Abstract
The formation of mature mRNAs in vertebrates involves the cleavage and polyadenylation of the pre-mRNA, 10–30 nt downstream of an AAUAAA or AUUAAA signal sequence. The extensive cDNA data now available shows that these hexamers are not strictly conserved. In order to identify variant polyadenylation signals on a large scale, we compared over 8700 human 3′ untranslated sequences to 157,775 polyadenylated expressed sequence tags (ESTs), used as markers of actual mRNA 3′ ends. About 5600 EST-supported putative mRNA 3′ ends were collected and analyzed for significant hexameric sequences. Known polyadenylation signals were found in only 73% of the 3′ fragments. Ten single-base variants of the AAUAAA sequence were identified with a highly significant occurrence rate, potentially representing 14.9% of the actual polyadenylation signals. Of the mRNAs, 28.6% displayed two or more polyadenylation sites. In these mRNAs, the poly(A) sites proximal to the coding sequence tend to use variant signals more often, while the 3′-most site tends to use a canonical signal. The average number of ESTs associated with each signal type suggests that variant signals (including the common AUUAAA) are processed less efficiently than the canonical signal and could therefore be selected for regulatory purposes. However, the position of the site in the untranslated region may also play a role in polyadenylation rate.
The 3′ untranslated regions (UTRs) of eukaryotic mRNAs contain regulatory elements affecting mRNA translation, stability, and transport. Mature 3′ UTRs are formed by polyadenylation of the pre-mRNA, a coupled reaction involving endonucleolytic cleavage followed by poly(A) synthesis. A significant fraction of mRNAs display multiple polyadenylation sites (Gautheret et al. 1998). The choice of poly(A) sites may influence the stability, translation efficiency, or localization of an mRNA in a tissue- or disease-specific manner (Edwalds-Gilbert et al. 1997). In the mammalian system, effective polyadenylation requires two main sequence components: a highly conserved AAUAAA signal located 10–30 nucleotide 5′ to the cleavage site and a more variable GU-rich element, 20–40 bases 3′ of the site (see Proudfoot 1991; Colgan and Manley 1997 for reviews). Although the AAUAAA signal is often considered to be present in 90% of the mRNAs and replaced by a AUUAAA variant in the other 10% (Wahle and Keller 1996; Colgan and Manley 1997), alternate signals are certainly present in a significant fraction of the 3′ ends (Claverie 1997; Gautheret et al. 1998; Tabaska and Zhang 1999; Graber et al. 1999).
The expressed sequence tag (EST) database, dbEST (Boguski et al. 1993), which contains highly redundant partial cDNAs, especially from the 3′ UTRs, is a rich source of information on mRNA 3′ ends. Analyzing clustered EST sequences, we previously identified multiple cases of alternate polyadenylation in mRNA (Gautheret et al. 1998). Based on a public EST collection now containing over 1.4 million human sequences, the present work focuses on the region immediately upstream of the cleavage sites, collecting statistics on the most frequent polyadenylation signals, their position in the UTR, and their frequency of use in UTRs with multiple cleavage sites. In order to compensate for the low accuracy of EST sequences, we selected ESTs with near perfect matches to UTR sequences from Genbank and used the Genbank sequence as the reference. Therefore, sequence errors are minimized. This study provided evidence for the existence of 10 variant polyadenylation signals that may be responsible for up to 14.9% of the mRNA 3′ ends. We then analyzed the distribution of noncanonical signals in UTRs with alternate poly(A) sites and assessed the processing efficiency of polyadenylation signals in function of their sequence and their position in the UTR. Significant biases were observed, with interesting consequences for the regulation of mRNA 3′ end formation.
RESULTS
The comparison of 8775 human UTR sequences to the 157,775 ESTs with a poly(A) or poly(T) extremity was performed using the criteria exposed in Methods to reduce experimental artifacts, including internal priming and partial matches from chimeric ESTs or confusion between ESTs from paralogous genes. This selected 4344 UTRs with at least one putative polyadenylation site. The number of polyadenylation sites per mRNA molecule is distributed as shown in Table 1. 3377 sequences (77.7%) have one putative poly(A) site, and 967 sequences (22.3%) have two sites or more. This figure supersedes our previous minimum estimate of 18.9% alternatively polyadenylated mRNAs (Gautheret et al. 1998). The total number of putative poly(A) sites observed is 5647. The 50-nucleotide fragment preceding each of these sites was collected, producing a database of 5647 sequences. Hexanucleotide frequencies in this 3′ fragment database were analyzed as described in Methods. Results are shown in Tables 2 and 3. The AAUAAA and AUUAAA polyadenylation signals are by far the most frequently found hexamers, present in 58.2% and 14.9% of the 3′ fragments, respectively. The remaining 26.8% of the 3′ fragments do not contain a usual polyadenylation signal. Analyzing 5-mer, 7-mer, and 8-mer frequencies did not identify any recurrent word other than combinations of the hexameric motifs, such as AAAUAAA (data not shown).
Table 1.
Number of mRNAs with Alternative Poly(A) Sites
| Number of poly(A) sites per mRNA | 1 | 2 | 3 | 4+ |
|---|---|---|---|---|
| Number of mRNAs | 3377 | 724 | 182 | 61 |
Table 2.
Most Significant Hexamers in 3′ Fragments: Clustered Hexamers
| Hexamer | Observed (expected)a | % sites | Pb | Position average ± SD | Locationc | |||
|---|---|---|---|---|---|---|---|---|
 |
3286 (317) | 58.2 | 0 | −16 ± 4.7 | 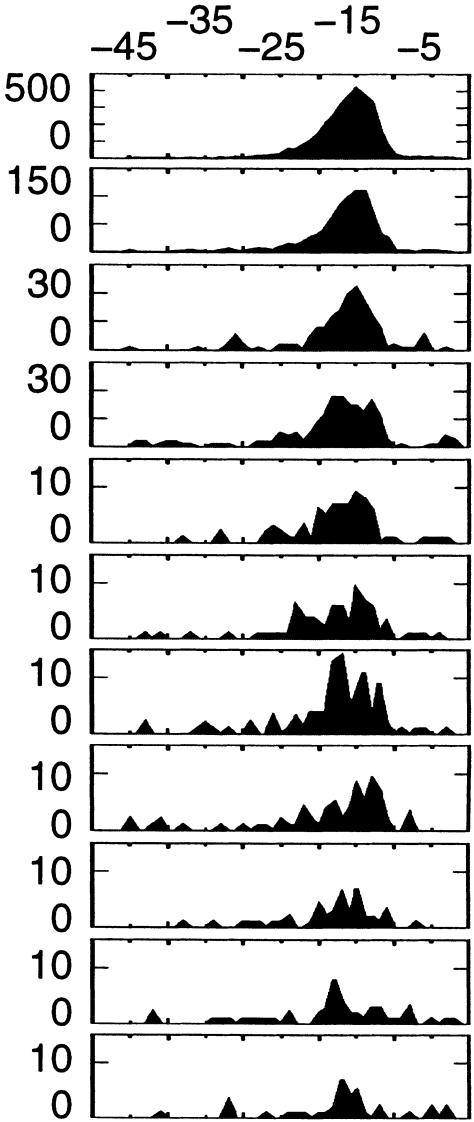 |
|||
| 843 (112) | 14.9 | 0 | −17 ± 5.3 | |||||
| 156 (32) | 2.7 | 6 × 10−57 | −16 ± 5.9 | |||||
| 180 (53) | 3.2 | 4 × 10−45 | −18 ± 7.8 | |||||
| 76 (23) | 1.3 | 1 × 10−18 | −17 ± 5.9 | |||||
| 72 (21) | 1.3 | 2 × 10−18 | −18 ± 6.9 | |||||
| 96 (33) | 1.7 | 2 × 10−19 | −18 ± 6.9 | |||||
| 70 (16) | 1.2 | 5 × 10−23 | −18 ± 8.7 | |||||
| 43 (14) | 0.7 | 1 × 10−9 | −18 ± 6.3 | |||||
| 49 (11) | 0.8 | 5 × 10−17 | −18 ± 8.9 | |||||
| 36 (11) | 0.6 | 1 × 10−08 | −17 ± 8.1 | |||||
Expected occurrences based on a random distribution in a database of same nucleotide composition.
Probability of reaching the observed frequency by chance based on a cumulative binomial distribution. All sequences containing the most significant hexamer were removed from the database before the next most significant hexamer was sought.
Location of the last base of hexamer. Position 0 is the putative poly(A) site.
Table 3.
Most Significant Hexamers in 3′ Fragments: Scattered Hexamers
| Hexamer | Observed (expected)a | % sites | Pb | Position average ± SD | Locationc | |||
|---|---|---|---|---|---|---|---|---|
 |
62 (10) | 1.1 | 9 × 10−28 | −19 ± 11 | 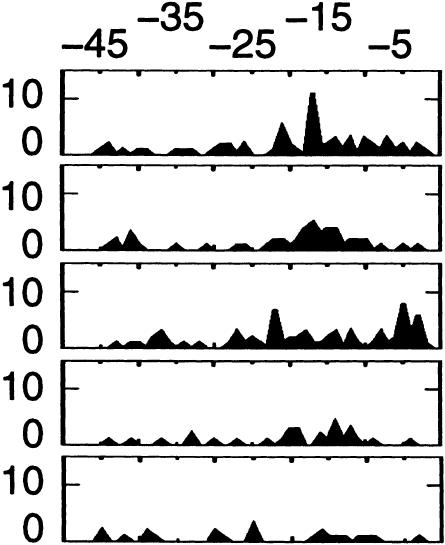 |
|||
| 49 (10) | 0.8 | 4 × 10−18 | −20 ± 10 | |||||
| 69 (20) | 1.2 | 3 × 10−18 | −17 ± 12 | |||||
| 29 (5) | 0.5 | 8 × 10−12 | −20 ± 10 | |||||
| 22 (3) | 0.3 | 9 × 10−12 | −24 ± 13 | |||||
Expected occurrences based on a random distribution in a database of same nucleotide composition.
Probability of reaching the observed frequency by chance based on a cumulative binomial distribution. All sequences containing the most significant hexamer were removed from the database before the next most significant hexamer was sought.
Location of the last base of hexamer. Position 0 is the putative poly(A) site.
Variant Signals
The right part of Table 2 shows the distribution of hexamer positions over the 3′ fragment (position of the sixth nucleotide of hexamer is plotted). The AAUAAA and AUUAAA hexamers are clearly clustered around −15/−16 nt upstream of the putative poly(A) site, as expected from experimentally validated signals (Chen and Shyu 1995). In a preliminary analysis, several motifs with high P-values were found scattered along the 3′ segment. The absence of spatial preference with respect to the poly(A) site suggested that these motifs were not involved in any specific interaction with the polyadenylation machinery. Since our primary focus was on polyadenylation-related motifs, we first sought spatially “clustered” motifs. We did so based on the standard deviation (SD) around the mean motif position (see Methods). The list of significant motifs in Table 2 comprises only those motifs with P < 10−5 and SD < 9 nt. Variant hexamers are also clustered around positions −15/−20. The most significant motifs with SD > 9 nt are shown separately (Table 3). The first two motifs in this table most frequently occur near −15/−20, albeit less obviously than the previous motifs.
Even though the 50-nt UTR fragments used for hexamer searches are from Genbank rather than EST sequences, one may argue that unexpected hexamers could result from sequencing errors in the UTR sequences, especially when these hexamers have a single base difference from the common AAUAAA signal. This hypothesis can be rejected on the basis of the very good agreement between UTR and EST sequences. A control analysis that required a 99% similarity between UTR and EST sequences (instead of 95%) produced nearly the same proportion of noncanonical hexamers (data not shown). Further, alignments of UTRs with their corresponding ESTs were inspected visually for agreement at the level of noncanonical hexamers. AAUACA hexamers (70 UTR sequences; Table 2) and AAUAGA hexamers (43 UTR sequences; Table 2) were confirmed by at least one EST in 92% and 93% of the cases, respectively.
Hexamers AGUAAA, UAUAAA, CAUAAA, GAUAAA, AAUAUA, AAUACA, AAUAGA, and ACUAAA are significantly overrepresented near the polyadenylation site, and their spatial distribution (Table 2) closely follows that of known poly(A) signals (Chen and Shyu 1995). Both facts strongly suggest that these motifs are widespread polyadenylation signals in human mRNA. The penultimate motif (AAAAAG) is actually a statistical artifact caused by the high rate of the AAGAAA motifs within this region (Table 3, see below). Although the motifs shown in Table 3 are more scattered along the 3′ segment, the AAGAAA and AAUGAA hexamers display minor but distinguishable peaks at position −15/−20, which is best explained by their role as a polyadenylation signal. Combined together, and neglecting statistical noise and sequence errors, the 10 variant motifs could account for 14.9% of the putative mRNA 3′ ends, which potentially represents a considerable number of mRNA forms in the whole transcriptome.
Positional Preferences
Messenger RNAs with two or more putative poly(A) sites represent 967 mRNAs (22.3%) and 2270 poly(A) sites (40.2%) in our study. Using this large data set, we can now analyze on a large scale alternatively polyadenylated mRNAs for possible biases in poly(A) signal sequence and position.
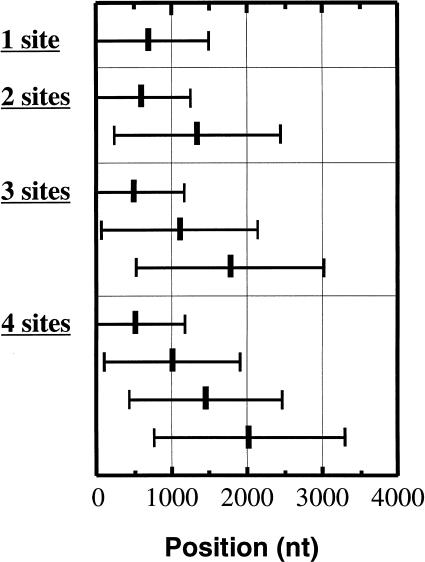
Figure 1 presents the average position of putative polyadenylation sites on 3′ UTRs as a function of the number of observed alternative sites. The high standard deviations (error bars) indicate that locations of poly(A) sites are highly variable. Indeed, the observed distribution resembles that expected from a random selection of n points in the same sequence set. When four random points are picked in our sequence set (with the fourth point taken at the end of the sequence), the average positions and standard deviations of the first to fourth sites are 518 ± 472, 1064 ± 1023, 1635 ± 1231, and 2059 ± 1220, which is very similar to the result shown in Figure 1 with 4 polyA sites. Multiple sites are interspersed on average every 600 bp on the 3′ UTR. This average number, however, could be affected by the presence of yet unidentified sites in UTRs. What appears to be the “first” site may actually be the second one, and so forth.
Figure 1.
Average position of observed polyadenylation sites on 3′ UTRs in function of the number of observed alternate sites. From top to bottom: mRNAs with a single poly(A) site identified (3377 RNAs), mRNAs with two poly(A) sites identified (724 RNAs), mRNAs with three poly(A) sites identified (182 RNAs), and mRNAs with four poly(A) sites identified (43 RNAs). Position 1 on the X-axis is the first base following the Stop codon. Error bars indicate standard deviations.
Table 4 presents, for mRNAs with a given number of putative poly(A) sites, the average number of sites per molecule containing AAUAAA, AUUAAA, or other signals. “Signals” are understood here as in Table 2–3, that is, found in the 50-nt segment upstream of an EST-supported poly(A) site and in the absence of a more frequent signal. For instance, mRNAs with three poly(A) sites have on average 1.30 AAUAAA signals and 0.58 AUUAAA signals. It appears that, as the number of poly(A) sites in an mRNA molecule increases, the proportion of canonical AAUAAA signals decreases (see the ratio AAUAAA/AUUAAA on the last line). In other words, mRNAs with multiple poly(A) sites tend to use a higher proportion of noncanonical signals. This is true for all noncanonical signals, including the common AUUAAA.
Table 4.
Average Number of Poly(A) Sites with Each Type of Polyadenylation Signal, per mRNA Molecule
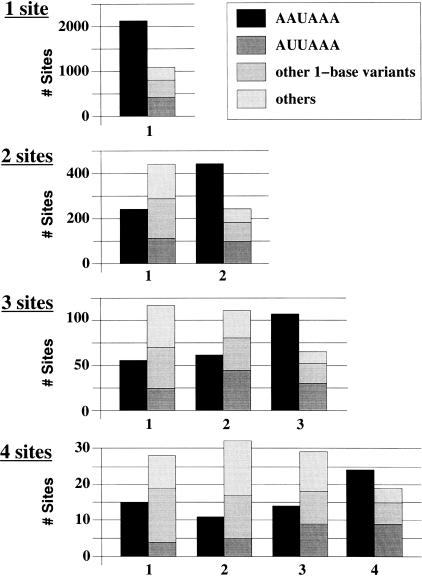
We then counted occurrences of each type of polyadenylation signal at different sites on the UTR (Fig. 2). There is a striking difference between the 3′-most distal site and other sites closer to the Stop codon. The 3′ distal site predominantly uses a canonical signal, while all other sites predominantly use noncanonical signals, particularly one-base variants of the AAUAAA sequence. Unidentified signals (“Others” in Fig. 2), which represent a significant fraction of the poly(A) sites closer to the Stop codon, should be taken cautiously because they could result from internally primed ESTs that have escaped our filtering procedure. In any case, putting aside other signals, the one-base variants of the AAUAAA signal are more represented than the canonical signal in sites proximal to the Stop codon.
Figure 2.
Distribution of polyadenylation signal types at each site on the UTR. From top to bottom: mRNAs with a single poly(A) site identified, mRNAs with two poly(A) sites identified, mRNAs with three poly(A) sites identified, and mRNAs with four poly(A) sites identified. Poly(A) sites are numbered from 5′ to 3′.
Processing Efficiency
Highly expressed mRNAs are commonly expected to result in a higher number of ESTs than weakly expressed ones. However, because normalization procedures have been applied to most EST libraries, artificially reducing EST levels for certain types of mRNAs, biases in EST counts are not always meaningful. In this context, can we use EST counts as a rough estimate of the polyadenylation rate at various sites? Here, we will not compare the expression of different mRNAs but, instead, the efficiency of different types of poly(A) sites, whatever mRNA or EST library is considered. Answers to this question should less be affected by biases induced by library construction protocols.
Table 5 shows the mean numbers of ESTs observed associated with each putative poly(A) signal (hereafter called “revealing” ESTs). For instance, putative poly(A) sites with an AAUAAA hexamer are supported on average by 5.4 ESTs. The number of revealing ESTs is higher with the AAUAAA signal than with any other signal. This effect cannot be attributed to some canonical signals associated to abundantly expressed genes, as it is also observed when both types of signals are found on the same gene. For instance, mRNAs having both an AAUAAA and a noncanonical signal in their UTR have nearly twice as many ESTs associated to the canonical signal on average (data not shown). This strongly suggests that sites with noncanonical signals are processed less efficiently than those with a canonical signal. Interestingly, the common AUUAAA signal falls in the same range as the less frequent variant AGUAAA.
Table 5.
Number of Revealing ESTs per Poly(A) Site for Each Putative Polyadenylation Signal
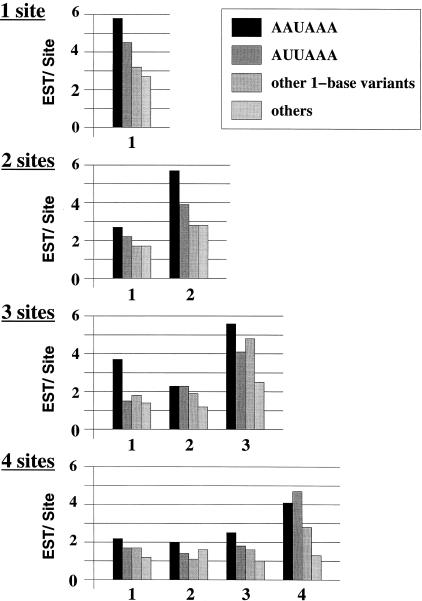
We finally asked how processing efficiency varied with the position of poly(A) sites in alternatively polyadenylated mRNAs. Histograms in Figure 3 give the number of revealing ESTs associated on average with each polyadenylation site in mRNAs with one, two, three, and four observed polyadenylation sites. Sites with canonical or other poly(A) signals are distinguished. The hierarchy of canonical and noncanonical signals with respect to polyadenylation rate is maintained independently of the cleavage position. However, the 3′-most distal cleavage sites generally have more revealing ESTs than sites closer to the Stop codon, suggesting that 3′-terminal sites are processed more efficiently. A possible pitfall in this conclusion would be the presence of erroneous 3′ ends among sites closer to the Stop codon. Such incorrect poly(A) sites would have fewer associated ESTs, lowering average EST counts. If this were true, poly(A) sites with a canonical signal would not be lowered since they most likely correspond to true 3′ ends. However, when the signal closest to the Stop codon is AAUAAA, there are also fewer revealing ESTs, further suggesting a position dependency of poly(A) site processing efficiency.
Figure 3.
Number of revealing ESTs per poly(A) site, in function of the position of sites in the UTR. From top to bottom: mRNAs with a single poly(A) site identified, mRNAs with two poly(A) sites identified, mRNAs with three poly(A) sites identified, and mRNAs with four poly(A) sites identified. Poly(A) sites are numbered from 5′ to 3′.
DISCUSSION
The human polyadenylation signals identified in this study are summarized in Figure 4. Until recently, only a single-variant hexamer, AGUAAA, had been identified as a possibly recurrent signal in human mRNA (Gautheret et al. 1998; Tabaska and Zhang 1999). After this work was completed, a study by Graber et al. (1999) was published identifying variant polyadenylation signals in 3′ EST sequences from diverse species. The set of 4427 human ESTs selected in that study was analyzed independently of reference mRNA or genomic sequences, which probably raised the sequence error rate (only 53.2% of 3′ ends had a AAUAAA signal vs. 58.2% in our study). Nevertheless, these authors did use reference genomic sequences to analyze Drosophila ESTs and obtained a list of variant poly(A) signals very similar to ours (Graber et al. 1999). The effect of poly(A) signals' mutations on polyadenylation and cleavage rates has been studied experimentally in vivo (Sheets et al. 1990). Comparing in silico and in vitro results, Graber et al. noted that the natural frequency of variant signals in Drosophila was closely related to in vitro polyadenylation rate (Graber et al. 1999). This striking observation also applies to the human poly(A) signals.
Figure 4.

The 12 putative human polyadenylation signals and their 90% consensus sequence (N = any nucleotide). The consensus does not take into account the relative frequency of signals. Positions conserved in more than 90% of the variants are highlighted.
With respect to in vivo studies, a literature search for the 10 variant signals reveals that most have been occasionally reported as forming “unusual” poly(A) signals in mammalian or mammalian virus mRNAs (Table 6). The agreement between our results and the literature is excellent: those naturally occurring polyadenylation signals that do not figure in our list are either weakly active, deleterious, or found only in plants. Interestingly, the AAUAGA motif was reported as functional solely in flatworms (Wahlberg and Johnson 1997), while its presence in human β-globin mRNA in replacement of the canonical signal is a known cause of β-thalassemia (Jankovic et al. 1990; van Solinge et al. 1996). Mutations in poly(A) signals causing α- and β-thalassemia result in elongated mRNAs (Orkin et al. 1985; Smetanina et al. 1996), meaning the poly(A) signal either is not functional or is used inefficiently. The situation is similar for the AAGAAA and AAUGAA motifs. AAGAAA is reportedly an active polyadenylation signal in a mammalian mRNA (Anand et al. 1997), but this motif is also commonly used in replacement of canonical signals in order to inactivate polyadenylation sites in DNA viruses (Moore et al. 1988; Wilusz and Shenk 1988). Likewise, AAUGAA is a potentially deleterious polyadenylation signal (Jankovic et al. 1990; Yuregir et al. 1992), but is nevertheless functional in two mammalian mRNAs (Martins et al. 1995; Battersby et al. 1999). This is no reason to believe that the AAUAGA, AAGAAA, or AAUGAA signals we observed are inactive since all correspond to experimentally identified (in the form of ESTs) mature mRNA terminations. However, their possibly deleterious effects suggest that either their function is context dependent (e.g., external factors might inactivate them) or their efficiency is intrinsically different than that of a canonical signal.
Table 6.
Functional and Deleterious Noncanonical Polyadenylation Signals Reported in the Literature
| Signal | Context |
|---|---|
| AGUAAA | Functional in 6 mammalian mRNAs (Cheng et al. 1986; Hsu et al. 1988; Nagashima et al. 1988; Yoshimura et al. 1994; Wang et al. 1996; Zhang et al. 1998) and 2 mammalian virus mRNAs (Klemenz et al. 1981; Nasseri et al. 1987). |
| UAUAAA | Functional in hepatitis B virus (Simonsen and Levinson 1983), Epstein Barr virus (Silver Key and Pagano 1997), 1 plant pararetrovirus (Sanfacon 1994), and 1 mammalian mRNA (suggested; Martins et al. 1995). |
| CAUAAA | Functional in hepatitis B virus mRNA (Hilger et al. 1991), 2 mammalian mRNAs (Wetsel et al. 1987; Faber et al. 1991), and 1 Xenopus mRNA (Rabbitts and Morgan 1992). |
| GAUAAA | Functional in 1 mammalian mRNA (Epstein et al. 1986) and 4 related nematode mRNAs (Yan et al. 1998). |
| AAUAUA | Functional in 1 mammalian mRNA (Hu et al. 1994), 1 insect mRNA (Larochelle and Suter 1995), and 1 plant mRNA (Ishikawa et al. 1997). |
| AAUACA | Functional in 5 mammalian mRNAs (Suzuki et al. 1990; Taylor et al. 1990; Herve et al. 1991; Myohanen et al. 1991; Parthasarathy et al. 1997), and 1 insect mRNA (Tokishita et al. 1997). |
| AAUAGA | Functional in 1 Platyhelminthe (flatworm) mRNA (Wahlberg and Johnson 1997); deleterious in human β-globin mRNA (Jankovic et al. 1990; van Solinge et al. 1996). |
| ACUAAA | Functional in 1 mammalian mRNA (Trowsdale and Kelly 1985). |
| AAGAAA | Functional in 1 mammalian mRNA (Anand et al. 1997); deleterious (used as an artificial polyadenylation signal suppressor) in mammalian adenovirus mRNA (Moore et al. 1988; Wilusz and Shenk 1988). |
| AAUGAA | Functional in 2 mammalian mRNAs (Martins et al. 1995; Battersby et al. 1999), and 2 plant mRNAs (Wu et al. 1993; de Freitas et al. 1994). Deleterious (α- and β-thalassemia) in human globin mRNA (Jankovic et al. 1990; Yuregir et al. 1992). |
| UAUAUA | Weakly functional in 1 mammalian DNA virus (papilloma virus; Andrews and DiMaio 1993). |
| AAUAAC | Weakly functional in 1 mammalian DNA virus (parvovirus) mRNA (Ozawa et al. 1987). |
| AAUAAG | Functional in 1 plant mRNA (Graham et al. 1985); deleterious (cause of α- and β-thalassemia) in human globin mRNA (Higgs et al. 1983; Rund et al. 1993). |
The principal components of the polyadenylation machinery in mammals are the two cleavage factors CFI and CFII; the poly(A) polymerase (PAP), and two factors involved in RNA sequence recognition: CstF (Cleavage Stimulation Factor), which binds the downstream GU-rich region, and CPSF (Cleavage/Polyadenylation Specificity Factor), which binds the polyadenylation signal. Given the variability of polyadenylation signals, can we suggest the existence of several cognate CPSFs? Probably not. All the observed signals are single-base variants of the canonical AAUAAA hexamer. Positions 3, 4, and 6 are highly conserved, while positions 1, 2, and 5 are tolerant to point mutations (Fig. 4). Combinations of two or more mutations have not been observed at a significant level. For instance, although AUUAAA is observed 843 times (Table 2), we did not find the prefix AUU associated with any of the other possible suffixes (ACA, AUA, AGA, or GAA). This suggests a model where a unique polyadenylation machinery is tolerant to a limited level of mutation in its regular signal.
The mRNAs with multiple poly(A) sites tend to use noncanonical polyadenylation signals (including the common AUUAAA) more often than mRNAs with a single poly(A) site (Table 4). Why would variant signals be selected in these mRNAs? The prevailing hypothesis for the occurrence of variant polyadenylation signals is that variation of control sequences mediates variation in polyadenylation rate, thus regulating gene expression (Edwalds-Gilbert et al. 1997; Graber et al. 1999). Expressed sequence tag counts, used as a measure of polyadenylation rate, provide in silico evidence in favor of this hypothesis. Table 5 and Figure 3 show that poly(A) sites with a noncanonical signal (including AUUAAA) were usually revealed by a lower number of ESTs than poly(A) sites with an AAUAAA signal. This observation is true independently of the number and position of the sites on the mRNA (Fig. 3) and cannot be explained by a bias in EST library construction or in our poly(A) site selection procedure. This suggests that variant signals are not processed as efficiently as the AAUAAA signal. This differential rate is of functional interest for mRNAs with multiple poly(A) sites since it provides a means to regulate synthesis of specific mRNA forms. The mRNAs with multiple sites may then use noncanonical (presumably weaker) signals because it is easier to regulate alternative polyadenylation with these weak signals. An additional form of regulation could be that 3′-terminal sites are processed more efficiently, as suggested by results in Figure 3. Current models for the binding of the polyadenylation machinery to its targets on the 3′ UTR—hexameric signal, GU-rich region, cleavage site (Colgan and Manley 1997)—do not help to explain this phenomena.
Another factor probably contributes to a higher polyadenylation rate at 3′ terminal sites. When observing the distribution of signals in alternatively polyadenylated mRNAs (Fig. 2), we noticed that AAUAAA signals, which are generally processed more efficiently, are more frequent at 3′-terminal sites. We may predict from this body of expression data that the major form of alternatively polyadenylated mRNAs will in general be the longest one. This high rate of long versus short 3′ UTRs might denote a better stability of the longer mRNA form. However, long 3′ UTRs are not necessarily more stable than shorter ones, especially since they often contain destabilization signals (Gautheret et al. 1998). This predominance of long forms may thus suggest the future discovery of stabilization signals in extended 3′ UTR fragments.
CONCLUSION
Ten variant polyadenylation signals characterized by a significant overrepresentation in EST-supported mRNA 3′ ends and by a peak of occurrence around position 15–17 (last base of signal) upstream of the putative poly(A) site have been identified. This information on poly(A) signal variation, combined with that of other polyadenylation control elements, should be incorporated in gene-detection programs, the performances of which are very poor in delineating 3′ UTRs. The consensus sequences or position weight matrices used for polyadenylation signal detection (Salamov and Solovyev 1997; Tabaska and Zhang 1999) can be adapted to agree with these observations. Similarly, statistics on the differential use of alternate sites can be incorporated in these programs.
On the biological side, two interesting questions are now raised. First, only 88% of the mRNA 3′ ends studied contained a characteristic poly(A) signal variant, leaving 678 putative 3′ ends with no detectable polyadenylation signal. A fraction of these may be artifactual 3′ ends (e.g., internally primed) that went through our selection procedure, but we cannot exclude that radically different signals or mechanisms may be used for the polyadenylation of this class of mRNAs. A detailed study of these unusual mRNAs, their function and pattern of expression, must be carried on to address this question. The second issue is that of the regulation of polyadenylation rate at different sites on the same mRNA. Is there a higher processing efficiency at the 3′-most poly(A) site, as our results suggest? Which unknown mechanism could produce this effect? While extensive experimental data is available on the processing of polyadenylation control elements in a single-site context, little is known about the effect of the relative position of multiple polyadenylation signals (including the downstream GU-rich region). This question is closely related to that of the kinetics and mechanisms of control sequence recognition by the polyadenylation machinery.
METHODS
Human 3′ UTR sequences were taken from UTRdb-nr release 10 (Pesole et al. 2000), a nonredundant database of eukaryotic UTRs generated by parsing the feature keys in the EMBL database. UTRdb can be retrieved from ftp://area.ba.cnr.it/pub/embnet/database/utr.
We compared the 8775 human UTRs to ESTs from dbEST (July 1999 release), using a variant of the sequence comparison procedure presented previously (Gautheret et al. 1998). Based on the gapped BLAST program (Altschul et al. 1997), this procedure seeks 3′ ESTs corresponding to mature mRNA 3′ ends. A typical dbEST match to an mRNA or UTR sequence contains a mixture of 5′ and 3′ ESTs, spurious hits from low complexity or repeated sequences, chimeric ESTs and ESTs resulting from internal priming. Our goal was to identify in this mixture those ESTs resulting from bona fide mRNA 3′ ends.
As a first criterion, and since actual 3′ ESTs are not consistently annotated in the database, we selected ESTs with a poly(T) or poly(A) extremity of length 10 or more. This filter retained only 157,775 of the original 1,561,241 human ESTs. Untranslated region sequences were masked for common human repeats, low complexity, and vector sequences. We then imposed ESTs to match the template mRNA sequence with at least 95% identity (a level of mismatch required to accommodate errors in EST sequences), encompassing the entire length of the EST sequence except for allowed 25-nt and a 5-nt mismatches at the EST 5′ and 3′ sides, respectively, as revealed by the boundaries of the BLAST hit. This last requirement dismisses about 23% of the ESTs, comprising probable chimeric ESTs, ESTs produced from alternatively spliced RNAs, and ESTs exhibiting lane tracking errors or high error rates in the terminal region. Poly(A) and poly(T) trailers were removed from EST sequences before running BLAST to ensure these tails did not create additional dangling regions. Internal priming, that is, cDNA primers hybridized to internal poly(A) stretches instead of the actual poly(A) tail, was assessed by seeking adenine stretches in the UTR region flanking the 3′ extremity of the EST. Six or more consecutive adenines, or eight adenines in a 10-nt window, were considered as a possible source of internal priming, and the corresponding EST sequence was discarded. Finally, the use of UTRs instead of complete mRNAs as query sequences eliminated the risk of identifying false 3′ ends in coding regions.
Any EST respecting the above constraints was considered indicative of a polyadenylation site at the 3′ end of the match. When several putative polyadenylation sites occurred in a region of 30 nt or less, we retained the site represented by the highest number of ESTs. Each potential polyadenylation site was recorded (mRNA, position, number of revealing ESTs), and the 50-nt segment preceding the site in the UTR was extracted (3′ fragment database) and searched for recurrent sequence motifs.
Significant 6-nt patterns were identified by comparing hexamer frequencies in the 3′ fragment database to those expected by chance from its nucleotide composition. Probabilities were computed assuming a cumulative binomial distribution (Press et al. 1992). Significant hexamers were collected iteratively as follows. After the most significant hexamer (lowest P-value) was identified, all 3′ fragments containing this motif were removed from the database before the next most frequent hexamer was sought. This procedure ensured that sequences overlapping the most frequent motifs (such as AUAAAN or NAAUAA for AAUAAA) were not improperly selected. The spatial distribution of motifs along the 50-nt segment was also considered in our selection of significant hexamers. The mean position of each motif in the 50-mer was computed, and the standard deviation (SD) around this average was used as a measure of scattering. Motifs with SD > 9 nt (empirical value) were considered as “scattered” and less likely to form a polyadenylation signal.
Acknowledgments
We thank Stéphane Audic for his advice and for sharing useful Perl Scripts with us.
Footnotes
E-MAIL gauthere@igs.cnrs-mrs.fr; FAX 33 4 91 16 45 49.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Batista FD, Tkach T, Efremov DG, Burrone OR. Multiple transcripts of the murine immunoglobulin epsilon membrane locus are generated by alternative splicing and differential usage of two polyadenylation sites. Mol Immunol. 1997;34:175–183. doi: 10.1016/s0161-5890(96)00110-1. [DOI] [PubMed] [Google Scholar]
- Andrews EM, DiMaio D. Hierarchy of polyadenylation site usage by bovine papillomavirus in transformed mouse cells. J Virol. 1993;67:7705–7710. doi: 10.1128/jvi.67.12.7705-7710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, Teague P, Goodwin GM, Harmar AJ. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72:1384–1388. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for expressed sequence tags. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Cheng JF, Raid L, Hardison RC. Isolation and nucleotide sequence of the rabbit globin gene cluster psi zeta-alpha 1-psi alpha. Absence of a pair of alpha-globin genes evolving in concert. J Biol Chem. 1986;261:839–848. [PubMed] [Google Scholar]
- Claverie JM. Computational methods for the identification of genes in vertebrate genomic sequences. Human Mol Genet. 1997;6:1735–1744. doi: 10.1093/hmg/6.10.1735. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- de Freitas FA, Yunes JA, da Silva MJ, Arruda P, Leite A. Structural characterization and promoter activity analysis of the gamma-kafirin gene from sorghum. Mol Gen Genet. 1994;245:177–186. doi: 10.1007/BF00283265. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: mean to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein P, Means AR, Berchtold MW. Isolation of a rat parvalbumin gene and full length cDNA. J Biol Chem. 1986;261:5886–5891. [PubMed] [Google Scholar]
- Faber PW, van Rooij HC, van der Korput HA, Baarends WM, Brinkmann AO, Grootegoed JA, Trapman J. Characterization of the human androgen receptor transcription unit. J Biol Chem. 1991;266:10743–10749. [PubMed] [Google Scholar]
- Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: a large scale analysis by EST clustering. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JS, Pearce G, Merryweather J, Titani K, Ericsson LH, Ryan CA. Wound-induced proteinase inhibitors from tomato leaves. II. the cDNA-deduced primary structure of pre-inhibitor II. J Biol Chem. 1985;260:6561–6164. [PubMed] [Google Scholar]
- Herve D, Rogard M, Levi-Strauss M. Molecular analysis of the multiple Golf alpha subunit mRNAs in the rat brain. Brain Res Mol Brain Res. 1991;32:125–134. doi: 10.1016/0169-328x(95)00070-9. [DOI] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hilger C, Velhagen I, Zentgraf H, Schroder CH. Diversity of hepatitis B virus X gene-related transcripts in hepatocellular carcinoma: a novel polyadenylation site on viral DNA. J Virol. 1991;65:4284–4191. doi: 10.1128/jvi.65.8.4284-4291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SL, Marks J, Shaw JP, Tam M, Higgs DR, Shen CC, Shen CK. Structure and expression of the human theta I globin gene. Nature. 1988;331:94–96. doi: 10.1038/331094a0. [DOI] [PubMed] [Google Scholar]
- Hu ZZ, Buczko E, Zhuang L, Dufau ML. Sequence of the 3′-noncoding region of the luteinizing hormone receptor gene and identification of two polyadenylation domains that generate the major mRNA forms. Biochim Biophys Acta. 1994;1220:333–337. doi: 10.1016/0167-4889(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Yoshimura K, Tamoi M, Takeda T, Shigeoka S. Alternative mRNA splicing of 3′-terminal exons generates ascorbate peroxidase isoenzymes in spinach (Spinacia oleracea) chloroplasts. Biochem J. 1997;328:795–800. doi: 10.1042/bj3280795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic L, Efremov GD, Petkov G, Kattamis C, George E, Yang KG, Stoming TA, Huisman TH. Two novel polyadenylation mutations leading to β(+)-thalassemia. Br J Haematol. 1990;75:122–126. doi: 10.1111/j.1365-2141.1990.tb02627.x. [DOI] [PubMed] [Google Scholar]
- Klemenz R, Reinhardt M, Diggelmann H. Sequence determination of the 3′ end of mouse mammary tumor virus RNA. MolBiol Rep. 1981;7:123–126. doi: 10.1007/BF00778742. [DOI] [PubMed] [Google Scholar]
- Larochelle S, Suter B. The drosophila melanogaster homolog of the mammalian MAPK-activated protein kinase-2 (MAPKAPK-2) lacks a proline-rich N-terminus. Gene. 1995;163:209–214. doi: 10.1016/0378-1119(95)00279-f. [DOI] [PubMed] [Google Scholar]
- Martins AS, Greene LJ, Yoho LL, Milsted A. The cDNA encoding canine dihydrolipoamide dehydrogenase contains multiple termination signals. Gene. 1995;161:253–257. doi: 10.1016/0378-1119(95)00268-b. [DOI] [PubMed] [Google Scholar]
- Moore CL, Chen J, Whoriskey J. Two proteins crosslinked to RNA containing the adenovirus L3 poly(A) site require the AAUAAA sequence for binding. EMBO J. 1988;7:3159–3169. doi: 10.1002/j.1460-2075.1988.tb03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myohanen S, Kauppinen L, Wahlfors J, Alhonen L, Janne J. Human spermidine synthase gene: structure and chromosomal localization. DNA Cell Biol. 1991;10:467–474. doi: 10.1089/dna.1991.10.467. [DOI] [PubMed] [Google Scholar]
- Nagashima M, McLean JW, Lawn RM. Cloning and mRNA tissue distribution of rabbit cholesteryl ester transfer protein. J Lipid Res. 1988;29:1643–1649. [PubMed] [Google Scholar]
- Nasseri M, Hirochika R, Broker TR, Chow LT. A human papilloma virus type 11 transcript encoding an E1–E4 protein. Virology. 1987;159:433–439. doi: 10.1016/0042-6822(87)90482-x. [DOI] [PubMed] [Google Scholar]
- Orkin SH, CHeng TC, Antonarakis SE, Kazazian HH. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy L, Parthasarathy R, Vadnal R. Molecular characterization of coding and untranslated regions of rat cortex lithium-sensitive myo-inositol monophosphatase cDNA. Gene. 1997;191:81–87. doi: 10.1016/s0378-1119(97)00045-0. [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Licciulli F, Larizza A, Makalowski W, Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2000;28:193–196. doi: 10.1093/nar/28.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in C. Cambridge University Press; 1992. In: p. 229. [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rabbitts KG, Morgan G. Alternative 3′ processing of xenopus alpha-tubulin mRNAs; efficient use of a CAUAAA polyadenylation signal. Nucleic Acids Res. 1992;20:2947–2953. doi: 10.1093/nar/20.12.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund D, Filon D, Oppenheim A, Abramov A. Silent carrier beta-thalassaemia due to a severe β-globin mutation interacting with other genetic elements. Eur J Pediatr. 1993;574:574–576. doi: 10.1007/BF01954083. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Recognition of 3′-processing sites of human mRNA precursors. Comp Appl Biosci. 1997;13:23–28. doi: 10.1093/bioinformatics/13.1.23. [DOI] [PubMed] [Google Scholar]
- Sanfacon H. Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology. 1994;198:39–49. doi: 10.1006/viro.1994.1006. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver Key SC, Pagano JS. A noncanonical poly(A) signal, UAUAAA, and flanking elements in Epstein-Barr virus DNA polymerase mRNA function in cleavage and polyadenylation assays. Virology. 1997;234:147–159. doi: 10.1006/viro.1997.8647. [DOI] [PubMed] [Google Scholar]
- Simonsen CC, Levinson AD. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983;3:2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetanina NS, Oner C, Baysal E, Oner R, Bozkurt G, Altay C, Gurgey A, Adekile AD, Gu LH, Huisman TH. The relative levels of alpha 2-, alpha 1-, and zeta-mRNA in HB H patients with different deletional and nondeletional alpha-thalassemia determinants. Biochim Biophys Acta. 1996;1316:176–182. doi: 10.1016/0925-4439(96)00024-5. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yamamoto K, Sinohara H. Molecular cloning and sequence analysis of full-length cDNA coding for mouse contrapsin. J Biochem (Tokyo) 1990;108:344–346. doi: 10.1093/oxfordjournals.jbchem.a123204. [DOI] [PubMed] [Google Scholar]
- Tabaska J, Zhang MQ. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- Taylor RG, Lambert MA, Sexsmith E, Sadler SJ, Ray PN, Mahuran DJ, McInnes RR. Cloning and expression of rat histidase. Homology to two bacterial histidases and four phenylalanine ammonia-lyases. J Biol Chem. 1990;265:18192–18199. [PubMed] [Google Scholar]
- Tokishita S, Shiga Y, Kimura S, Ohta T, Kobayashi M, Hanazato T, Yamagata H. Cloning and analysis of a cDNA encoding a two-domain hemoglobin chain from the water flea Daphnia magna. Gene. 1997;189:73–78. doi: 10.1016/s0378-1119(96)00836-0. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Kelly A. The human HLA class II alpha chain gene DZ alpha is distinct from genes in the DP, DQ and DR subregions. EMBO J. 1985;4:2231–2237. doi: 10.1002/j.1460-2075.1985.tb03919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Solinge WW, Lind B, van Wijk R, Hart HC, Kraaijenhagen RJ. Clinical expression of a rare β-globin gene mutation co-inherited with haemoglobin E-disease. Eur J Clin Chem Clin Biochem. 1996;34:949–954. doi: 10.1515/cclm.1996.34.12.949. [DOI] [PubMed] [Google Scholar]
- Wahlberg MH, Johnson MS. Isolation and characterization of five actin cDNAs from the cestode Diphyllobothrium dendriticum: a phylogenetic study of the multigene family. J Mol Evol. 1997;44:159–168. doi: 10.1007/pl00006132. [DOI] [PubMed] [Google Scholar]
- Wahle E, Keller W. The biochemistry of polyadenylation. TIBS. 1996;21:247–250. [PubMed] [Google Scholar]
- Wang W, Acland GM, Aguirre GD, Ray K. Cloning and characterization of the cDNA and gene encoding the gamma-subunit of cgmp-phosphodiesterase in canine retinal rod photoreceptor cells. Gene. 1996;181:1–5. doi: 10.1016/s0378-1119(96)00319-8. [DOI] [PubMed] [Google Scholar]
- Wetsel RA, Ogata RT, Tack BF. Primary structure of the fifth component of murine complement. Biochemistry. 1987;26:737–743. doi: 10.1021/bi00377a013. [DOI] [PubMed] [Google Scholar]
- Wilusz J, Shenk T. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Wu L, Ueda T, Messing J. 3′-end processing of the maize 27 kDa zein mRNA. Plant J. 1993;4:535–544. doi: 10.1046/j.1365-313x.1993.04030535.x. [DOI] [PubMed] [Google Scholar]
- Yan Y, Smant G, Stokkermans J, Qin L, Helder J, Baum T, Schots A, Davis E. Genomic organization of four beta-1,4-endoglucanase genes in plant-parasitic cyst nematodes and its evolutionary implications. Gene. 1998;220:61–70. doi: 10.1016/s0378-1119(98)00413-2. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Suemizu H, Taniguchi Y, Arimori K, Kawabe N, Moriuchi T. The human plasma glutathione peroxidase-encoding gene: organization, sequence and localization to chromosome 5q32. Gene. 1994;145:293–297. doi: 10.1016/0378-1119(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Yuregir GT, Aksoy K, Curuk MA, Dikmen N, Fei YJ, Baysal E, Huisman TH. Hb H disease in a turkish family resulting from the interaction of a deletional alpha-thalassaemia-1 and a newly discovered poly A mutation. Br J Haematol. 1992;80:527–532. doi: 10.1111/j.1365-2141.1992.tb04568.x. [DOI] [PubMed] [Google Scholar]
- Zhang YL, Akmal KM, Tsuruta JK, Shang Q, Hirose T, Jetten AM, Kim KH, O'Brien DA. Expression of germ cell nuclear factor (GCNF/RTR) during spermatogenesis. Mol Reprod Dev. 1998;50:93–102. doi: 10.1002/(SICI)1098-2795(199805)50:1<93::AID-MRD12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]