larger than 5 are predicted to bind significantly better to both MHC class II alleles of donor A (DRB1*0101 and DRB1*1301) than peptides with a ratio below 5.
Fig. 2.
Identification and relative quantification of MHC class II-bound peptides. A, Volcano plot representation showing reproducibly detected peptide ions across duplicate analyses. MoDCs from donor A (DRB1*0101, 1301) were used. FVIII-treated cells were immunoprecipitated using either an anti-MHC class II antibody or an isotype control antibody. Experiments were performed in duplicate. SIEVE was used to compare intensities of individual peptides eluted from L243 or isotype-control Sepharose. Peptide clusters highlighted in the box on the right hand side were considered as MHC class II-bound peptides (fold change ≥5). Only peptides that were sequenced by MS/MS are depicted. FVIII-derived peptides are indicated in red. The sequence and ratio of all FVIII peptides are shown in panel (B).
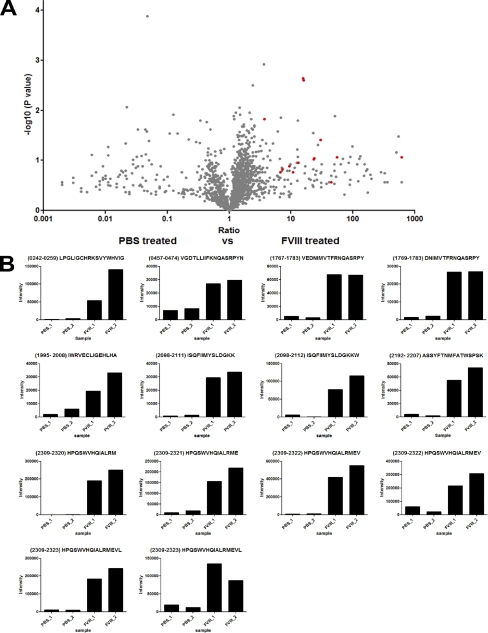
Differential expression analysis of PBS-treated moDCs versus FVIII-treated moDCs, using SIEVE 1.2 software, revealed that all FVIII peptides were identified as specific for the FVIII-treated cells as seen in Figs. 3A and 3B. This validates the correct identification of FVIII peptides. Peak intensities from individual FVIII peptides obtained for duplicate samples of PBS- and FVIII-treated moDCs revealed that FVIII peptides are markedly less present in samples derived from PBS-treated moDCs (Fig. 3B).
Fig. 3.
Identification and relative quantification of MHC class II-bound FVIII peptides. A, Volcano plot representation showing reproducibly detected peptide ions across duplicate analyses. MoDCs from donor A (DRB1*0101, 1301) were used. Cells were treated with either 100 nm FVIII or with PBS. All samples were immunoprecipitated using an anti-MHC class II antibody. Experiments were performed in duplicate. SIEVE was used to compare intensities of individual peptides obtained from FVIII and PBS treated cells. Only peptides that were derived from MS2 spectra are depicted. Peptides identified as FVIII-derived peptides are indicated in red. B, Peak intensities of all identified FVIII peptides are shown for experimental and control samples. It should be noted that the sets of FVIII peptides that were identified in the analysis presented in Fig. 3B when compared with Fig. 2B are not completely identical. The reason for the observed differences between Fig. 2B and 3B is that these particular peptides were filtered out by the algorithms employed by SIEVE during the comparison, due to local spectral differences. Manual analysis of the raw data files revealed that all peptides depicted in Fig. 2B and 3B were indeed present in the duplicate samples prepared from FVIII-treated cells using L243 (MHC class II specific) Sepharose.
Analysis of MHC Class II-Presented Peptides
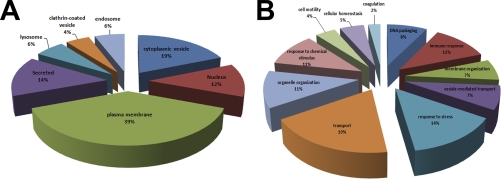
Fig. 2A clearly shows that most peptides that are presented on MHC class II are not derived from FVIII. We therefore analyzed which other proteins are presented as well. All peptides in Fig. 2A with a ratio larger than 5 were subsequently identified using Sequest and annotated based on subcellular localization (Fig. 4A) and function (Fig. 4B), using the online functional annotation tool DAVID v6.7 (http://david.abcc.ncifcrf.gov)(24, 25). Many peptides presented on HLA-DR are derived from proteins that reside in compartments, which are part of the MHC class II-presentation pathway, such as plasma membrane, cytosol, and lysosomes. However, proteins from other cellular compartments, such as the nucleus (12%), and secreted vesicles (14%) are also presented. Many of the endogenously presented peptides are derived from proteins that are functionally involved in intracellular transport (19%), immune response (12%) and stress response (14%) (Fig. 4B).
Fig. 4.
Subcellular and functional and categorization of source proteins. All proteins of which peptides with a ratio ≥5 were identified in Fig. 2A (specific MHC class II-bound peptides) were annotated based on subcellular localization or function using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resource. A, A pie chart shows the percentages of proteins found presented on MHC class II according to their subcellular localization. B, A pie chart shows the functional association of these proteins.
Analysis of MHC Class II-Presented FVIII Peptides
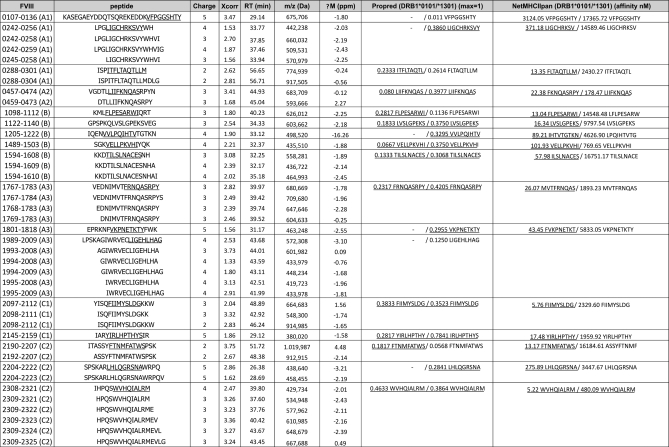
In the previous paragraph we have established that limited numbers of moDCs suffice to detect a significant part of the repertoire of peptides presented on HLA-DR after endocytosis of FVIII. We used moDCs derived from four different donors to further study the repertoire of naturally presented FVIII peptides. All FVIII-derived peptides identified comply with the XCorr scores described in the Experimental procedures. In addition, FVIII peptides were excluded when they were identified as aspecific binders according to the method described in Fig. 2A. FVIII peptides could be divided into sets of peptides with overlapping sequences that were comprised of differently processed variants of the same core peptide sequence. The occurrence of these sets of peptides is likely caused by the trimming of MHC-bound peptides by endo- and exopeptidases whereas the core peptide sequence that binds to the groove is protected from proteolytic degradation (1). To illustrate this, Fig. 5 shows the complete list of FVIII peptides from donor A. Two different MHC prediction algorithms, Propred and NetMHCIIpan, were chosen for comparison, with respect to binding to MHC class II alleles DRB1*0101 and DRB1*1301 (23, 26).
Fig. 5.
FVIII-derived MHC class II ligands identified from donor. A The first column shows residue numbers and corresponding domain of the FVIII molecule. The following columns show amino acid sequence, charge, XCorr value as provided by Sequest, retention time (RT), mass-to-charge ratio (m/z) and mass tolerance (ΔM). The two last columns show predicted binding scores to the MHC class II alleles of the donor. Binding scores to corresponding MHC class II alleles of the donor were calculated using Propred and NetMHCIIpan epitope prediction programs. Binding thresholds were defined as ≤500 nm for NetMHCIIpan and as top 3% natural binders for Propred (≥0.023 for DRB1*0101 and ≥0.295 for DRB1*1301). The binding motif of peptides that are predicted to bind above the binding threshold are underlined.
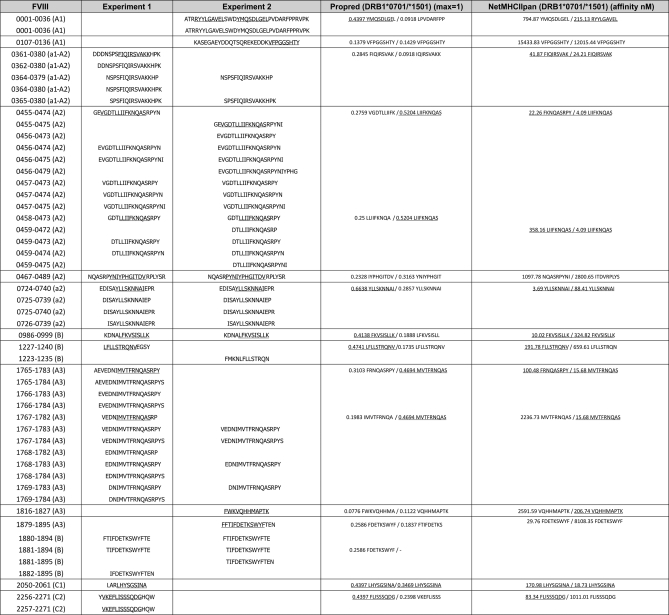
To determine the reproducibility of our findings, duplicate experiments were performed using moDCs obtained from donor B from two different blood draws, with a time period of 2 months in between the different blood draws. The duplicate experiments yielded similar sets of FVIII derived peptides (Fig. 6). All the core peptide sequences of which three or more variants are found could be identified in both experiments. In total 10 different core sequences were found in the first experiment and 11 in the second experiment. These sequences were also included in peptides that are only present once or twice. No more than two core sequences could be found in the first experiment only and three sequences were unique for the second experiment. All these were core sequences of which only one or two variants were found. All other core sequences were found both in the first and in the second experiment.
Fig. 6.
Presentation of FVIII peptides in duplicate experiments from donor B. Table of FVIII-derived MHC class II ligands identified from donor B. The first column shows residue numbers and corresponding domain of the FVIII molecule. The following columns show amino acid sequences of the first and second experiment. The two last columns show predicted binding scores to the MHC class II alleles of the donor. Binding scores to corresponding MHC class II alleles of the donor were calculated using Propred and NetMHCIIpan epitope prediction programs. Binding thresholds were defined as ≤500 nm for NetMHCIIpan and as top 3% natural binders for Propred (≥0.353 for DRB1*0701 and ≥0.329 for DRB1*1501). The binding motif of peptides that are predicted to bind above the binding threshold are underlined.
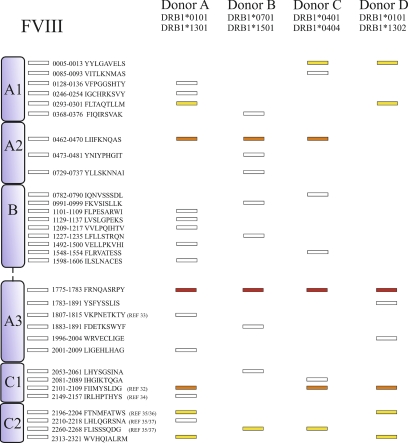
Overall, between 930 (donor A) and 270 (donor D) MHC class II-associated peptides could be identified; 702 and 882 peptides were identified for donor B and C, respectively. Thirty-two unique core FVIII peptide sequences could be recovered from all donors together. These sequences are distributed through all the domains of the FVIII molecule. The total amount of FVIII core peptide sequences was 17 for donor A, 10 for donor B, 9 for donor C and 8 for donor D (Fig. 7). One core sequence was presented on moDCs derived from all four donors. Two core peptides were common in three out of four donors and five core peptides were common between two out of four donors. All other core peptides were donor-specific. The complete list of FVIII-derived peptides from donor A and B, listed in Fig. 5 and Fig. 6, respectively, also depict the binding score of each peptide for both DRB1 alleles as calculated using the Propred MHC class II prediction server (23) and NetMHCpan (26).
Fig. 7.
Distribution of FVIII core peptides in four different donors. Different core peptides are presented by different donors. Results displayed in this figure. were obtained from single experiments for all donors. FVIII-derived MHC class II peptides are represented as rectangles for each individual donor. Indicated in yellow are sequences that are common between two donors. Indicated in orange are sequences common between three donors. Indicated in red is a sequence that is common in all four donors. The different domains and the location of the peptides are depicted schematically. Some sequences have been marked with a reference number, indicating that they have been identified previously as CD4+ T-cell epitope in the corresponding publication.
DISCUSSION
In this study we report the analysis of MHC class II-presented peptides on moDCs. Previous reports of MHC class II-presented peptides were performed on cell lines such as B-cell lymphomas, or on tissue-derived material (5, 12). Although most of these studies used 109 cells for sample preparation, we performed our experiments with 5 × 106 moDCs per incubation. Our findings show that naturally presented MHC class II peptides can be successfully obtained from antigen pulsed DC derived of 50 ml of blood. To ensure specificity and reproducibility we performed control experiments, in which moDC cell lysates were incubated with Sepharose coupled with an isotype control antibody. Indeed, many peptides were found present in the anti-MHC class II immunoprecipitation as well as in the control situation, as seen in Fig. 2A. We regarded a peptide as a true MHC class II-presented peptide when it was five times more abundant in the MHC class II samples as compared with control samples. Using this threshold, almost all FVIII peptides were MHC class II-presented peptides. FVIII peptides that did not fit these criteria were also positively identified in control samples using Sequest peptide identification software. These were excluded from our analysis.
As the data in Figs. 2A and 3A clearly indicate, immunoprecipitation of MHC class II-bound peptides from moDC cell shows that many endogenous proteins are presented on MHC class II. Fig. 3A shows that many peptides are presented even when cells were incubated with PBS. Fig. 2A shows that 51% of all peptides identified were specific for MHC class II. Most of these peptides are derived from endogenous proteins, and not FVIII. Not only proteins that reside in or are targeted to the endosomal and lysosomal pathway are presented on MHC class II, but also proteins from other organelles, such as the nucleus, and cytosol are presented. Sampling of intracellular compartments, called autophagy, is a constant process responsible for the lysosomal degradation of intracellular proteins, which can subsequently be presented on MHC class II (8). Professional APCs, such as moDCs used in this study, can enhance the formation of autophagosomes upon triggering of pattern-recognition receptors, such as toll-like receptor 4 (TLR4) and NOD2 (27–29). LPS-induced triggering of TLR4 during maturation of moDCs is likely to promote autophagy under our experimental conditions. Another possible explanation for the high percentage of endogenous proteins presented on MHC class II is the turnover of moDCs during culture. The efficacy of moDC generation from monocytes is about 50%. The other cells undergo apoptosis and can be endocytosed by moDCs. Some of the MHC class II-presented endogenous proteins identified in this study have been shown to be presented on MHC class II previously, such as CD14, α-2-macroglobulin, low-density lipoprotein-related protein 1, macrophage mannose receptor, and CD74 (invariant chain) (13) (data not shown). The identification of CD74 in all four donors provides an internal control for the peptide isolation procedure. CD74-derived peptides were expected to be found in each donor as CD74 is associated with newly synthesized MHC class II molecules and occupies the binding groove in the ER to prevent MHC class II loading in the ER (30). In the endo-lysosomal pathway CD74 is exchanged by other peptides or presented on MHC class II.
We observed that moDC derived from four healthy, HLA-typed volunteers all presented FVIII peptides after endocytosis of FVIII. Duplicate experiments from different blood draws (Fig. 6) suggest that there is a low level of variability in the presentation of FVIII peptides between independent experiments using cells from the same donor. As shown in Figs. 5 and 6, FVIII peptides could be divided into sets of peptides with overlapping sequences, which were comprised of differently processed variants of the same core peptide sequence. Almost all FVIII core peptides that were found in this study were predicted to bind to either one or both DRB1 alleles present in the donor in which the core peptides was found. Two MHC class II binding prediction-algorithms, Propred and NetMHCIIpan, were chosen based on the fact that they are reportedly the two most accurate algorithms available (31) and on the fact that other algorithms, such as SYFPEITHI, do not have predictors for all MHC class II haplotypes present in our set of donors.
The comparisons between the peptide identification and the two prediction models does not only confirm that the peptides that were found fit into the binding groove of the corresponding MHC class II molecules, but in addition it also stresses the need for proper identification of potential T-cell epitopes. The prediction values shown for donor A and B in Fig. 5 and 6 clearly show that, although all peptides are predicted to bind to at least one of both donor DRB1-molecules, there are many discrepancies between the two different prediction algorithms used. For example, in Fig. 5, peptide 1205–1222 QIENVVLPQIHTVTGTKN does not bind to DRB1*0101 and binds quite well to DRB1*1301 according to Propred. According to NetMHCIIpan, however, this peptide binds much better to DRB1*0101 (89 nm) than to DRB1*1301 (4.6 μm). Similar to this peptide, many other peptides can be found in these two figures in which the two algorithms are in disagreement. There are also many examples of peptides in which the exact 9-amino-acid sequence motif that is supposed to bind into the groove of the DRB1 molecule is different according to one prediction algorithm in comparison to the other. Furthermore, there are many regions in the FVIII sequence that are predicted to bind with high affinity to donor DRB1 molecules, which were not identified in this study. In summary, these observations demonstrate that prediction algorithms alone are not sufficient to accurately predict potential CD4+ T-cell epitopes in FVIII.
Analysis of the FVIII peptide repertoire identified for the four donors revealed that eight out of 32 core sequences were presented by multiple donors (Fig. 7). This suggests that FVIII contains a number of potential HLA-promiscuous ligands. One such promiscuous ligand, consisting of core sequence FIIMYSLDG, was described previously as an immunodominant epitope using T-cell stimulation assays (32). Most of the peptides found, however, were donor-specific. It is important to raise the question whether these peptides are clinically relevant. There are other core sequences that were identified in this study, which have been reported previously as sequences against which CD4+ T cells were found. A FVIII peptide containing region 1807–1815 was able to induce CD4+ T-cell proliferation in a hemophilia A patient with FVIII inhibitors (33). Region 2149–2157 is another core sequence that was reported previously. A CD4+ T-cell clone recognizing this region of FVIII was isolated (34). Proliferation of this clone has been used by another group as a general readout for FVIII endocytosis by dendritic cells (22). Region 2196–2204 was identified as a region against which CD4+ T cells are directed both in a study with hemophilic mice (35) and in a study with a hemophilia A patient (36). This sequence was identified using DRB1*0101 tetramers. Both donors that present this peptide in our study are DRB1*0101 as well. Two other C2 peptides identified in this study were found previously to induce T-cell proliferation (35, 37). The fact that most peptides found were donor-specific suggests that these peptides are presented in a HLA-specific context. It is important, however, to keep in mind that the donors are not related and are potentially heterogeneous with respect to the expression of genes involved in antigen presentation. The identification of putative FVIII-derived CD4+ T-cell epitopes is important to understand the formation of FVIII-neutralizing antibodies in hemophilia A. As mentioned earlier, there are studies that have reported that CD4+ T cells isolated from hemophilia A patients can proliferate in a FVIII-specific manner (32–34, 38, 39). More recently, MHC-class II tetramers have been used to assess the exact epitope, HLA-restriction and the phenotype of FVIII-specific CD4+ T cells (36, 40). These studies have implied a role for CD4+ T cells in inhibitor formation, but there is still very limited data on the repertoire of FVIII peptides that is presented by APCs from donors with different HLA haplotypes. There are various prediction models available that calculate the binding affinity of linear peptides for MHC class II variants using mathematics- or structure-based algorithms (23, 41–45), such as the ones used in this study. These prediction models are mathematics- or structure-based methods. They can predict binding motifs with considerable accuracy, but are restricted in the sense that they are based on known peptide-MHC interactions.
An alternative method to identify the binding properties of antigen-derived peptides to MHC variants is to measure the binding of synthetic peptides to recombinant MHC molecules (46). Combined, these computational and in vitro methods accurately define sequences within an antigen that can bind MHC molecules, but these data do not completely reflect the different processes that are involved in antigen presentation. Processes that are important for antigen presentation on MHC class II are the route of endocytosis, cleavage by endosomal and lysosomal proteases, and antigenic competition. In this study we have taken these processes into consideration by investigating which FVIII peptides are naturally presented on MHC class II in moDCs.
The methods described in this paper can be used to determine the repertoire of naturally presented peptides in antigen-pulsed APC. In this study we show that our approach can be used to obtain information on the repertoire of naturally presented peptides of FVIII. Potentially, this knowledge can be used to design novel less immunogenic FVIII variants that lack promiscuously presented peptides such as FRNQASRPY (A3 domain), FIIMYSLDG (C1 domain), or LIIFKNQAS (A2 domain). We and others show peptide analysis of antigen-pulsed moDCs allows for the identification of naturally presented peptides derived from clinically relevant antigens (47). Identification of HLA promiscuous sequences within clinically relevant antigens can potentially assist in the design of more efficient vaccines or novel tolerization strategies.
Footnotes
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- moDC
- monocyte-derived dendritic cell
- APC
- antigen-presenting cell
- FVIII
- factor VIII
- PBMC
- peripheral blood mononuclear cell
- PBS
- phosphate-buffered saline.
REFERENCES
- 1. Trombetta E. S., Mellman I. (2005) Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23, 975–1028 [DOI] [PubMed] [Google Scholar]
- 2. Burgdorf S., Kurts C. (2008) Endocytosis mechanisms and the cell biology of antigen presentation. Curr. Opin. Immunol. 20, 89–95 [DOI] [PubMed] [Google Scholar]
- 3. Chicz R. M., Urban R. G., Gorga J. C., Vignali D. A., Lane W. S., Strominger J. L. (1993) Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 178, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lechler R., Aichinger G., Lightstone L. (1996) The endogenous pathway of MHC class II antigen presentation. Immunol. Rev. 151, 51–79 [DOI] [PubMed] [Google Scholar]
- 5. Dongre A. R., Kovats S., deRoos P., McCormack A. L., Nakagawa T., Paharkova-Vatchkova V., Eng J., Caldwell H., Yates J. R., 3rd, Rudensky A. Y. (2001) In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur. J. Immunol. 31, 1485–1494 [DOI] [PubMed] [Google Scholar]
- 6. Nedjic J., Aichinger M., Mizushima N., Klein L. (2009) Macroautophagy, endogenous MHC II loading and T cell selection: the benefits of breaking the rules. Curr Opin Immunol. 21, 92–97 [DOI] [PubMed] [Google Scholar]
- 7. Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., Brock R., Driessen C., Rammensee H. G., Stevanovic S. (2005) Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmid D., Pypaert M., Münz C. (2007) Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peakman M., Stevens E. J., Lohmann T., Narendran P., Dromey J., Alexander A., Tomlinson A. J., Trucco M., Gorga J. C., Chicz R. M. (1999) Naturally processed and presented epitopes of the islet cell autoantigen IA-2 eluted from HLA-DR4. J. Clin. Invest. 104, 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seeger F. H., Schirle M., Gatfield J., Arnold D., Keilholz W., Nickolaus P., Rammensee H. G., Stevanović S. (1999) The HLA-A*6601 peptide motif: prediction by pocket structure and verification by peptide analysis. Immunogenetics 49, 571–576 [DOI] [PubMed] [Google Scholar]
- 11. Krogsgaard M., Wucherpfennig K. W., Cannella B., Hansen B. E., Svejgaard A., Pyrdol J., Ditzel H., Raine C., Engberg J., Fugger L., Canella B. (2000) Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85–99 complex. J. Exp. Med. 191, 1395–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fissolo N., Haag S., de Graaf K. L., Drews O., Stevanovic S., Rammensee H. G., Weissert R. (2009) Naturally presented peptides on MHC I and II molecules eluted from central nervous system of multiple sclerosis patients. Mol Cell Proteomics. 8, 2090–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wahlström J., Dengjel J., Persson B., Duyar H., Rammensee H. G., Stevanović S., Eklund A., Weissert R., Grunewald J. (2007) Identification of HLA-DR-bound peptides presented by human bronchoalveolar lavage cells in sarcoidosis. J. Clin. Invest. 117, 3576–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fortier M. H., Caron E., Hardy M. P., Voisin G., Lemieux S., Perreault C., Thibault P. (2008) The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 205, 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de, Verteuil D., Muratore-Schroeder T. L., Granados D. P., Fortier M. H., Hardy M. P., Bramoullé A., Caron E., Vincent K., Mader S., Lemieux S., Thibault P., Perreault C. (2010) Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol Cell Proteomics 9, 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Niel G., Wubbolts R., Ten Broeke T., Buschow S. I., Ossendorp F. A., Melief C. J., Raposo G., van Balkom B. W., Stoorvogel W. (2006) Dendritic cells regulate exposure of MHC class II at their plasma membrane by oligoubiquitination. Immunity 25, 885–894 [DOI] [PubMed] [Google Scholar]
- 17. Mannucci P. M., Tuddenham E. G. (2001) The hemophilias–from royal genes to gene therapy. N. Engl. J. Med. 344, 1773–1779 [DOI] [PubMed] [Google Scholar]
- 18. Lollar P. (2004) Pathogenic antibodies to coagulation factors. Part one: factor VIII and factor IX. J. Thromb. Haemost. 2, 1082–1095 [DOI] [PubMed] [Google Scholar]
- 19. Reding M. T., Lei S., Lei H., Green D., Gill J., Conti-Fine B. M. (2002) Distribution of Th1- and Th2-induced anti-factor VIII IgG subclasses in congenital and acquired hemophilia patients. Thromb. Haemost. 88, 568–575 [PubMed] [Google Scholar]
- 20. Ten Brinke A., Karsten M. L., Dieker M. C., Zwaginga J. J., van Ham S. M. (2007) The clinical grade maturation cocktail monophosphoryl lipid A plus IFNgamma generates monocyte-derived dendritic cells with the capacity to migrate and induce Th1 polarization. Vaccine 25, 7145–7152 [DOI] [PubMed] [Google Scholar]
- 21. Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 22. Dasgupta S., Navarrete A. M., Bayry J., Delignat S., Wootla B., André S., Christophe O., Nascimbeni M., Jacquemin M., Martinez-Pomares L., Geijtenbeek T. B., Moris A., Saint-Remy J. M., Kazatchkine M. D., Kaveri S. V., Lacroix-Desmazes S. (2007) A role for exposed mannosylations in presentation of human therapeutic self-proteins to CD4+ T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 104, 8965–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh H., Raghava G. P. (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17, 1236–1237 [DOI] [PubMed] [Google Scholar]
- 24. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- 25. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 26. Nielsen M., Lundegaard C., Blicher T., Peters B., Sette A., Justesen S., Buus S., Lund O. (2008) Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput. Biol. 4, e1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Y., Jagannath C., Liu X. D., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. (2007) Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., Green D. R. (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 [DOI] [PubMed] [Google Scholar]
- 29. Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010) NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 [DOI] [PubMed] [Google Scholar]
- 30. Roche P. A., Cresswell P. (1990) Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature 345, 615–618 [DOI] [PubMed] [Google Scholar]
- 31. Lin H. H., Zhang G. L., Tongchusak S., Reinherz E. L., Brusic V. (2008) Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 12, S22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones T. D., Phillips W. J., Smith B. J., Bamford C. A., Nayee P. D., Baglin T. P., Gaston J. S., Baker M. P. (2005) Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J. Thromb. Haemost. 3, 991–1000 [DOI] [PubMed] [Google Scholar]
- 33. Reding M. T., Okita D. K., Diethelm-Okita B. M., Anderson T. A., Conti-Fine B. M. (2004) Epitope repertoire of human CD4(+) T cells on the A3 domain of coagulation factor VIII. J. Thromb. Haemost. 2, 1385–1394 [DOI] [PubMed] [Google Scholar]
- 34. Jacquemin M., Vantomme V., Buhot C., Lavend'homme R., Burny W., Demotte N., Chaux P., Peerlinck K., Vermylen J., Maillere B., van der Bruggen P., Saint-Remy J. M. (2003) CD4+ T-cell clones specific for wild-type factor VIII: a molecular mechanism responsible for a higher incidence of inhibitor formation in mild/moderate hemophilia A. Blood 101, 1351–1358 [DOI] [PubMed] [Google Scholar]
- 35. Pratt K. P., Qian J., Ellaban E., Okita D. K., Diethelm-Okita B. M., Conti-Fine B., Scott D. W. (2004) Immunodominant T-cell epitopes in the factor VIII C2 domain are located within an inhibitory antibody binding site. Thromb. Haemost. 92, 522–528 [DOI] [PubMed] [Google Scholar]
- 36. James E. A., Kwok W. W., Ettinger R. A., Thompson A. R., Pratt K. P. (2007) T-cell responses over time in a mild hemophilia A inhibitor subject: epitope identification and transient immunogenicity of the corresponding self-peptide. J. Thromb. Haemost. 5, 2399–2407 [DOI] [PubMed] [Google Scholar]
- 37. Ettinger R. A., James E. A., Kwok W. W., Thompson A. R., Pratt K. P. (2010) HLA-DR-restricted T-cell responses to factor VIII epitopes in a mild haemophilia A family with missense substitution A2201P. Haemophilia 16, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reding M. T., Okita D. K., Diethelm-Okita B. M., Anderson T. A., Conti-Fine B. M. (2003) Human CD4+ T-cell epitope repertoire on the C2 domain of coagulation factor VIII. J. Thromb. Haemost. 1, 1777–1784 [DOI] [PubMed] [Google Scholar]
- 39. Reding M. T., Wu H., Krampf M., Okita D. K., Diethelm-Okita B. M., Key N. S., Conti-Fine B. M. (1999) CD4+ T cell response to factor VIII in hemophilia A, acquired hemophilia, and healthy subjects. Thromb. Haemost. 82, 509–515 [PubMed] [Google Scholar]
- 40. Ettinger R. A., James E. A., Kwok W. W., Thompson A. R., Pratt K. P. (2009) Lineages of human T-cell clones, including TH17/TH1 cells, isolated at different stages of anti-factor VIII immune responses. Blood. 114, 1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sturniolo T., Bono E., Ding J., Raddrizzani L., Tuereci O., Sahin U., Braxenthaler M., Gallazzi F., Protti M. P., Sinigaglia F., Hammer J. (1999) Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 17, 555–561 [DOI] [PubMed] [Google Scholar]
- 42. Bui H. H., Sidney J., Peters B., Sathiamurthy M., Sinichi A., Purton K. A., Mothé B. R., Chisari F. V., Watkins D. I., Sette A. (2005) Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics 57, 304–314 [DOI] [PubMed] [Google Scholar]
- 43. Nielsen M., Lundegaard C., Lund O. (2007) Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics 8, 238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen M., Lund O. (2009) NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics 10, 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang P., Sidney J., Dow C., Mothé B., Sette A., Peters B. (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 4, e1000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. James E. A., Moustakas A. K., Bui J., Nouv R., Papadopoulos G. K., Kwok W. W. (2009) The binding of antigenic peptides to HLA-DR is influenced by interactions between pocket 6 and pocket 9. J. Immunol. 183, 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mutschlechner S., Egger M., Briza P., Wallner M., Lackner P., Karle A., Vogt A. B., Fischer G. F., Bohle B., Ferreira F. (2010) Naturally processed T cell-activating peptides of the major birch pollen allergen. J Allergy Clin Immunol 125, 711–718 [DOI] [PubMed] [Google Scholar]