Abstract
Many cellular processes are carried out by large macromolecular assemblies. We systematically analyzed large macromolecular assemblies in the cytoplasm of mouse macrophages (RAW264.7 cell line), cells with crucial roles in immunity and inflammation. Fractionation of the cytoplasmic fraction was performed using sucrose density gradient centrifugation, and individual fractions were subjected in parallel to (i) identification of constituent proteins by mass spectrometry and (ii) structural visualization by electron microscopy. Macromolecular assemblies present in the fractions were analyzed by integrating available data using bioinformatic approaches. We identified 368 unique proteins in our sample. Among these are components of some well-characterized assemblies involved in diverse cellular processes and structures including translation, proteolysis, protein folding, metabolism, and the cytoskeleton, as well as less characterized proteins that may correspond to additional components of known assemblies or other homo- or hetero-oligomeric structures. Single-particle analysis of electron micrographs of negatively stained samples allowed the identification of clearly distinguishable two-dimensional projections of discrete protein assemblies. Among these, we can identify small ribosomal subunits and preribosomal particles, the 26S proteasome complex and small ringlike structures resembling the molecular chaperone complexes. In addition, a broad range of discrete and different complexes were seen at size ranges between 11 to 38 nm in diameter. Our procedure selects the assemblies on the basis of abundance and ease of isolation, and therefore provides an immediately useful starting point for further study of structure and function of large assemblies. Our results will also contribute toward building a molecular cell atlas.
Rapid progress is being made in understanding how cells function at different scales from individual molecules to the entire cell. At the molecular level, biophysical and genetic techniques yield structure-function relationships for individual molecules. Genome sequencing and proteomics studies are leading to cellular inventories of these macromolecules. At the cellular level, cell biology and interactomic studies are revealing the spatial and temporal organization of cellular interactions and signaling networks. To understand how the cellular system functions as a whole—the aim of systems biology—we need to integrate the data corresponding to these different levels under a single framework. However, significant gaps exist in our ability to relate the different levels of cellular function to each other.
Central to cellular functions are interactions between macromolecules. Interaction patterns reveal functional modules that correspond to either stable complexes or transient modules that remodel in response to signals. Over 80% of proteins from the cellular proteome are involved in these interactions, and the resulting complexes or assemblies play a central role in virtually every biological process including transcription, translation, cellular transport, metabolism, and signaling (1). Identification of the components in these assemblies and the structural and functional characterization of how they interact to generate a biological function is critical to understanding the mechanisms of biological processes. High-throughput methods have allowed a comprehensive mapping of interactions, for example by identifying binary interactions using yeast two-hybrid (Y2H)1 assays (2) or by characterizing tagged protein complexes using co-affinity purification followed by mass spectrometry (3). At a structural level however, multichain complex structures remain poorly represented in protein structure databases, and structural genomics efforts, employing the high-resolution structure determination techniques of x-ray crystallography and NMR, have currently focused on individual proteins. Single-particle electron microscopy analysis routinely provides medium resolution structures of complexes, as its throughput improves. Ultimately, a complete understanding of the function of the cell requires a molecular atlas with a spatial arrangement of the proteome. One technique that could help deliver this data is cryo-electron tomography, as it may be able to bridge the resolution gap that currently exists among structural studies at the molecular and cellular levels (4). The interpretation of cryotomograms relies on the knowledge of the molecular inventory of the system under study, as well as a library of the structures of individual components determined by the complementary high- to medium-resolution methods. As a step toward this goal, Han et al. have recently combined mass spectrometry-based proteomics and electron microscopy to characterize macromolecular assemblies in the bacterium Desulvibrio vulgaris (5).
In this study, we used a combination of techniques to complement the existing interactomic and structural approaches, and provide a direct link among the cellular inventory of macromolecular assemblies and their structures. Rather than focus on a specific known assembly of interest using molecular tagging, our experimental approach involved the separation of many macromolecular assemblies using sucrose density gradient centrifugation, followed by the analysis of individual fractions in parallel by (i) proteomic identification of constituent proteins by mass spectrometry, and by (ii) structural visualization using electron microscopy. The putative assemblies were identified by integrating available data using bioinformatic approaches. We used the RAW264.7 macrophage cell line as our model system. Macrophages are cells with crucial roles in both immunity and inflammation, and therefore expected to be a rich source of macromolecules with therapeutic potential (6). To this end, we limited our study to the cytoplasmic fraction, cytoplasm being the largest compartment of eukaryotic cells, where many important biological processes occur, including translation and protein synthesis, cell growth, cell division, protein degradation, cellular trafficking and transport, signal transduction, and cytoskeletal organization. Our studies complement existing approaches in the effort toward creating a molecular atlas of the cell.
EXPERIMENTAL PROCEDURES
Cell Lysis and Fractionation
Near-confluent adherent RAW264.7 cells (5 × 108) were harvested from 10 cm × 10 cm Sterilin dishes by draining the medium and replacing it with ice-cold phosphate-buffered saline (PBS). After 5 min, the cells detached and were removed by pipeting, the pooled cells pelleted at 400 × g for 5 min and rewashed three times with PBS. Finally, they were resuspended in 20 ml hypotonic wash buffer (HWB) comprising 10 mm HEPES pH 7.5, 10 mm KC1, 0.1 mm EDTA, with one tablet of “Complete” protease inhibitors per 50 ml (Roche).
Cells were lysed by nitrogen decompression (Cell Disruption Bomb, Parr Instrument Company, IL) after a 30 min incubation at 4 °C under 350 psi of nitrogen. Nuclei were spun out of the lysate at 1000 × g and the supernatant pooled and centrifuged at 14,000 × g in 15 ml Corex tubes in a Beckman JA-20 rotor for 20 min at 4 °C to remove unlysed cells, nuclei, mitochondria, and large cell fragments. The supernatant, containing the membrane fraction and cytoplasm, was ultracentrifuged at 50,000 rpm for 1 h at 4 °C in a TLA100.3 rotor (Beckman) on a two-step sucrose cushion (50 and 20%) to isolate the bulk of the high molecular mass complexes and membrane vesicles. Fractions of 0.2 ml were taken and a small amount run on SDS-PAGE to check that the separation had been effective. Fractions were assayed using Bradford reagent (Bio-Rad) to estimate the total yield of protein. The proteins that penetrated the 20% sucrose layer through to the interface between the 20 and 50% layers were pooled, concentrated at 4 °C by centrifugal ultrafiltration (Millipore, Billerica, MA) to 100 μl and layered onto a 10–50% analytical linear sucrose gradient made in HWB by a freeze-thaw cycle in 12.5-ml polyallomer centrifuge tubes.
Velocity sedimentation was carried out in an SW41 rotor for 9 h at 35,000 rpm at 4 °C. The centrifuge tubes were removed, punctured at the base and the gradient collected in ∼500 μl fractions. These fractions were assayed for protein and a small amount removed for electron microscopy and SDS-PAGE, after which they were stored at −80 °C.
Separation of Protein Complexes on SDS-PAGE
Approximately 20 μl of each fraction was concentrated ∼8× by centrifugal ultrafiltration (Millipore) and run on 4–12% gradient SDS-PAGE gel (NuPAGE, Novex, Invitrogen) at 90 V.
Sample Preparation for Mass Spectrometry
Sample preparation for in-gel trypsin digestion and peptide mass fingerprinting was performed according to (7). SDS was extracted from gel slices by incubation twice for 30 min each at 4 °C (with agitation) in a 500 μl mixture of acetone, triethylamine, acetic acid, and water (85:5:5:5). The gel slices were then destained and washed three times for 10 min at room temperature with 250 μl per tube of wash solution (100 mm ammonium bicarbonate (pH 8) with 30% acetonitrile). Gel slices were covered with 40 μl 10 mm dithiotreitol (DTT) in 50 mm ammonium bicarbonate for 15 min at 37 °C (with agitation). DTT was then removed and replaced with 20 μl of 100 mm iodoacetamide in 50 mm ammonium bicarbonate to alkylate the proteins. Samples were incubated for 30 min at 37 °C in the dark, before being washed three times for 10 min each with 100 mm ammonium bicarbonate containing 30% acetonitrile. After the last wash, the gel was cut into small (∼1 mm) pieces, washed for 10 min with ultrapure water (500 μl) and dried completely in a SpeedVac for 30 min. Dried gel fragments were stored at −80 °C prior to trypsin digestion. They were re-hydrated with 20 μl of a solution of 50 mm ammonium bicarbonate (pH 8) containing trypsin at final concentration of either 25 ng/μl (1 μl of 0.5 mg/ml trypsin in 10 mm HCl per 20 μl 50 mm ammonium bicarbonate) or 12.5 ng/μl. The digest solution was stored for 1 h at 4 °C to allow the trypsin to diffuse into the gel. Another 20 μl of the trypsin solution was added for a final volume of 40 μl per tube (to fully cover the gel fragments), the tubes were agitated, briefly centrifuged to pellet the gel fragments and stored for additional 1 h at 4 °C.
Digestion was carried out in a thermal cycler with a hot lid either overnight at 37 °C or for 2 h at 37 °C and then for 2 h at 50 °C, before storage at 4 °C. Digests were then briefly centrifuged and acidified with 10 μl of 1% (v/v) formic acid and the peptides prepared for matrix-assisted laser desorption ionization (MALDI) analysis using Zip Tips with C18 resin (Millipore) according to the manufacturer's instructions.
MALDI-Time-of-Flight (TOF) Analysis
Eluted peptides were combined with the matrix (α-cyano-4-hydroxycinnamic acid (CHCA)) at a 1:1 ratio and 1 μl of mixture was spotted onto a MALDI plate and analyzed by Voyager-DE STR MALDI-TOF mass spectrometry (Applied Biosystems, Foster City, CA). All MS spectra were recorded in positive reflector mode using multiple laser intensities. The voltage acceleration was set to 20 000 V and the grid was set to 67%, with a delay time of 350 ns. Each sample was analyzed by applying a minimum of 500 laser shots per spectrum.
Peptide Fingerprinting
Data Explorer 3.0 (Applied Biosystems) was used to extract monoisotopic peptide masses for protein identification. Internal calibration was carried out based on trypsin auto-catalysis peptides. PeakErazor 2.01 software (8) was used to remove any contaminants such as trypsin and keratin masses. Peptide masses were submitted to the Mascot Peptide Mass Fingerprint web server (www.matrixscience.com, Mascot Server 2.2 (31/08/2006)) (9) for protein identification with the following search criteria: MSDB 20060831 database (3239079 sequences), Mus musculus (house mouse) taxonomy (90914 sequences), 0.2 Da peptide mass tolerance, 0 to 1 maximum missed cleavage site, carbamidomethylation of cysteines as fixed and methionine oxidation as variable modifications. Only identifications with Mascot scores over 60 and with credible and nonredundant peptide sets were used. All annotated spectra for each PMF identification have been deposited in the Tranche repository (https://proteomecommons.org/index.jsp), under the hash codes Dd5tQvDFA2NjWSPsgOLI5XFLEEKvotPh6DiXE3NVzLdIFDvXdLMbfTKPwR93VZPyGyxiDhZNYA79ENnCnAOMyIlm854AAAAAAAABtQ = = (spectra in mzXML file format) and 0t/jdfbdbSQmsTd9hb+e+ZJgRzfQ+eafnxnXJBHfcxYCYbfPaIfda9b4Ny2mq56C8lUO2OhSfRlG6zXrXQsB81QZmdAAAAAAAAABtA = = (annotated spectra in PDF file format).
Bioinformatics Analysis
To characterize the identified proteins functionally and structurally, a range of different available bioinformatic tools and databases was employed, including Protein Information Resource (http://pir.georgetown.edu) (10), String (http://string.embl.de) (11), Reactome (http://www.reactome.org/) (12), UniProt/SwissProt (http://www.uniprot.org) (13), National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov), Protein Data Bank (PDB) (http://www.rcsb.org) (14), and GNF (Genomics Institute of the Novartis Research Foundation) SymAtlas (http://symatlas.gnf.org/SymAtlas) (15). Microaaray analysis of mRNA expression (15) was analyzed in the RAW264.7 cell line and was classified as weak, moderate, and high if the normalized expression values (a ratio of expression in a particular sample as compared with the median of all samples in tested data-set) (15) were less than five, between 5 and 10, and more than 10, respectively. The expression of mRNAs was also compared (i) among sets of macrophage (bone marrow macrophages, TEPM (thioglycollate-elicited peritoneal macrophages) versus nonmacrophage (3T3-L1 (mouse embryonic fibroblast - adipose like), C2C12 (mouse myoblast), NIH 3T3 (mouse embryonic fibroblast), Baf3 (mouse bone marrow-derived pro-B-cell)) cells, and (ii) among sets of macrophage-rich (bone marrow, spleen) versus nonmacrophage-rich (skeletal muscle, cerebral cortex, pancreas, heart) mouse tissues. The mRNAs were classified as macrophage-restricted if the normalized expression values (15) were higher in macrophage cells or macrophage-rich tissues than other cells/tissues, if the normalized expression values in macrophages were higher than five, and if the normalized expression values in other cells/tissues were lower than one; macrophage-enriched, if the normalized expression values were higher in RAW264.7 cells or macrophage-rich tissues than other cells/tissues, if the normalized expression values in macrophages were higher than five, and if the normalized expression values in other cells and tissues were lower than five; and macrophage-depleted, if the normalized expression values were lower in RAW264.7 cells or macrophage-rich tissues than other cells and tissues, if the normalized expression values in macrophages were lower than five, and if the normalized expression values in other cells/tissues were higher than five.
Electron Microscopy
A 5-μl aliquot of each fraction was applied to a glow-discharged (Med 020, Bal-Tec, Balzers, Liechtenstein) copper grid (200 mesh/inch) coated with a parlodion plastic support film stabilized with an additional thin film of carbon. After 1 min adsorption, the grids were washed three times with MilliQ water and negatively stained with 2% aqueous uranyl acetate solution for 45 s and subsequently air-dried. Individual data-sets (30–40 micrographs) from each fraction were recorded on a Jeol JEM1011 electron microscope (JEOL, Tokyo, Japan) operating at 80-kV acceleration voltage equipped with a side-mounted slow-scan Morada CCD camera controlled by iTEM acquisition software (Olympus Soft Imaging Solutions, Münster, Germany). Each dataset was collected at a eucentric defocus setting of between 0 μm and −0.5 μm and a nominal magnification of 120,000× corresponding to a pixel size at the specimen level (after binning) of 9.19 Å. All data were 2 × 2 binned, either during acquisition (fractions 2, 4, 7, 9–29) or prior to processing (fractions 1, 3, 5, 6, 8). The final galleries of class averages were cropped to a uniform box size of 64 × 64 pixels.
The EMAN software package (16) was used for image processing and single-particle analysis. Individual particles were selected either manually by boxer (EMAN) or semi-automatically using SwarmPS (17). For each sucrose gradient fraction, all of the particle images boxed out across all micrographs were combined into a single dataset, resulting in a single dataset of boxed out particle projection images for each sucrose gradient fraction. The Ctfit procedure was used to determine optimum parameters for filtering out unwanted high (beyond the first zero of the averaged whole datatset Fourier transform) and low frequency information. Parameters were adjusted for each individual dataset and ranged from 1/25 Å to 1/40 Å and 1/200 Å to 1/300 Å, for low and high pass filters, respectively. Filtered datasets were precentered and multivariate statistical analysis, classification, alignment and calculation of final class averages were performed by the refine2d.py command (eight iterations).
RESULTS
Purification and Fractionation of Macromolecular Assemblies
In this study, we employed a fast and simple biochemical fractionation of the cytoplasmic fraction of mouse macrophages to isolate large assemblies in their native state. Cell lysis by nitrogen decompression allowed the isolation of the cytoplasm, free from the release of lysosomal and endosomal proteases. Native protein assemblies were further separated according to their molecular mass by ultracentrifugation through an analytical sucrose gradient. Twenty-nine fractions were collected from the sucrose gradient and subjected in parallel to a proteomic identification of proteins in each fraction, and to the structural visualization of assemblies in each fraction by electron microscopy.
Proteomic Characterization of Protein Subunits of Macromolecular Assemblies
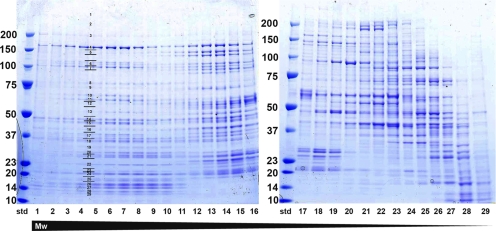
Each of the 29 fractions was concentrated and applied to a 4–12% gradient SDS-PAGE gel to separate individual proteins according to size (Fig. 1). We excised the gel bands from the gel, performed in-gel trypsin digestion, extracted and concentrated the tryptic peptides, and analyzed the peptides by MALDI-TOF MS. Monoisotopic peptide masses extracted from the recorded spectra were submitted to the Mascot Peptide Mass Fingerprint web server (9) for protein identification. We analyzed 792 gel bands, 363 of which yielded 709 protein identifications. Many of the identified proteins were present in multiple fractions, resulting in a total of 368 unique proteins (supplemental Table S1). On average, we identified 24 proteins per fraction.
Fig. 1.
SDS-PAGE gel of individual fractions after sucrose gradient fractionation of the cytoplasmic fraction of mouse macrophages. Protein complexes present in the cytoplasmic fraction were separated according to their size by ultracentrifugation over sucrose gradient. After concentration of each of the 29 fractions, 20 μl was applied to SDS-PAGE, in order to separate proteins present in each particular fraction (fraction 1 represents fraction at the bottom of the gradient, fraction 29 represents the fraction at the top of the gradient; std, molecular mass standards with masses indicated). For the proteomic analysis, every band from SDS-PAGE gel was excised (as indicated as an example for fraction 4) and subjected to in-gel trypsin digestion followed by peptide mass fingerprinting by MALDI-TOF.
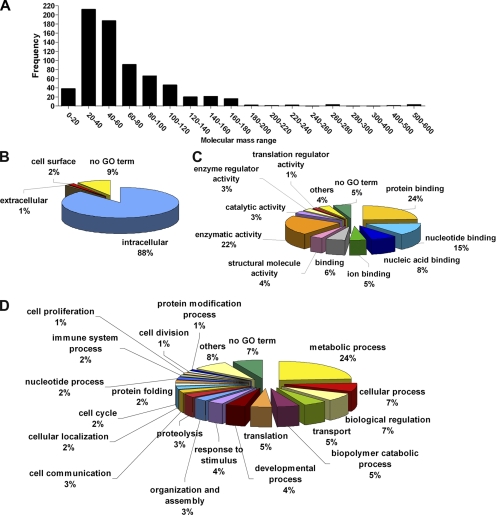
The chosen protein purification and identification strategies allowed us to identify proteins covering a broad range of molecular masses ranging from 10 to 600 kDa, with the majority in the 20–60 kDa range (Fig. 2A). We analyzed the proteins in terms of cellular localization and cellular and molecular function as defined by gene ontology (GO) nomenclature. Nine percent of the proteins identified had no GO term and so were excluded from the following analysis. The remaining 91% could be identified by GO. Eighty-eight percent of the proteins were classified as intracellular, validating our purification strategy of the cytoplasmic fraction (88/91 corresponds to 96.7% purity; Fig. 2B). Only two of the identified proteins are designated exclusively as extracellular by GO. One of these, the macrophage migration inhibitory factor (MIF), however is known to have intracellular functions in addition to extracellular ones (e.g. (18); not annotated by GO). The reasons for detecting the other protein, the extracellular matrix protein FRAS1 (Fraser syndrome 1), are less clear: it is a transmembrane extracellular matrix protein reported only in basal membranes of epithelial cells (19). A further four proteins are designated exclusively as cell surface proteins according to GO. All these four proteins (CD209 antigen-like protein B/DC-SIGH related protein 1, CD28 antigen, integrin αL and cell adhesion molecule-related/down-regulated by oncogenes) are transmembrane receptor-like proteins and may have been captured in our sample during the internalization of plasma membrane vesicles, or they correspond to the 3.3% portion corresponding to contaminant proteins (Fig. 2B).
Fig. 2.
Molecular mass distribution and functional classification of identified proteins. A, Number of proteins (frequency) in a particular molecular mass range (kDa). B, Subcellular location of the identified proteins as classified by GO terms. C, Molecular function of the identified proteins as classified by GO terms. D, Cellular function of the identified proteins as classified by GO terms.
Classification in terms of cellular process and molecular function revealed that the identified proteins, as expected, covered a broad range of functions. In terms of cellular processes, the most highly represented are metabolic processes, transport, translation, proteolysis, and cell cycle (Fig. 2C). In terms of molecular function, the most abundant are protein binding, enzymatic activity and nucleotide and nucleic acid binding (Fig. 2D).
Identification of Assemblies and Their Constituents
The 29 analyzed fractions (Fig. 1) contained macromolecular assemblies in the range of about 500 kDa to 1.7 MDa (as inferred from particle sizes in electron micrographs; see below). All the identified proteins would therefore be expected to form either homo-oligomers or to interact with other proteins present in the same fraction. We used the Reactome knowledgebase (12) to map the identified proteins to known macromolecular complexes. One-hundred-two of the 368 proteins could be mapped to one or more of 297 characterized complexes. Examples of the identified complexes include the ribosomal subunits, the proteasome, centrosomes, spliceosomal complexes, various cytoskeletal complexes, and enzymes such as enolase and glycogen synthase.
Of the remaining 265 proteins, 98 are known or predicted to interact with other proteins in the data-set, as assessed by the String database of functional interactions (11), forming 998 binary interactions. After removing redundancy in protein and gene designations, 124 proteins remain that have not been reported to be part of complexes or to interact with other proteins in our list. These proteins may therefore either correspond to new constituents of known complexes, form large homo-oligomers or form new macromolecular complexes that had not been previously identified. Our approach therefore provides a powerful tool for the isolation and characterization of previously uncharacterized macromolecular assemblies.
Electron microscopy and single-particle analysis
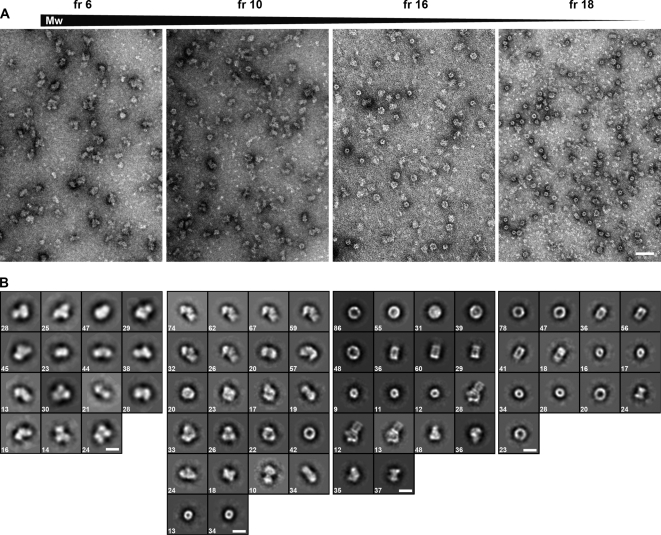
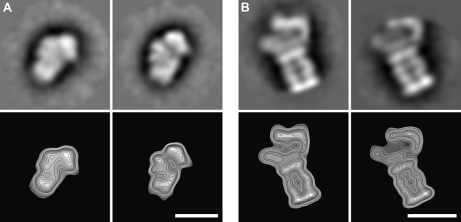
The individual fractions were subjected to electron microscopy to visualize the assemblies structurally. From each fraction, we collected 30–50 electron micrographs and selected between 2000 and 8000 particles. Using EMAN image processing software (16), we performed reference-free classification (multivariate statistical analysis) to separate the particles into different classes, and calculate an average image for each class (Fig. 3, supplemental Table S2). As expected, electron micrographs show that the large molecular mass fractions (bottom of the gradient) contained large particles, whereas low molecular mass fractions (top of the gradient) contained smaller particles. The only exception corresponded to the very top fraction of the gradient, which contained relatively large particles and presumably represented a mixture of particles that did not enter the gradient. The size of the particles ranged from 15 to 34 nm. Relative to the size of the small ribosomal subunit (molecular mass of 1.4 MDa), we estimated the molecular mass range of the observed protein assemblies to be between 500 kDa and 1.7 MDa. The final average images of the classes allowed us to relate each of the projection images with the projections of reference protein structures, such as the 26S proteasome complex and the small ribosomal subunit (Fig. 4).
Fig. 3.
Structures of assemblies in different fractions as visualized by electron miscroscopy. A, Electron micrographs of representative sucrose gradient fractions (from left to right: fr 6, fraction 6; fr 10, fraction 10; fr 16, fraction 16; fr 18, fraction 18) after negative staining. Scale bar: 50 nm. B, Galleries of selected final two-dimensional averages of putative macromolecular complexes (corresponding to different classes) from the fractions shown in (A). Scale bars: 20 nm. Numbers in the lower left corner indicate the number of particles from which the final average was calculated. Fraction 18 contains 20S proteasome particles.
Fig. 4.
Assemblies with known structures can be observed in sucrose gradient fractions. The calculated final class averages (upper panels) and electron-density contour maps (lower panels) are shown after single-particle analysis of selected particles. Based on the proteins identified in the relevant fraction and the comparison with the published structures, the assemblies may correspond to: A, The small ribosomal subunit (30); B, The 26S proteasome complex (31). Scale bars: 20 nm.
Structural Novelty
Sequence comparisons with proteins in the Protein Data Bank show that about 31% of the identified proteins have a known three-dimensional structure (>80% sequence identity with >80% sequence coverage), whereas 29% of the proteins have no known structure (<30% sequence identity or <10% sequence coverage when >30% sequence identity). For the remaining proteins, a structure of a homolog is known for 12% of the proteins (at sequence identity 30–80% and >80% sequence coverage), the structure is known for only a part of the protein for 13% of the proteins (>80% sequence identity with 10–80% sequence coverage), or the structure of a homolog is known for only a part of the protein for 15% of the proteins (30–80% sequence identity with 10–80% sequence coverage).
Expression in Macrophages
Microarray data (15) allow us to estimate the expression level in RAW264.7 macrophages of the mRNAs corresponding to 87% of the identified proteins (those that were probed in these arrays). The mRNAs corresponding to 80% of the identified proteins are expressed at weak levels, 7% at moderate levels, 2% at high levels, and the expression of 11% was not detected in these experiments. We also checked the expression of the proteins in other cell lines and in various tissues (see the Experimental Procedures section). The mRNAs corresponding to 68% of the proteins are expressed at similar levels in macrophages and nonmacrophage cells, 7% are enriched in macrophages, 7% are restricted to macrophages, and 3% have lower expression in macrophages (no expression information is available for the remaining 15%). Similar statistics are obtained if we compare macrophage-rich and nonmacrophage-rich tissues. The mRNAs corresponding to 75% of the proteins are expressed at similar levels in tissues that are macrophage-rich and nonmacrophage-rich, 5% are enriched in macrophage-rich tissues, 6% are restricted to macrophage-rich tissues and 5% have lower expression in macrophage-rich tissues (no expression information is available for the remaining 12%). In summary, these data suggest that only a small portion of the observed assemblies are macrophage-specific, and our results have relevance for mammalian cells in general.
DISCUSSION
The primary methods that have been used for high-throughput identification of protein interactions include the Y2H approach and co-affinity purification followed by mass spectrometry. Y2H reveals binary interactions, and the presence of complexes needs to be inferred computationally from interaction network diagrams. Co-affinity purification such as tandem affinity purification (TAP) (20), on the other hand, focuses on specific complexes and identifies their components. In addition to the need to prepare, test and transfect the tagged bait protein, the tag can sometimes interfere with normal complex formation. Here we employed a biochemical fractionation technique to purify high molecular weight complexes in their native state using a sucrose gradient, to eliminate the majority of cellular proteins of smaller molecular masses and fractionate stable complexes. This approach eliminates any concerns about tag interference as well as the need to transfect cells and select the tagged proteins. In particular, the method was intended to rapidly identify the presence of macromolecular complexes that are suitable candidates for single-particle analysis by virtue of being (a) relatively abundant, (b) stable enough to isolate with little difficulty, and (c) not structurally characterized to date. Apart from the expected well-known complexes, we identified a range of protein components that comprise candidates for further single-particle analysis. The strength of the method consequently lies in its ability to select suitable candidates to work with, rather than being a targeted approach to a specific biology. Although our work was in progress, a similar approach has been applied to the bacterium Desulvibrio vulgaris (5).
Conversely, the limitations of this approach as applied here are (i) limited sensitivity (it is expected that only high-abundance assemblies can be detected, although it is amenable to increasing the sensitivity if desired by scaling up), and (ii) the assemblies are not purified to homogeneity, so that several assemblies are present in each fraction. The identification of individual assemblies therefore relies on the use of complementary data obtained by other approaches, which includes data available in databases and in the literature. In the future, further fractionation by sucrose gradients or alternative methods such as size-exclusion chromatography or blue-native PAGE could be used to facilitate the purification of individual complexes. It is further possible that one or more of the components of a complex may dissociate during the handling, and may not be present in the correct fraction. The dissociation may be countered by the use of the gentle GRAFIX cross-linking method (e.g. (21)) that is compatible with the sucrose gradient approach used here, and which has been statistically demonstrated to preserve structural integrity of isolated assemblies well.
A distinguishing feature of our approach is that we can directly probe the fractionated assemblies structurally using electron microscopy and single-particle analysis techniques. Information is therefore provided on the size and shapes of the fractionated assemblies, which can be related directly to the individual proteins identified in a particular fraction by mass spectrometry-based proteomics. In favorable cases single-particle analysis also allows the recognition of individual assemblies by comparison to assemblies having known structures, and “in silico purification” of individual assemblies. It also allows the refinement of multiple structures from mixed particle populations (16, 22). We show examples of assemblies with known three-dimensional structures that can be recognized in electron micrographs, and further work promises to yield structural information on novel assemblies, but this will require further fractionation and data processing steps. Further work will also be required to achieve three-dimensional reconstruction of individual assemblies in our system; this goal has been achieved in a less complex system by Han et al. (5).
The major well-characterized assemblies detected in our sample, as recorded in the Reactome knowledgebase (12) include the components of the proteasome, tRNA synthetases, the ribosome, eukaryotic initiation and elongation factors, heterogeneous nuclear ribonucleoproteins and spliceosomal complexes, the centrosome and cell-cycle associated complexes, the cytoskeleton, the clathrin-associated adaptor protein complex, chaperone complexes, and the microsomal triglyceride transfer protein. There are also several metabolic enzyme complexes including glycogen synthase, glucose-6-phosphate isomerase, aldolase A, triose-phosphate isomerase, enolase, pyruvate kinase, lactate dehydrogenase, transketolase, glucose-6-phosphate dehydrogenase, ribonucleotide reductase, bifunctional purine biosynthesis protein PURH, and S-adenosylmethionine synthetase.
Less well-characterized complexes include an array of ribonucleoprotein complexes, and complexes containing proteins for which little functional information is available. This technique therefore offers the possibility of probing the function of these proteins by virtue of their associations with other proteins and by the identification of the molecular architecture of the complexes that contain them. Several other proteins in our sample have been reported to form binary interactions (String database (11)) with other proteins in the sample and may therefore represent additional components of the complexes listed above, or may be part of less characterized complexes. For example, several eukaryotic translation initiation and elongation factors are expected to be associated with the ribosome, and the deubiquitinating enzyme ubiquitin carboxyl-terminal esterase L5 is expected to be associated with the proteasome. As another example, the tumor suppressor adenomatosis polyposis coli shows associations with several proteosomal subunits.
Several proteins do not show associations with other proteins in the dataset according to Reactome and String analysis, but could represent further components of known assemblies listed above, or form homo- or hetero-oligomeric complexes. For example, fatty acid synthase (23) and CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, dihydroorotase) (24), multi-enzyme polypeptides involved in fatty acid and pyrimidine nucleotide biosynthesis, respectively, are known to form large homo-oligomeric structures. Another intriguing candidate, CML66 was identified as a tumor antigen involved in the immune regulation of chronic myelogenous leukemias (25). Subsequent work has shown that it is involved in tumor proliferation invasion and metastasis (26).
We used the cytoplasmic fraction of RAW264.7 macrophages as our model system for the present study. Only a small proportion of identified proteins are considerably enriched in macrophages as compared with other cells, therefore our study is pertinent to the structural organization of the cytoplasm of mammalian cells in general. However, a small proportion of the identified proteins may represent macrophage-specific assemblies. For example, the macrophage-enriched identified proteins include Fyn-binding protein (Fyb), an adaptor protein that interacts with actin-binding proteins, and the actin-binding protein plastin-2/lymphocyte cytosolic protein 1, which may both represent components of complexes that regulate actin dynamics in macrophages (27, 28). Interestingly, two proteasome subunits appear macrophage-enriched also, supporting the idea that differing subunit compositions of the proteasome may play distinct roles in cell biology (29).
In summary, we have used a combination of approaches to survey large macromolecular assemblies in the cytoplasm of macrophage cells. The approach included the fractionation of the cytoplasmic fraction using sucrose density gradient centrifugation, and individual fractions were subjected in parallel to the identification of constituent proteins by mass spectrometry, and structural characterization by electron microscopy. Macromolecular assemblies in the molecular mass range between 0.5 and 1.7 MDa were isolated, and included 124 proteins not reported previously to be associated with large assemblies. Several particles matched projection images of known reference structures. Our approach is complementary to other approaches that contribute toward building a molecular cell atlas.
Acknowledgments
We thank Kate Schroder, Jane Lattin, and Stuart Kellie for help with the analysis of expression in macrophages, Brad Chapman for help with PyMs XML and David Hume and the Kobe and macrophage structural biology group members for helpful discussions. BK was an Australian Research Council (ARC) Federation Fellow and is a National Health and Medical Research Council (NHMRC) Research Fellow.
Footnotes
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- GO
- gene ontology
- HWB
- hypotonic wash buffer
- Mw
- molecular weight (mass)
- TAP
- tandem affinity purification
- Y2H
- yeast two hybrid.
REFERENCES
- 1. Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 2. Parrish J. R., Gulyas K. D., Finley R. L., Jr. (2006) Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17, 387–393 [DOI] [PubMed] [Google Scholar]
- 3. Köcher T., Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 4. Nickell S., Kofler C., Leis A. P., Baumeister W. (2006) A visual approach to proteomics. Nat. Rev. Mol. Cell Biol. 7, 225–230 [DOI] [PubMed] [Google Scholar]
- 5. Han B. G., Dong M., Liu H., Camp L., Geller J., Singer M., Hazen T. C., Choi M., Witkowska H. E., Ball D. A., Typke D., Downing K. H., Shatsky M., Brenner S. E., Chandonia J. M., Biggin M. D., Glaeser R. M. (2009) Survey of large protein complexes in D. vulgaris reveals great structural diversity. Proc. Natl. Acad. Sci. U.S.A. 106, 16580–16585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hume D. A. (2006) The mononuclear phagocyte system. Curr. Opin. Immunol. 18, 49–53 [DOI] [PubMed] [Google Scholar]
- 7. Zischka H., Gloeckner C. J., Klein C., Willmann S., Swiatek-de Lange M., Ueffing M. (2004) Improved mass spectrometric identification of gel-separated hydrophobic membrane proteins after sodium dodecyl sulfate removal by ion-pair extraction. Proteomics 4, 3776–3782 [DOI] [PubMed] [Google Scholar]
- 8. Hjernø K., Højrup P. (2004) PeakErazor: A Windows-based program for improving peptide mass searches. In: Kamp R. M., Calvete J. J., Choli-papadopoulou T. eds. Methods in Proteome and Protein Analysis, pp. 359–370, Springer Verlag, New York, NY [Google Scholar]
- 9. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 10. Wu C. H., Yeh L. S., Huang H., Arminski L., Castro-Alvear J., Chen Y., Hu Z., Kourtesis P., Ledley R. S., Suzek B. E., Vinayaka C. R., Zhang J., Barker W. C. (2003) The Protein Information Resource. Nucleic Acids Res. 31, 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D'Eustachio P. (2009) Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37, D619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Consortium U. (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berman H., Henrick K., Nakamura H., Markley J. L. (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35, D301–D303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 17. Woolford D., Ericksson G., Rothnagel R., Muller D., Landsberg M. J., Pantelic R. S., McDowall A., Pailthorpe B., Young P. R., Hankamer B., Banks J. (2007) SwarmPS: rapid, semi-automated single particle selection software. J. Struct. Biol. 157, 174–188 [DOI] [PubMed] [Google Scholar]
- 18. Kleemann R., Hausser A., Geiger G., Mischke R., Burger-Kentischer A., Flieger O., Johannes F. J., Roger T., Calandra T., Kapurniotu A., Grell M., Finkelmeier D., Brunner H., Bernhagen J. (2000) Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 408, 211–216 [DOI] [PubMed] [Google Scholar]
- 19. Petrou P., Makrygiannis A. K., Chalepakis G. (2008) The Fras1/Frem family of extracellular matrix proteins: structure, function, and association with Fraser syndrome and the mouse bleb phenotype. Connect. Tissue Res. 49, 277–282 [DOI] [PubMed] [Google Scholar]
- 20. Bürckstümmer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 21. Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Lührmann R., Stark H. (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 22. Brink J., Ludtke S. J., Kong Y., Wakil S. J., Ma J., Chiu W. (2004) Experimental verification of conformational variation of human fatty acid synthase as predicted by normal mode analysis. Structure 12, 185–191 [DOI] [PubMed] [Google Scholar]
- 23. Leibundgut M., Maier T., Jenni S., Ban N. (2008) The multienzyme architecture of eukaryotic fatty acid synthases. Curr. Opin. Struct. Biol. 18, 714–725 [DOI] [PubMed] [Google Scholar]
- 24. Lee L., Kelly R. E., Pastra-Landis S. C., Evans D. R. (1985) Oligomeric structure of the multifunctional protein CAD that initiates pyrimidine biosynthesis in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 82, 6802–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang X. F., Wu C. J., McLaughlin S., Chillemi A., Wang K. S., Canning C., Alyea E. P., Kantoff P., Soiffer R. J., Dranoff G., Ritz J. (2001) CML66, a broadly immunogenic tumor antigen, elicits a humoral immune response associated with remission of chronic myelogenous leukemia. Proc. Natl. Acad. Sci. U.S.A. 98, 7492–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q., Li M., Wang Y., Zhang Y., Jin S., Xie G., Liu Z., Wang S., Zhang H., Shen L., Ge H. (2008) RNA interference targeting CML66, a novel tumor antigen, inhibits proliferation, invasion and metastasis of HeLa cells. Cancer Lett. 269, 127–138 [DOI] [PubMed] [Google Scholar]
- 27. Yuan M., Mogemark L., Fällman M., Mogemark L., Fällman M. (2005) Fyn binding protein, Fyb, interacts with mammalian actin binding protein, mAbp1. FEBS Lett. 579, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 28. Janji B., Giganti A., De Corte V., Catillon M., Bruyneel E., Lentz D., Plastino J., Gettemans J., Friederich E. (2006) Phosphorylation on Ser5 increases the F-actin-binding activity of L-plastin and promotes its targeting to sites of actin assembly in cells. J. Cell Sci. 119, 1947–1960 [DOI] [PubMed] [Google Scholar]
- 29. Dhungana S., Merrick B. A., Tomer K. B., Fessler M. B. (2009) Quantitative proteomics analysis of macrophage rafts reveals compartmentalized activation of the proteasome and of proteasome-mediated ERK activation in response to lipopolysaccharide. Mol. Cell Proteomics 8, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nickell S., Mihalache O., Beck F., Hegerl R., Korinek A., Baumeister W. (2007) Structural analysis of the 26S proteasome by cryoelectron tomography. Biochem. Biophys. Res. Commun. 353, 115–120 [DOI] [PubMed] [Google Scholar]
- 31. Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]