Abstract
Sorghum is an important target for plant genomic mapping because of its adaptation to harsh environments, diverse germplasm collection, and value for comparing the genomes of grass species such as corn and rice. The construction of an integrated genetic and physical map of the sorghum genome (750 Mbp) is a primary goal of our sorghum genome project. To help accomplish this task, we have developed a new high-throughput PCR-based method for building BAC contigs and locating BAC clones on the sorghum genetic map. This task involved pooling 24,576 sorghum BAC clones (∼4× genome equivalents) in six different matrices to create 184 pools of BAC DNA. DNA fragments from each pool were amplified using amplified fragment length polymorphism (AFLP) technology, resolved on a LI-COR dual-dye DNA sequencing system, and analyzed using Bionumerics software. On average, each set of AFLP primers amplified 28 single-copy DNA markers that were useful for identifying overlapping BAC clones. Data from 32 different AFLP primer combinations identified ∼2400 BACs and ordered ∼700 BAC contigs. Analysis of a sorghum RIL mapping population using the same primer pairs located ∼200 of the BAC contigs on the sorghum genetic map. Restriction endonuclease fingerprinting of the entire collection of sorghum BAC clones was applied to test and extend the contigs constructed using this PCR-based methodology. Analysis of the fingerprint data allowed for the identification of 3366 contigs each containing an average of 5 BACs. BACs in ∼65% of the contigs aligned by AFLP analysis had sufficient overlap to be confirmed by DNA fingerprint analysis. In addition, 30% of the overlapping BACs aligned by AFLP analysis provided information for merging contigs and singletons that could not be joined using fingerprint data alone. Thus, the combination of fingerprinting and AFLP-based contig assembly and mapping provides a reliable, high-throughput method for building an integrated genetic and physical map of the sorghum genome.
[The sequence data described in this paper have been submitted to the GenBank data library under accession no. AF218263.]
Integrated genetic and physical genome maps are extremely valuable for map-based gene isolation, comparative genome analysis, and as sources of sequence-ready clones for genome sequencing projects. Various methods have been developed for assembling physical maps of complex genomes. One of the best characterized approaches uses restriction enzymes to generate large numbers of DNA fragments from genomic subclones (Brenner and Livak 1989; Gregory et al. 1997; Marra et al. 1997). These DNA fingerprints are compared to identify related clones, and to assemble overlapping clones in contigs. The utility of fingerprinting for ordering a complex genome is limited, however, due to variation in DNA migration from gel to gel, the presence of repetitive DNAs, unusual distribution of restriction sites and skewed clone representation. Moreover, fingerprinting, unless combined with other methods, does not link genomic clones directly to genetic maps. Therefore, most high-quality physical maps of complex genomes have been constructed using a combination of fingerprinting and PCR-based or hybridization-based methods (Marra et al. 1997; Cao et al. 1999; Vollrath and Jaramillo-Babb 1999; Zhu et al. 1999).
Over the past two and one-half years we have been constructing an integrated genetic and physical map of the sorghum genome. Sorghum was selected for genome map construction for several reasons. Firstly, the sorghum genome is small (750 Mbp) relative to most other grasses of commercial importance, with the exception of rice which has a genome size of 430 Mbp (Arumuganathan and Earle 1991). Secondly, sorghum is tolerant to harsh environments and has a diverse germplasm collection (∼40,000 accessions), making this species an excellent system for the analysis of the genes contributing to environmental stress tolerance and other traits (Doggett 1988). Third, sorghum is closely related to corn, one of the best plant genetic systems, from which it diverged only 15–20 million years ago (Doebley et al. 1990). Genome sequencing, mapping, and related analyses indicate that although noncoding sequences in sorghum and maize have diverged significantly, gene order and gene sequences in these two species are highly conserved making comparative analysis useful (Avramova et al. 1996; Tikhonov et al. 1999). Finally, several sorghum genetic maps have been constructed (Chittenden et al. 1994; Boivin et al. 1999; Peng et al. 1999), and a high-quality sorghum BAC library was available as a starting point for the construction of a physical map (Woo et al. 1994).
In this article, we describe a combination of methods that, when fully implemented, will allow construction of an integrated genetic and physical map of the sorghum genome. The project started with the construction of an additional sorghum BAC library (Tao and Zhang 1998), and the characterization of the two sorghum BAC libraries for chloroplast, mitochondrial, centromeric, rDNA, and subtelomeric sequences. The entire set of 26,000 BAC clones was fingerprinted and contigs based on the fingerprint data were assembled. In addition, a high-throughput PCR-based screening method was developed which combines sixfold BAC DNA pooling and amplified fragment length polymorphism (AFLP) technology. This methodology allowed us to identify BAC clones containing genetic markers, thereby linking the DNA-based physical map to the sorghum genetic map. This combination of approaches provides a low cost, efficient way to build high-quality integrated genetic and physical genome maps.
RESULTS
BAC Library Characterization
Two sorghum BAC libraries were used in this study. DNA for both libraries was derived from the elite sorghum genotype BTx623. The first library of 13,440 clones was constructed by Woo et al. (1994) from total sorghum DNA that had been partially restricted with HindIII. Woo et al. (1994) reported an average insert size of 157 kbp in this library, although inserts up to 340 kbp were observed. A second BAC library containing 12,576 clones was constructed for this study from nuclear DNA partially restricted with EcoRI (Tao and Zhang 1998). EcoRI was selected to increase the overall genome coverage of the two libraries. The average insert size in the EcoRI-based library was 140 kbp.
To survey the level of contaminating organellar DNA and identify clones containing repetitive DNA elements in the sorghum HindIII and EcoRI BAC libraries, the BAC clones were arrayed on high density filters and hybridized to 32P-labeled organellar and repetitive DNA probes. To determine the level of chloroplast contamination within the two BAC libraries, six plastid genes, estimated to be equally spaced around the plastid genome, were selected as probes. Approximately 10.5% of the clones in the HindIII library (1404/13,440 inserts) and 3.3% of the clones in the EcoRI library (152/4608 inserts) contained plastid DNA (data not shown). The reduced fraction of chloroplast clones in the EcoRI library reflects the use of nuclear DNA for library construction (Tao and Zhang 1998). Both libraries were also analyzed for the presence of mitochondrial DNA sequences by probing with a pool of three different sorghum mitochondrial genes. Less than 0.1% of the clones (10/12,288 inserts) hybridized to this probe mixture, indicating little contamination from mitochondrial DNA (data not shown). Gel analysis further revealed 7.5% of the clones (∼1950) lacked genomic inserts. Accounting for the clones with no inserts and those of mitochondrial and chloroplast origin, ∼22,200 BAC clones remained for physical map construction. With an average BAC insert size of 148.5 kbp and a sorghum genome size of 750 Mbp (Arumuganathan and Earle 1991), the collection provides ∼4 times the coverage of the genome. Therefore, there is a 98% chance of finding any specific region of the sorghum genome from this combined collection of BACs.
Repetitive DNA sequences can be a source of error in contig assembly. Therefore, the two sorghum BAC libraries were screened for three known classes of repetitive DNA elements (rDNA, centromeric, and subtelomeric sequences). Centromere-specific repetitive DNA elements have been identified in sorghum (Jiang et al. 1996; Miller et al. 1998a,b) and some of these elements show significant sequence identity to Ty3/gypsy-like retrotransposons (Miller et al. 1998a). The Sau3A9 repetitive element is present in the centromeres of sorghum as well as many other cereals (Jiang et al. 1996), whereas the Sau3A10 element is limited to the centromeres of the genus Sorghum (Miller et al. 1998b). The Sau3A10 element is estimated to comprise between 1.6%–1.9% of the sorghum genome and is arranged in long tandem arrays, interspersed with other DNA repeat sequences, including Sau3A9 (Miller et al. 1998b). When Sau3A10 was used to probe the two BAC libraries, 6.4% (395/6144 inserts) of clones from the HindIII library and 4.6% (282/6144 inserts) of clones from the EcoRI library hybridized to this probe. When the Sau3A9 element was used as probe, 3.6% (222/6144 inserts) and 3.1% (192/6144 inserts) of the clones in the HindIII and EcoRI libraries hybridized, respectively. The majority of the clones recognized by the Sau3A9 probe also hybridized to the Sau3A10 probe.
Telomeres are DNA-protein complexes found at the termini of eukaryotic chromosomes. The DNA portion of the telomere is comprised of a heptameric sequence (CCCTAAA) arranged in tandem repeats many kilobases in length (Richards and Ausubel 1988; Burr et al. 1992). To determine the percent of BAC clones in the sorghum libraries containing subtelomeric repeats, a sequence from a subtelomeric repeat was obtained by amplification of sorghum DNA with the telomere-specific primer (CCCTAAA)7 followed by flanking PCR (Siebert et al. 1995). This probe hybridized to 2.8% of the clones in the BAC library (171/144 clones). Additional experiments will be required to determine if the clones identified by this probe are derived from telomeres because these telomeric repeats are also found in the centromeric regions of some plant species (Richards et al. 1991; Presting et al. 1996).
Ribosomal RNA genes (rDNA) in sorghum are organized as direct tandem repeats of several thousand rDNA monomer units encoding the 5.8S, 17S, and 26S rRNAs (Springer et al. 1989). Clones containing ribosomal gene sequences comprised 2.3% of the EcoRI library (107/4608 inserts) but less than 0.1% (2/3072 inserts) of the HindIII library. The discrepancy in the percentage of clones containing rDNA between the two libraries is likely due to the presumed absence of HindIII sites in the rDNA monomer and thus the inability to clone inserts containing the rDNA tandem repeats with HindIII. The absence of HindIII sites within rDNA regions is also reflected in the fingerprinting data. BAC clones from the EcoRI library that contained rDNA repeats did not produce a fingerprint pattern following digestion with HindIII and HaeIII and subsequent labeling of the HindIII termini (data not shown). All of the BAC clones identified with these three classes of DNA repetitive elements were noted in the database and the information used during contig assembly.
DNA Fingerprint Analysis and Contig Assembly
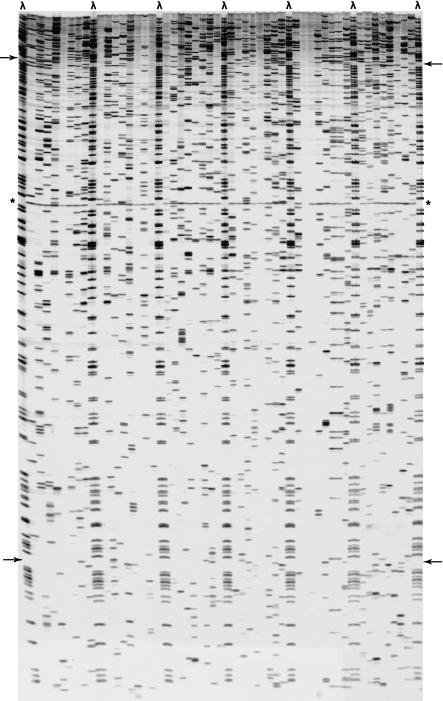
DNA fingerprints of the approximately 26,000 BAC clones present in the two libraries described above were collected as the first step in contig assembly. For DNA fingerprint analysis, the strategy developed at the Sanger Centre (Sulston et al. 1988, 1989; Soderlund et al. 1997, 1998) and modified by Tao et al. (1995) was used. An autoradiogram of a representative 33P-labeled DNA fingerprinting gel is shown in Figure 1. We have previously shown that the fingerprinting protocol used here is highly reproducible since fingerprinting the same BAC clone multiple times yielded identical fingerprint patterns (Klein et al. 1998). The overall efficiency for this procedure was ∼86% (imaged fingerprints/BAC clones analyzed). Of the 14% of BAC clones not producing usable fingerprints, ∼2% were due to failed DNA isolations, 3.5% resulted from poor quality fingerprints and the remainder were either clones without inserts (7.5%) or clones containing rDNA inserts (1%). The fingerprint data was analyzed using the software program Image V3.5. An average of 40 DNA fragments was analyzed per BAC clone. Bands at the top and bottom of the gel (area above and below arrows in Fig. 1) were ignored in the band calling process because of band compression and distortion, respectively.
Figure 1.
Autoradiograph of a typical 33P-labeled DNA fingerprint gel. Each lane represents the fingerprint analysis from an individual sorghum BAC clone digested with HindIII and HaeIII followed by labeling at the HindIII terminus. Labeled λ/Sau3A standard was loaded in the first and every ninth lane (λ). The asterisk marks the position of the common cloning vector band appearing in all BAC clone lanes. Arrows at the top and bottom of the image delineate the region of the gel that was used for band calling in Image 3.5.
Following image analysis, contig assembly was performed using the program FPC V4.5. The tolerance and cut-off values for automated contig assembly were determined empirically using the FPC V4 User's Manual and User's Guide for reference (Soderlund 1999). The tolerance (i.e., the maximum distance, measured in tenths of a millimeter, which two bands from two different clones can differ and still be considered the same band), was determined by viewing a set of related clones with similar fingerprints in the FPC fingerprint window and varying the tolerance between three and nine. The effect of the change in tolerance was visualized by highlighting a clone at each tolerance and comparing bands of the selected clone with the same bands in the other clones. From this analysis, a fixed tolerance of seven was chosen for this fingerprinting project. To determine the appropriate cut-off value or “Sulston score” (i.e., the threshold value representing the maximum allowable probability of a chance match between any two clones), the cut-off value was varied and the effect on the chloroplast contig was examined. All of the chloroplast-containing BACs should assemble into one contig at the correct cut-off value. Using this criterion, a cut-off value of 5 × 10−14 was chosen. The results of automated contig assembly at a tolerance of seven and a cut-off of 5 x 10−14 are shown in Table 1. This initial set of core contigs was analyzed for correct clone order by running the consensus bands (CB) algorithm on each individual contig using the calc function at a cut-off value of 10−14. In addition, clones of a contig were viewed in the FPC fingerprint window to help verify order as described in the FPC User's Guide (Soderlund 1999). Contigs that were split into two or more disconnected contigs following the calc routine were disassembled, if necessary, and the nonoverlapping clones moved to new contigs or to contig 0 (singletons).
Table 1.
Automated Contig Assembly of Sorghum bicolor Project
| Contig Sizeb | No. of Contigs | |
|---|---|---|
| 5 × 10−14 | 10−10 | |
| ≥25 | 7 | 30 |
| 10 to 24 | 135 | 191 |
| 3 to 9 | 1938 | 2058 |
| 2 | 1265 | 1087 |
| Singletons | 6701 | 2485 |
| Total of contigs | 3345c | 3366 |
Total clones = 22,233; tolerance = 7; bury ∼0.1, minimum bands = 8; average clone length = 39.7a
Average number of bands in a clone.
Total number of clones in a contig.
Average number of clones/contig = 4; average contig length = 58 bands.
Average number of clones/contig = 5; average contig = 62 bands.
Following verification of the set of core contigs, the cut-off value was raised in fivefold increments from 5 × 10−14 to 10−10 to look for possible merges between existing contigs, to add singletons to existing contigs, and to create new contigs from the group of singletons. During this analysis, the number of singleton clones decreased from 6701 to 2485 and the number of contigs in each size class increased (Table 1). After assembly at a cut-off value of 10−10, there were 3366 contigs each containing an average of 5 BACs. The largest contig resulting from this analysis consisted of 100 BACs and spanned an estimated 1.84 Mbp (data not shown).
Following contig assembly and verification at an initial tolerance of seven and cut-off value of 5 × 10−14, the overlap between clones within a contig was greater than 80%. At a cut-off valueof 10−10, the overlap between clones added to existing contigs was still greater than 60%. However, at this stringency, some contigs could be merged with more than one other contig or singleton indicating that some incorrect merges would be made at this cutoff. Moreover, if the cut-off value was increased (up to 10−6) in an attempt to build even larger contigs, an increasing number of branch points and thus false merges were observed. The contig branch points were not due to clones containing rDNA, centromeric, or subtelomeric sequences but could be caused by other families of repeats found in the sorghum genome (i.e., transposable elements). In any case, the construction of larger contigs could not be accomplished with great accuracy using DNA fingerprinting alone. Furthermore, fingerprinting provided no information on the localization of BAC contigs on the sorghum genetic map; therefore, additional information such as genetic marker-content mapping was required.
BAC DNA Pooling Strategy and Genetic Marker-Content Mapping
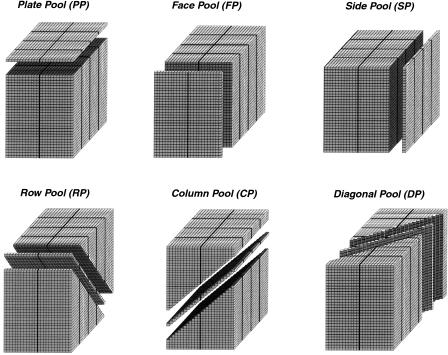
To permit efficient screening of BAC clones for PCR-based genetic markers, a pooling strategy was designed to allow 4–5× genome equivalents of DNA to be screened for the presence of one or more clones containing the same PCR product. The pooling approach described here is based on theoretical considerations (Barillot et al. 1991; Bruno et al. 1995), empirical testing, and the requirements of practical implementation. The BAC libraries were pooled according to the scheme shown in Figure 2. The strategy involved arranging 256 microtiter plates containing 24,576 BAC clones into a three-dimensional stack consisting of 32 layers or plates by 24 columns by 32 rows. The stack was pooled in six distinct ways to generate 184 unique pools (see Fig. 2 and Methods). Since the BAC libraries were constructed randomly and deliberately oversampled the genome (∼4× redundancy), a simple pooling strategy using only three pool types would be inadequate to unambiguously identify an individual BAC responsible for a PCR signal. Therefore, our pooling strategy utilized three additional pool types to provide redundancy and to help identify BACs containing unique DNA sequences.
Figure 2.
Sixfold BAC DNA pooling strategy. Two hundred-fifty six 96-well microtiter plates containing 24,576 individual BAC clones were arranged in a three-dimensional stack consisting of 32 layers (or plates) × 24 columns × 32 rows. The stack was pooled on the six unique coordinate axes as shown to generate a total of 184 DNA pools.
Although the stack dimensions are somewhat constrained by the use of 96-well microtiter plates organized in 8 row x 12 column arrays, a number of different stack configurations for a library of the size used here were possible. To determine the stack geometry necessary for efficient screening of our libraries, computer simulations were performed using a constant genome size of 760 Mbp and an average BAC insert size of 157 kbp, but varying the stack geometry. From these simulations it was determined that in a stack with dimensions of 32 × 24 × 32, ∼87% of PCR signals could be correctly associated with their corresponding BAC clones. Furthermore, the simulations predicted that ∼72.5% of the markers would identify between 2–6 BACs, and over 90% of the time the coordinates of these BACs could be assigned reliably.
Testing of BAC DNA Pools
The quality of the BAC DNA pools, and their utility for identifying BACs containing PCR-based markers, was tested using primers that amplify sorghum SSRs and STSs. Primers for 36 STSs and 48 SSRs, spaced across the sorghum genome, were used for PCR analysis of the BAC DNA pools. The pools shown in Figure 3A consisted of either 768 (PP) or 1024 (SP) unique clones and, in most cases, a single BAC clone accounted for the PCR signal associated with the positive pools. On average, 2.6 BAC clones were identified with each STS and SSR marker analyzed. To confirm the accuracy of the data obtained from the BAC pools, all clones identified as positive were individually tested for the presence of the marker (data not shown). This analysis revealed a false-positive rate of ∼3–5%. Of the 94 BAC clones identified as positive for an STS marker, 3 did not contain the given marker, whereas 5 out of 106 clones identified as positive for an SSR marker did not contain the marker. In addition, several BAC clones which were marked in the data output file as potential positives did, in fact, contain an STS (8 clones) or SSR (14 clones) marker. These false-negative clones were occluded in the stack (i.e., shared an x, y, or z coordinate with at least one other candidate BAC clone) and were observed most notably when the marker was present in eight or more pools of a given pool type (data not shown).
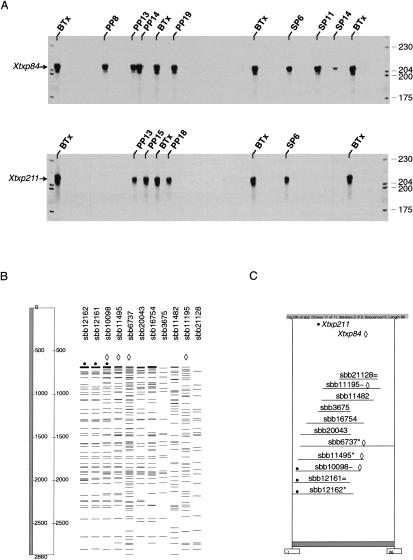
Figure 3.
PCR-based screening of BAC DNA pools for SSR markers. (A) BAC DNA plate pools (PP) and side pools (SP) were analyzed for the presence of two SSRs, Xtxp84 and Xtxp211. BAC DNA pools positive for a given SSR are identified above the respective lane. Plant genomic DNA (BTx623) was used as PCR template in control lanes labeled BTx. Fluorescent-labeled molecular weight standards (LI-COR) were loaded in the first and last lanes and their sizes (bp) are indicated to the right of the gels. (B) FPC V4.5 fingerprint analysis window displaying the representative fingerprint patterns of individual BAC clones from ctg190. Four BAC clones in ctg190 were identified as positive for Xtxp84, and three BACs were identified as positive for Xtxp211 (·) following analysis of the BAC DNA pools from A. (C) FPC V4.5 contig window displaying a horizontal representation of ctg190 that contains BAC clones positive for Xtxp84 and Xtxp211. The characters (*, = and ∼) to the right of individual clone names represent canonical, equal, and approximate clones, respectively (Soderlund et al. 1998). The shaded bar below the contig display indicates the length of the contig, measured in number of bands.
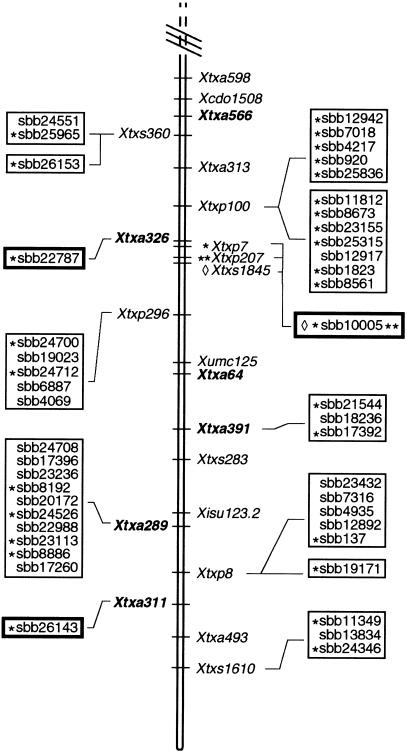
When the PCR results shown in Figure 3A were analyzed, four BAC clones were positive for SSR, Xtxp84, and three clones were positive for SSR, Xtxp211. These results were in complete agreement with those obtained by fingerprint analysis (Fig. 3, B and C). The four clones positive for Xtxp84 had very similar DNA fingerprints, as did the three clones containing Xtxp211 (Fig. 3B). In fact, one clone (sbb10098) contained both Xtxp84 and Xtxp211, and all six BAC clones exhibited a significant degree of overlap and were placed into the same BAC contig following automated contig assembly (Fig. 3C). These results are supported by the genetic mapping results obtained with the recombinant inbred line (RIL) population where both Xtxp84 and Xtxp211 map in close proximity to one another on sorghum linkage group (LG) B (G. Hart, pers. comm. and Fig. 6).
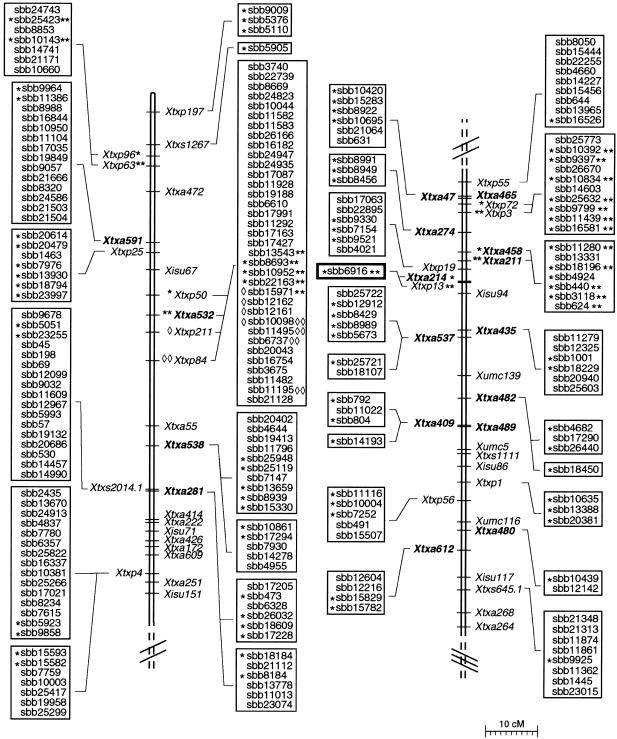
Figure 6.
Integrated genetic and physical map of LG B from S. bicolor BTx623 x IS3620C recombinant inbred population. LG B is shown in three parts from the upper end to the bottom end. All SSR and RFLP markers shown were previously mapped to LG B (Peng et al. 1999; Kong et al. 2000; G. Hart, pers. comm.). AFLP markers (labeled Xtxa along LG B) were mapped in the present study. BACs were linked to SSRs (Xtxp markers), STSs (Xtxs markers) or AFLPs (bold-type Xtxa markers) by PCR-based screening of the BAC DNA pools. The individual BAC clones positive for each genetic marker are indicated by symbols next to the clone name. BAC clones that were placed into the same contig by DNA fingerprint analysis are grouped in boxes. Genetic markers linked to two different contigs or one contig and a BAC singleton clone indicate those BAC clones positive for a common marker but not placed into the same contig by DNA fingerprint analysis.
Although the results obtained from the PCR-based screening of the BAC pools with Xtxp84 and Xtxp211 and the corresponding fingerprint data were in complete agreement, this was not the case with all 84 markers analyzed (data not shown). For example, there were cases where two or more clones were identified with the same marker but fingerprinting failed to place the clones in one contig. Because all of the clones identified as positive for an STS or SSR were individually confirmed, the clones, although related, may overlap to only a small extent. At the high stringency used for fingerprint-based contig assembly (i.e., cut-off value of 10−10), clones with minimal overlap will not be placed in the same contig. Alternatively, if the region of the sorghum genome containing the STS or SSR marker has undergone duplication, then the positive BAC clones may not be part of the same contig. In these cases, additional information will be required for accurate BAC ordering.
High-throughput PCR-based Contig Assembly
To construct a saturated STS-based map of the sorghum genome (i.e., a marker every ∼0.3 Mbp) would require approximately 2500 STS markers. However, the cost of obtaining such a large number of STS markers is currently too high to consider for sorghum and many other plant species. What is required is a low cost, high-throughput PCR-based method that identifies overlapping BAC clones and links them to the sorghum genetic map. AFLP mapping utilizes the simultaneous amplification and screening of sets of 25–100 genomic DNA fragments (Vos et al. 1995). A large number of the DNA fragments amplified by this procedure are unique (based on two restriction sites, the number of specific selective bases added to each primer, and the specific size of the amplified DNA fragment). The DNAs amplified during AFLP analysis are, therefore, ideal for identifying BAC clones that contain the same unique PCR-amplifiable DNA sequence. For clarity, we call these unique amplified DNA fragments, SAS-DNAs (simultaneously amplified singleton DNAs). A subset of the SAS-DNAs will be polymorphic in different genetic backgrounds and can, therefore, be mapped as AFLPs on the sorghum genetic map. These AFLP/SAS-DNAs can provide numerous links between the sorghum genetic and physical BAC-based genome maps.
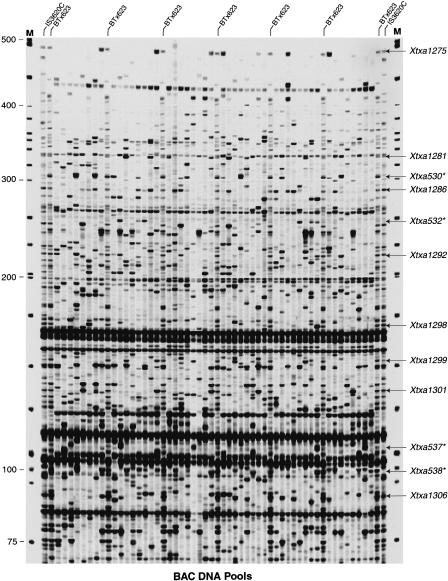
To obtain a system for high-throughput contig assembly and mapping, we combined our sixfold BAC pooling strategy with AFLP technology. On average, between 25–100 different SAS-DNA fragments, ranging in size from 25 to 500 bp, were amplified from sorghum genomic DNA using AFLP technology (Fig. 4, lanes labeled IS3620C and BTx623). Based on computer simulations, as well as results obtained from the STS-content mapping described above, BACs containing a unique SAS-DNA fragment should be identified (resolved) if present in eight or fewer copies in the pooled library. As can be seen in Figure 4, ∼10–40 SAS-DNA bands fitting this description were amplified from the BAC DNA pools with 1 primer pair (Xtxa markers) and hence, the addresses of individual BACs containing these SAS-DNA markers could be determined. In addition to the unique SAS-DNAs, some DNA fragments were amplified in all BAC DNA pools but not in sorghum genomic DNA (Fig. 4). These DNAs are likely derived from the vector or bacterial genomic DNA that is present in the BAC DNA pools. Other DNA bands were amplified in nearly all of the BAC DNA pools as well as from sorghum genomic DNA. These products most likely originate from repetitive sequences scattered throughout the sorghum genome.
Figure 4.
PCR-based screening of BAC DNA pools for SAS-DNA markers using AFLP technology. AFLP templates were prepared from BAC DNA PPs (1–32) and SPs (1–16) and selectively amplified with fluorescent-labeled EcoRI + TGA and MseI + CGG. Labeled products were analyzed on a LI-COR DNA sequencer. AFLP template from genomic DNAs (IS3620C and BTx623) were run as controls and are indicated above the respective lanes. Arrows to the right of the gel show selected SAS DNA markers that were analyzed in the DNA pools. Asterisks to the right of a subset of the markers indicate those SAS DNAs that revealed polymorphisms between BTx623 and IS3620C and could be mapped as AFLPs on the sorghum genetic map. Fluorescent-labeled molecular weight markers (LI-COR) were run in lanes marked M and their sizes (bp) are shown to the left of the gel.
In the first cycle of analysis, the BAC pools were screened with 32 different AFLP primer combinations. A total of 891 SAS-DNA markers were identified or an average of 28 markers per primer pair. This number of DNA bands per gel could easily be resolved and analyzed (i.e., Fig. 4). Each SAS-DNA marker identified an average of 2.7 BACs; similar to results obtained with STS and SSR markers. Therefore, SAS-DNA markers identify overlapping BACs in a manner similar to other single copy markers. Overall, the first set of 32 primer pairs identified approximately 2400 BACs and organized a subset of these BACs into ∼700 contigs.
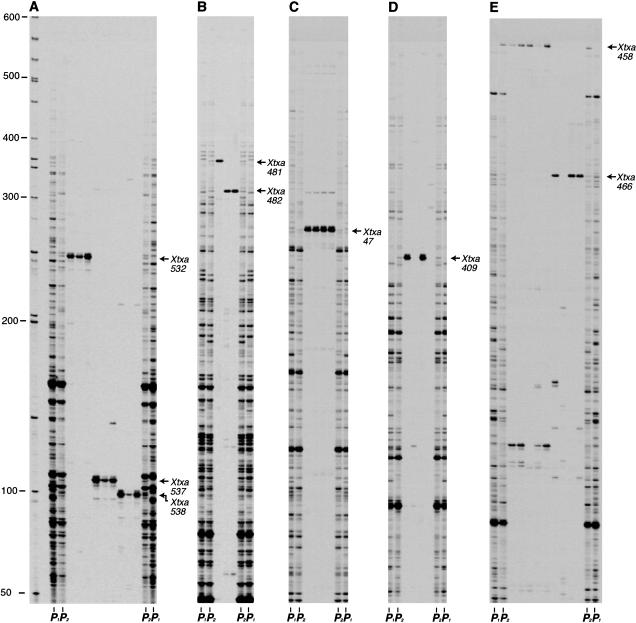
Detailed analysis of 101 BACs identified by 32 SAS-DNA markers was carried out to further characterize this method (Fig. 5). Of the 32 SAS-DNA markers examined, 23 markers identified two or more BACs with predicted overlaps that could be confirmed with fingerprint analysis and by analysis of the individual clones (Fig. 5). In total, the overlaps among 65 of the 101 individual BACs identified by SAS-DNA analysis were confirmed by fingerprinting, providing a high degree of confidence in these contigs. In addition, seven of these 23 SAS-DNA markers also identified a BAC clone from another contig or from the pool of singletons that could not be confirmed by fingerprint analysis. There were also five SAS-DNA markers that identified one BAC from each of two contigs created by fingerprint analysis, or one BAC in a contig and one singleton BAC. These 12 SAS-DNA markers that identified BAC clones in more than one contig were valuable since they identified putative links between contigs and/or singleton BAC clones that were not predicted by fingerprint analysis. Finally, four of the 32 SAS-DNA markers identified only one BAC clone that was located in the pool of singletons. Analysis of the individual clones for the presence of SAS-DNAs revealed an overall false-positive rate of 15%, with the false-positive clones most prevalent when the marker was present in eight or more DNA pools. Therefore, to eliminate false-positives, the incorporation of individual BACs into contigs as well as the merging of contigs containing a common SAS-DNA marker was only allowed once order was supported by at least two independent analyses (i.e., fingerprinting and SAS-DNA analysis).
Figure 5.
Analysis of individual BAC clones for SAS-DNA/AFLP markers. BAC clones identified as positive for SAS-DNA/AFLP markers following analysis of the BAC DNA pools were individually tested for the presence of the marker. AFLP templates were prepared from individual BAC clones as well as from genomic DNA (IS3620C, lanes marked P1; BTx623, lanes marked P2). AFLP templates were selectively amplified with fluorescent-labeled EcoRI + TGA/MseI + CGG (A); EcoRI + TGA/MseI + CTG (B); EcoRI + CAA/MseI + CAA (C); EcoRI + CAA/MseI + CCT (D); and EcoRI + CAA/MseI + CGT (E). Fluorescent-labeled products were run on a LI-COR DNA sequencer. Numbers to the left of panel A indicate the sizes (bp) of molecular weight standards. SAS-DNA/AFLP markers identified in the individual BAC clones and BTx623 genomic DNA are indicated to the right of each panel.
AFLP Linkage Analysis
AFLPs have been used effectively as a high-throughput genetic marker system (Alonso-Blanco et al. 1998; Qi et al. 1998; Boivin et al. 1999; Vuylsteke et al. 1999; Young et al. 1999). Because the SAS-DNA markers used for contig assembly are based on AFLP technology (Vos et al. 1995), any SAS-DNA marker that corresponds to a polymorphic AFLP genetic marker will provide a direct link between the sorghum genetic and physical maps. Analysis of a sorghum RIL population (BTx623 x IS3620C) (Peng et al. 1999) with the 32 unique AFLP primer combinations used to identify SAS-DNAs identified 532 AFLP genetic markers (data not shown). Of these, 258 (48.5%) were amplified from BTx623 DNA corresponding to an average of 25–26 markers for each of the 10 sorghum LGs that comprise the genetic map. Of the AFLP markers, 104 amplified from BTx623 DNA were integrated at LOD <3 into a framework genetic map of the RI population (provided by G. Hart, TAMU, College Station, TX) along with 70 AFLPs amplified from IS3620C DNA. In addition, another 114 markers amplified from BTx623 were placed onto this framework map at a LOD >3 to aid in the generation of a saturated sorghum genetic map. These 218 genetic markers amplified from BTx623 were utilized for integration of the sorghum genetic and physical maps.
When the AFLP genetic markers amplified from BTx623 DNA were cross-referenced to the physical map, >98% of the markers had a corresponding signal in the BAC DNA pools. Of these, ∼73% corresponded to SAS-DNA markers that had been previously resolved to identify BACs (data not shown). The remaining ∼25% of the SAS-DNA markers were not useful as links between the genetic and physical maps due to bacterial or vector contamination in the region of the marker, missing data points in at least one of the six pool types, or overrepresentation of the marker in the BAC pools.
A representative example of the results obtained using this methodology is displayed in Figure 6. A total of 23 AFLPs amplified from BTx623 DNA (bold-type Xtxa markers along LG B) and 13 AFLPs amplified from IS3620C DNA (plain-type Xtxa markers) were mapped to LG B. Of the 23 AFLPs amplified from BTx623, 19 corresponded to SAS-DNAs that identified BAC contigs or BAC singletons, thereby creating physical links at these genetic loci. Whereas a majority of the markers either identified BAC clones within a single contig (or one clone within the group of singletons), there were cases in which an AFLP marker was linked to two different contigs or to one contig and one singleton (Xtxa538, Xtxa281, Xtxa537, Xtxa482 and Xtxa409). The BAC clones identified by these markers did not exhibit enough overlap to be considered contiguous at the cut-off values used for fingerprint contig assembly, and therefore, must be confirmed using another independent approach. Finally, a subset of LG B STSs and SSRs (labeled Xtxs and Xtxp markers, respectively) that were assigned to BAC contigs and located on the sorghum genome map are shown in Figure 6.
DISCUSSION
The generation of integrated genetic and physical maps is a central effort of eukaryote genome research. This can be a difficult task in complex genomes, however, because of genome size and repetitive DNA. In addition, the polyploid nature of many plant genomes makes physical map construction in these genomes an even more daunting task. In this article, we describe a novel approach for physical map construction of complex genomes that combines a sixfold BAC DNA pooling strategy with AFLP technology. The methodology allowed the identification of overlapping BAC clones and simultaneously established links between BAC contigs and the genetic map. Furthermore, this approach utilized selective AFLP primers for amplification rather than sequence-specific STS primers, thus eliminating the need to obtain DNA sequence information and thereby lowering the cost of map construction. Using this methodology, in conjunction with classical DNA fingerprinting, we have begun construction of an integrated genetic and physical map of the sorghum genome.
Two sorghum BAC libraries containing ∼26,000 clones were used for physical map construction (Woo et al. 1994; Tao and Zhang 1998). The average insert size in the two libraries was estimated at 148.5 kbp. After removal of clones containing organellar DNA (∼1840 clones) and clones without inserts (∼1950 clones), the remaining 22,233 clones provide ∼4× coverage of the sorghum genome. Analysis of the BAC libraries for the presence of over 300 SSR, STS, and AFLP markers indicated that the combined libraries provide coverage of ∼98% of the genome. This is in agreement with the 98% frequency predicted by a Poisson distribution for recovery of any marker from a 4× library.
All 26,000 BAC clones were subjected to standard DNA fingerprinting using 33P-labeling and polyacrylamide sequencing gels as a first step in contig assembly (Sulston et al. 1988, 1989; Tao et al. 1995; Soderlund et al. 1997, 1998). An initial set of core contigs containing BAC clones exhibiting a significant degree of overlap was assembled by the software program FPC V4.5 (Table 1). Contigs were then merged and new contigs created from the group of singletons by successively raising the cut-off value (up to 10–10) followed by manual interaction with the program. Analysis of the fingerprinted clones using FPC allowed us to assemble a large number of BAC contigs at reasonable confidence with overlaps of at least 60%. After this analysis, the FPC database contained 2485 singletons and 3366 contigs (Table 1). On average, each contig contains 5 BAC clones with an average length of ∼62 bands. Assuming that each band is derived from a span of 3740 bp (148.5 kbp insert size ÷ 39.7 bands per BAC) then the average contig is ∼232 kbp and the longest contig assembled to date is 1.84 Mbp (data not shown).
Although FPC analysis of the fingerprint data provided a baseline for BAC ordering and contig assembly, the fingerprint analysis described here is limited in several ways. For example, at 4× genome coverage, it was difficult to assemble large contigs without including bridging clones that had minimal overlap. Ideally, genome coverage for fingerprint analysis should be ∼7–8×. Therefore, we have recently constructed a third sorghum BAC library using BamHI to increase the coverage to ∼8×. In addition, when analyzing the data at successively higher cut-off values to examine possible contig merges, the algorithm often identified multipoint branches during the process. In these cases, it was impossible to determine the correct two contigs to merge without additional information. The presence of repetitive elements in rDNA, centromeric or subtelomeric sequences did not cause branch points in our study, and others have also demonstrated that repetitive sequences do not normally cause false overlaps during FPC analysis (Tao and Zhang 1998; Zhu et al. 1999). Therefore, the observation of contig branches may be related to genome complexity.
The limitations inherent in fingerprinting complex genomes for the construction of physical maps led us to utilize PCR-based methods for assembling and mapping BAC contigs. Our goal was to obtain at least 2500 links between the sorghum genetic and physical maps, or an average of one link every 300 kbp. One approach to accomplish this was to identify BAC clones containing RFLP and SSR markers. However, our current RFLP/SSR-based genetic map does not contain the required number of markers to accomplish this task and the cost and time needed to generate such a large number of STSs and/or SSRs is prohibitive for this project. Therefore, we developed a method based on AFLP technology that would allow overlapping BAC clones to be identified while simultaneously generating markers that link the genetic and physical maps.
A DNA pooling strategy was developed that allows 4–5× genome equivalents of DNA to be screened efficiently for the presence of multiple clones containing the same PCR product. The DNA pooling strategy was also designed for use with multiplexed PCR assays that would allow parallel identification of numerous BAC contigs; each containing a different PCR-amplified marker. The pooling strategy implemented here consisted of constructing a three-dimensional stack containing 24,576 individual BAC clones and then pooling the BACs on six unique coordinate axes of the stack (Fig. 2). This resulted in a total of 184 pools each containing DNA from either 768 or 1024 individual BAC clones, which is well under the maximum number of clones per pool that can be screened using a PCR-based approach (Kim et al. 1996). The pooling approach allowed the identification of BAC clones harboring STS, SSR, or AFLP markers by screening the 184 DNA pools in a single step. Other strategies utilizing superpools and subpools in PCR-based screening approaches have been developed and used successfully to identify individual positive clones (Green and Olsen 1990; Asakawa et al. 1997). However, the use of superpools followed by subpools was not compatible with our need to screen a redundant library representing a large genome simultaneously for numerous AFLP markers. Therefore, the strategy utilized in the present study, in which clones are pooled on six coordinate axes to generate a fixed set of DNA pools, permits the parallel screening of redundant libraries with multiple markers and subsequent identification of individual clones harboring these markers using a minimal number of PCR assays.
The results from screening the DNA pools for STSs and SSRs indicated that the pooling approach designed here provided a rapid and efficient means of identifying overlapping BAC clones containing a common genetic marker. Analysis of the pools for 48 SSRs and 36 STSs resulted in an average of 2.6 BAC clones identified for each PCR marker analyzed with a false-positive rate of 3% to 5%. The average number of positive BACs per marker was less than what was expected for a library containing ∼4× genome equivalents. Some of this apparent discrepancy could be due to the lack of a signal in one or more pool types since our pooling strategy requires that a PCR signal be detected in all six unique pool types to be considered a true positive. However, these potentially false-negative clones can be marked and individually confirmed for the presence of the marker. False-negatives also arise if a clone is occluded within the stack. Upon screening individual BACs, we identified 22 out of 222 occluded clones (10%) that were, in fact, positive for the genetic markers analyzed. In general, we found that clones represented more than eight times in the pools are nearly always occluded, and require further analysis to confirm BAC relatedness. If necessary, contigs of BACs containing these markers can be identified using pools containing fewer BAC clones (i.e., subpools).
The sixfold BAC DNA pooling strategy facilitated the identification of overlapping BACs containing SSRs and STSs. However, with an initial goal of obtaining at least 2500 links between the sorghum genetic and physical maps, it was clear that standard STS-content mapping would not be a viable approach for our mapping project. Therefore, we combined our DNA pooling strategy with AFLP technology in order to achieve the necessary throughput within our budgetary constraints. The use of AFLP technology for identifying overlapping BAC clones containing a common marker has several advantages over STS-content mapping. First, the method is rapid because multiple markers can be mapped simultaneously. In this study, an average of 28 SAS-DNAs was amplified with each primer pair utilized, and all of the fragments were analyzed simultaneously in a single gel. The distribution of these 28 SAS-DNA fragments in the pools of BAC DNA was used to identify up to 28 small BAC contigs each containing a different SAS-DNA marker. As with STS and SSR content mapping, each SAS-DNA identified an average of ∼2.7 BAC clones. In the first cycle of analysis, 32 different AFLP primer combinations identified 891 unique SAS-DNA markers and organized ∼2400 BACs into ∼700 small contigs.
A second advantage of the present method is its efficiency and low cost. The selective primers used for AFLP amplification do not require information about DNA sequence; therefore, the cost for primer generation is low when compared to sequence-specific STS and SSR markers. In addition, a large number of selective primers can be utilized for AFLP amplification. In our case, three selective nucleotides were added to each primer giving us the ability to use 64 different EcoRI- and MseI-selective primers in 4096 different pairwise combinations. This high-throughput mapping approach was also facilitated by the use of a dual-dye LI-COR DNA sequencing system (LI-COR Inc., Lincoln, NE). Use of this system for data collection proved remarkably sensitive and cost-efficient. With this format, one fluorescent infrared dye (IRD)-labeled EcoRI primer ($295 for >20 nmoles) was used in combination with 16 different MseI primers to generate approximately 350 contigs. Because of the high sensitivity of the system, we have observed that ∼20 nmoles of IRD-labeled primer is sufficient for as many as 100,000 selective amplification reactions. This is more than enough reactions to screen the BAC DNA pools as well as the RIL mapping population with one labeled EcoRI primer and all 64 possible MseI primers. Moreover, when using a double LI-COR system, a total of 256 lanes of data (64 lanes/gel × 2 dyes × 2 gels) can be collected in a 4 h period. At this level of throughput, 2560 lanes of data corresponding to the screening of the BAC DNA pools with 10 different primer combinations were collected per week on the instruments.
A third advantage of the current approach is that it can be used to generate links between the BAC-based physical map and the sorghum genetic map. Since some of the SAS-DNA markers reveal polymorphisms in nearly any mapping population, they can be located on genetic maps as AFLP markers (Vos et al. 1995). In our study, ∼30% of the SAS-DNAs could be scored as AFLPs in the RIL mapping population derived from a cross between BTx623 and IS3620C. In the first cycle of analysis (i.e., 32 primer combinations), ∼190 SAS-DNA/AFLP links were established between the physical and genetic maps. The AFLPs were mapped onto the existing sorghum RFLP/SSR-based genetic map (Peng et al. 1999, Kong et al. 2000) without significant map distortion (at LOD > 3, map size increased ∼12%). The AFLP markers were distributed across each linkage group, with some clustering of markers in the central regions of each LG (Fig. 6). Clustering of AFLP markers has been seen in other genetic maps (Alonso-Blanco et al. 1998; Qi et al. 1998; Boivin et al. 1999; Vuylsteke et al. 1999; Young et al. 1999) and it is assumed that these clusters correspond to regions of the genome, perhaps around centromeres, which have relatively low amounts of recombination (Alonso-Blanco et al. 1998; Vuylsteke et al. 1999). If increased genome coverage is needed, PstI /MseI can be used to generate a different set of SAS-DNA and AFLP markers (Vuylsteke et al. 1999; Young et al. 1999). In contrast to EcoRI/MseI markers, PstI/MseI-generated AFLPs do not appear to cluster around centromeric regions due to the sensitivity of PstI to cytosine methylation (Vuylsteke et al. 1999; Young et al. 1999). In any case, the AFLP markers that are linked to the BAC contigs via SAS-DNA analysis provide a large number of connections between the sorghum genetic and physical maps.
Contigs organized using only fingerprint data or solely using PCR-based screening of BAC pools resulted in a low but significant error rate. To reduce this source of error, BACs were incorporated into the sorghum physical map only when their order or location was verified by two different analyses. Fortunately, ∼65% of the SAS-DNA markers identified two or more BACs whose predicted overlaps could be confirmed with fingerprint analysis (cut-off value = 10–10). Approximately 25% of these contigs were identified with SAS-DNA markers that could also be mapped as AFLPs and, therefore, directly placed on the integrated sorghum genetic and physical map. Another ∼12% of the SAS-DNAs identified singleton BACs, some of which also could be located on the genetic map as AFLPs (e.g., Fig. 6., sbb22787 with Xtxa326). However, singleton BACs were only incorporated into the map after the SAS-DNA was confirmed to be present in the isolated BAC. In some cases, several different but adjacent genetic markers identified the same singleton BAC (e.g., Fig. 6, sbb10005 with Xtxp7, Xtxp207 and Xtxs1845; sbb6916 with Xtxa214 and Xtxp13). This type of data was considered sufficient to localize these BACs on the sorghum genome map without further analysis.
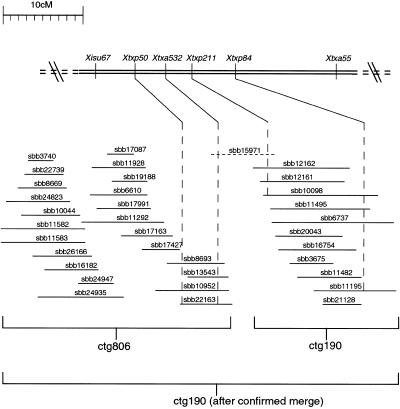
Approximately 30% of the SAS-DNA markers identified BACs that were located in two different contigs created by fingerprinting, or BACs located in a contig as well as in the pool of singleton BACs. These SAS-DNA markers are particularly valuable because they predict links between contigs or between contigs and singletons. The BAC clones predicted to contain these SAS-DNAs can be marked in the database for follow-up verification. This is important because fingerprint analysis could not reliably merge BAC contigs unless fingerprints overlapped by at least 60%. In contrast, SAS-DNA markers can identify related BACs that contain the same amplified DNA fragment yet have minimal overlap. An example of the utility of the combined analysis used in the current project is shown in Figure 7. SAS-DNA marker, Xtxa532, identified 4 BACs in ctg806 as well as the BAC singleton, sbb15971. The SSR marker, Xtxp211, also identified BAC singleton, sbb15971, as well as three BAC clones in ctg190. This information was sufficient to merge contigs 806 and 190 using the singleton clone, sbb15971, as a bridge. This placement was also consistent with the order of the four genetic markers in this region of the map (Xtxp50, Xtxa532, Xtxp211, and Xtxp84). Although this type of ordering is relatively infrequent at this stage of data collection, we expect similar reinforcing information to be available for large parts of the map once 10,000 SAS-DNA markers have been analyzed.
Figure 7.
Integrated genetic and physical map of the region of sorghum LG B containing markers Xtxp50, Xtxa532, Xtxp211, and Xtxp84. The three SSR markers, Xtxp50, Xtxp211, and Xtxp84, were previously mapped to this region of LG B (G. Hart, pers. comm.), whereas the AFLP marker, Xtxa532, was mapped between Xtxp50 and Xtxp211 in the present study. BACs linked to these four genetic markers by PCR-based screening of the BAC DNA pools are shown below the genetic map with the dashed lines extending from each marker through the respective, positive BAC clones. Four BAC clones in ctg806 were positive for Xtxa532 as well as the singleton BAC clone, sbb15971. Three BAC clones in ctg190 were positive for Xtxp211 as well as the singleton clone, sbb15971. Therefore, sbb15971 was used as a bridging clone to merge ctg190 and ctg806.
The first cycle of SAS-DNA analysis using 32 primer combinations identified ∼700 BAC contigs each containing between one and three unique markers. Our goal for the sorghum genome mapping project is to collect data on approximately 10,000 SAS-DNA markers providing one set of SAS-DNA linked BACs every 75 kbp on average. This depth of coverage would be similar to that provided by 41,000 STS markers in the ∼3 billion base pair human genome (Hudson et al. 1995; Deloukas et al. 1998). The ordering of ∼10,000 BAC contigs with SAS-DNAs will require 12–14 cycles of analysis (one cycle equals 32 primer combinations) on the LI-COR DNA sequencing system. The ability to collect data from one cycle in a 3- to 4-week period should allow ∼8,000 to 10,000 BAC contigs to be ordered in a 12- to 14-month period; and ∼2500 of these BAC contigs will also be linked to the sorghum genetic map.
Although this article has focused on the utility of AFLP technology and SAS-DNA markers for generating integrated genetic and physical maps, our objective is to create a map that will facilitate map-based gene isolation. Our research team, in collaboration with several other groups, is mapping genes that regulate flowering time, fertility restoration, disease resistance, and genes involved in plant response to environmental stress. In these projects, AFLP technology, in conjunction with bulked segregant analysis (Michelmore et al. 1991), is being used to identify regions of the sorghum genome encoding genes of interest. Even at this early stage of map development, a significant number AFLP markers found linked to a new locus have already been mapped, and in many cases a contig of linked BACs has been identified. This situation greatly accelerates the search for candidate genes as well as providing sequence-ready BACs for follow up genome sequencing.
METHODS
Plant Materials
A Sorghum bicolor population of 137 F6–8 recombinant inbred lines (RILs) obtained from a cross of BTx623 and IS3620C was used as the mapping population for construction of an AFLP linkage map. This RIL mapping population was previously used to establish an RFLP linkage map for sorghum containing over 300 RFLPs (Peng et al. 1999), and has recently been expanded to include more than 100 SSR loci (Kong et al. 2000; G. Hart, pers. comm.).
Genomic DNA Extraction
Total genomic DNA was extracted from 2- to 3-week-old seedlings using the procedure described in Williams and Ronald (1994) with modifications. Briefly, lyophilized leaf tissue (10–20 mg) was cut into small pieces and transferred to a 1.5 ml microcentrifuge tube. Extraction buffer (800 μl) containing 100 mM Tris pH 7.5, 10 mM EDTA pH 7.5, 700 mM NaCl, 12.5 mM potassium ethyl xanthogenate (PEX) was added. Leaf pieces were pressed to the bottom of the tube with a 1-ml pipette tip to aid in the release of nucleic acid from the tissue. Samples were incubated at 65° C for 1 h with occasional mixing. Following incubation, the supernatant was removed to a clean 1.5-ml microcentrifuge tube and centrifuged at 15000 g for 5 min. The supernatant (700 μl) was transferred to a 1.5-ml microcentrifuge tube containing 700 μl isopropanol and 70 μl of 3 M sodium acetate pH 5.2, mixed and incubated at -70° C for 15 min (or longer). The precipitated DNA was centrifuged at 15,000 g for 30 min, washed twice with 70% ethanol, air-dried and resuspended in 100 μl TE buffer. To aid in DNA resuspension, samples were incubated at 65° C for 10–15 min, centrifuged at 15,000 g for 5 min to remove insoluble material, and the supernatant transferred to a clean tube. The genomic DNA was quantified using a DYNA Quant 200 fluorimeter (Hoefer Pharmacia Biotech, San Francisco, CA).
AFLP Linkage Analysis
DNA Template Preparation and AFLP Reactions
Amplified fragment length polymorphisms (AFLPs) were generated using the protocol of Vos et al. (1995) with modifications. Genomic DNA of the parental lines and RI lines (500 ng) comprising the mapping population were completely and simultaneously restricted with EcoRI and MseI as described (Vos et al. 1995), except the incubation time was increased to 2.5 h. Restricted genomic DNA fragments were ligated to EcoRI and MseI adapters overnight at 37° C and the reaction mixture diluted to 500 μl with TE buffer and stored at −20° C.
Preamplification of the dilute template DNA was performed with AFLP primers having no (EcoRI + 0; GTAGACTGCGTACCAATTC) or one (MseI + 1; GATGAGTCCTGAGTAA-C) selective nucleotide. Twenty μl PCR reactions were performed containing 5 μl dilute template DNA, 30 ng each EcoRI + 0 and MseI + 1 primers, 0.4 U Taq polymerase (Promega Corp., Madison, WI), 1× Taq buffer (10 mM Tris-HCl pH 9.0, 0.1% triton X-100, 50 mM KCl), 2.5 mM MgCl2, and 200 μM dNTPs. Preamplification reactions were performed for 20 cycles of 30 sec at 94° C, 1 min at 56° C and 1 min at 72° C. Following preamplification, the reactions were diluted 10-fold with TE buffer and used as template for selective amplification. Selective amplification reactions were performed using primers with three selective nucleotides (EcoRI + CAA, EcoRI + TGA, all 16 possible primers of MseI + CNN) resulting in a total of 32 +3/+3 unique primer combinations. IRD-labeled EcoRI primers obtained from LI-COR Inc. (Lincoln, NE) were diluted to 1 μM according to the manufacturer's recommendation and stored at −20° C in the dark until ready for use. Selective AFLP reactions were performed in a final volume of 10 μl containing 2 μl dilute preamplified template DNA (50 pg), 15 ng MseI selective primer, 0.25–0.4 μl IRD-labeled EcoRI selective primer, 0.2 U Taq polymerase, 1× Taq buffer, 2.5 mM MgCl2, and 200 μM dNTPs. Selective amplification reactions were performed as follows: 1 cycle of 2 min at 94° C followed by 36 cycles of 30 sec at 94° C, 30 sec annealing step (see below), and 1 min at 72° C. The annealing temperature in the first cycle was 65° C and was subsequently reduced 0.7°C for each of the next 12 cycles and was then continued at 56° C for the remaining 23 cycles. Reactions were complete after a final extension of 5 min at 72° C.
Gel Analysis
The AFLP amplification products were analyzed using a LI-COR model 4200L-2 dual-dye automated DNA sequencing system. Following amplification, an equal volume (5 μl) of PCR products labeled using the IRD-700 nm EcoRI primer (EcoRI + CAA) was pooled with the products labeled with the IRD-800 nm EcoRI primer (EcoRI + TGA). Basic fusion dye (2 μl) (LI-COR) was added to each pooled sample and the samples were denatured for 2.5 min at 95° C. Each sample (1 μl) was loaded on a 6.5% polyacrylamide gel containing 7 M urea. Gels were cast using LI-COR 25-cm plates with 0.25-mm-thick spacers and comb. Electrophoresis was performed at a constant power of 40 W and a constant temperature of 47.5° C for 3 h.
Analysis of AFLP Images
The raw data from the LI-COR model 4200 sequencers is presented as an autoradiogram-like image that is stored in TIFF format. Band analysis was performed using Bionumerics software (Applied Maths BVBA, Kortrijk, Belgium). Following assignment of the bands from selected individuals to band classes; a comparative binary (+/−) table was generated which displayed polymorphic bands in all of the samples. The binary table was exported to Microsoft Excel (Microsoft, Tacoma, WA) where it was transformed and used for genetic mapping and other analyses.
Mapping of AFLP Markers
A framework linkage map of the BTx623 × IS3620C RI population, composed of a subset of the RFLPs (Peng et al. 1999) and SSRs (Kong et al. 2000; G. Hart, pers. comm.) was provided by Dr. G. Hart (TAMU, College Station, Texas). AFLP markers were placed on this framework map using the computer program MAPMAKER V2.0 for Macintosh. AFLP markers were initially assigned to one of the ten sorghum LGs using the MAPMAKER place command with a threshold of LOD of 3.0. The markers assigned to a LG were ordered with respect to the framework map using the try command, and the LOD score for each adjacent triplet was determined using the ripple command. An initial map of each LG was established at a LOD >3.0. Subsequently, a LOD <3 map of each LG was created utilizing all AFLP markers present in BTx623 DNA (source DNA for BAC library construction) to create a saturated genetic map to aid in physical map construction. At this point a drop marker test was performed on each LG and the data sets for AFLP markers with a high drop marker value were compared with those of neighboring markers to identify questionable data points. Where necessary, the gel images were rescored and errors corrected. Centimorgan values were calculated using the Kosambi mapping function (Kosambi 1944).
BAC Libraries
A BAC library of the sorghum inbred line, BTx623, was constructed at the Texas A & M University (TAMU) BAC Center (Woo et al. 1994). This library consists of 13,440 clones and was prepared with DNA isolated from protoplasts after partial digestion with HindIII. A second BAC library was constructed at the TAMU BAC Center for use in this study (Tao and Zhang 1998). BTx623 was again used as the source material for this library; however, the DNA was prepared from sorghum nuclei and partially restricted with EcoRI. The EcoRI BAC library contains 12,576 clones.
BAC Library Screening
High-density colony filters were prepared using a Biomek 2000 robotic workstation equipped with a high-density replicating system (HDR) (Beckman Coulter Inc., Fullerton, California). Each filter was inoculated with 1536 BAC clones using a 3 × 3 matrix pattern with a 384-pin HDR tool. Filters were inoculated and processed as described by Woo et al. (1994). Prehybridization was performed at 65° C for 2–3 h in 1 M NaCl, 10% dextran sulfate, 1% SDS and 1× Denhardt's solution. Following prehybridization, the labeled probe was added to a final concentration of 1 × 106 cpm probe/ml hybridization solution and hybridization continued for 14–16 h at 65° C. Filters were washed twice in 2× SSC/0.5% SDS and twice in 0.1× SSC/0.5% SDS. All washes were for 20–30 min each at 65° C. Following washing, the filters were exposed to X-ray film for 1–3 d.
DNA Probes
DNA fragments used to make probes for library screening were synthesized by standard PCR using gene specific primers that were derived from the gene sequences from GenBank. Six DNA fragments corresponding to barley chloroplast sequences were amplified using total genomic DNA isolated from barley. These included rbcL [X00630] nt 1059–2174; psbD [X07522] nt 1–1178; psbA [X07942] nt 991–2066; ndhB [X90650] nt 27–2245; psaC [L06607] nt 58–387; and psbB [X14107] nt 175–1608. Three fragments corresponding to sorghum mitochondrial sequences were amplified from sorghum genomic DNA including coxI [M14453] nt 681–2181; atp9 [U61165] nt 496–1221; and orf25 [U22069] nt 30–1037. A rDNA fragment was amplified from Arabidopsis DNA using gene-specific primers derived from 25S rRNA [X52320] nt 1181–3588. Amplification products were purified using a QIAquick PCR purification kit (Qiagen Inc, Valencia, California) according to the standard protocol supplied by the manufacturer. For plastid and mitochondrial probes, equal amounts of each purified gene-specific DNA fragment were pooled prior to radio-labeling. DNA probes were labeled with α-32P-dCTP by random priming (Feinberg and Volgelstein 1983).
TAR sequences linked to telomeric repeats were generated using a PCR-based method that allows walking in regions of unknown sequence flanking a region of defined sequence (Siebert et al. 1995). A telomere sequence-specific primer (CCCTAAA)7 was used for the amplification of several TAR PCR products. Several of these PCR products were cloned and sequenced. The non-telomeric-repeat sequence from one of these clones was used for the design of a primer that allowed amplification of additional unknown flanking sequence. One PCR product obtained in this fashion was cloned, sequenced, and radio-labeled as described above for use in hybridization experiments. The sequence of this TAR PCR product was deposited in GenBank under accession no. [AF218263].
DNA Fingerprinting of BAC Clones
BAC DNA Isolation and Restriction Enzyme Digestion
The inoculation of BAC clones and subsequent BAC DNA isolation from 96 deep-well plates was as previously described (Klein et al. 1998). Following purification, BAC DNA pellets were resuspended in 90-μl TE buffer containing 10 μg /ml of RNaseA. DNA yields ranged between 1.0 μg and 2.0 μg from an initial 1.5-ml culture. DNA fingerprinting was performed as described by Tao et al. (1995) with minor modifications. Briefly, 3 μl (30–60 ng) of BAC DNA was transferred to a 0.2-ml PCR tube on ice, to which was added an equal volume of a fingerprinting cocktail solution that consisted of 132.5 μl sterile water, 40.25 μl 10× multi-core buffer (Promega), 5.25 μl 0.5 mM ddGTP, 4 μl HindIII (50U/μl), 4 μl HaeIII (50U/μl), 5 μl AMV reverse transcriptase (10U/μl) and 2 μl α-33P-dATP. The reaction mixture was incubated for 1.5–2 h at 37° C. After digestion, the DNA was collected in the bottom of the tube by a brief spin and 3 μl of gel loading dye (98% v/v deionized formamide, 0.3% bromophenol blue, 0.3% xylene cyanol, 10 mM EDTA pH 8.0) was added. The restricted samples were subjected to electrophoresis on 4% polyacrylamide gels containing 8 M urea following denaturation for 10 min at 95° C. Lambda DNA that had been restricted with Sau3A and end-labeled with α-33P-dATP was loaded in the first and every ninth lane of the gel to serve as a marker for image analysis. Gels were run at 85 W for 2.5 h, dried and exposed to X-ray film for 2–4 days. Following autoradiography, the films were scanned on a UMAX Mirage D-16L scanner (UMAX Technologies Inc., Fremont, California) at 200 dpi and saved as TIFF files. TIFF files were transferred to a SUN ULTRA10 workstation (SUN Microsystems, Fremont, California) with a Solaris 2.6 operating system for band calling and contig assembly.
To identify the restriction bands in BAC DNA fingerprints, gel images saved as TIFF files were analyzed by the program Image 3.5 (Sulston et al. 1988, 1989). Bands at the top 3 cm and the bottom 7 cm of the image were ignored in the band-calling process because of band compression and band distortion, respectively. A specific marker file using bands from the λ/Sau3A restriction pattern was generated to normalize the mobility of all restriction fragments. A vector file was also created to filter out vector fragment(s) prior to contig assembly. Band calling was performed automatically in Image; however, all lanes were checked manually and band-calling errors corrected. The band data was then transferred to the program FPC V4.5 for automated contig assembly. Both Image and FPC were downloaded from http://www.sanger.ac.uk/Software (Soderlund et al. 1997). In the FPC analysis, a fixed tolerance of seven was used and contig assembly was initially performed at a cut-off value of 5 × 10−14. The cut-off value was subsequently raised from 5 × 10−14 to 10−10 for the addition of singletons to existing contigs and the merging of contigs.
BAC Pooling Strategy
Stack Design
For the pooling strategy used in the present study, 256 individual 96-well microtiter plates containing 24,576 BAC clones were arranged into a stack design. The 256 individual 96-well microtiter plates included clones from both the HindIII and EcoRI libraries. The stack consisted of 32 layers or plates with each layer containing eight 96-well plates. The eight plates in a layer were arranged in a 2 × 4 plate pattern. Because each 96-well plate is an array of 8 rows and 12 columns of wells, this 2 × 4 plate pattern resulted in each plate layer containing wells in a 32 row × 24 column array (768 wells/layer).
Every well in the stack has a unique address defined by its x, y, and z coordinates relative to the axes of the stack (see Fig. 2). Although the assignment of x, y, and z to a particular axis is arbitrary, for our purposes, the x axis extends left to right, as seen by an observer examining one side of the stack. A rank of wells parallel to the x axis is a row. The y axis extends away from the observer towards the horizon. A rank of wells parallel to the y axis is a column. Finally, the z axis is mutually perpendicular to x and y and defines vertical position (plate or layer number) within the stack. The origin of the stack is defined as the far-left corner of the topmost layer with its coordinates being 1,1,1.
Pooling Strategy
The stack of microtiter plates was sampled in six distinct ways to generate the BAC pools (Fig. 2). Each pool represents the intersection of a plane with the stack (with the plane passing through the wells). Planes intersecting the stack parallel to three, mutually perpendicular, surfaces define three of the pool types. Plate pools (PP) were prepared from each layer or plate of the stack. All BACs in a plate pool share the same layer (z) coordinate. The front surface of the stack facing an observer was termed the face. Planes parallel to this surface defined face pools (FP). Each face pool consists of BACs with the same y coordinate (i.e., they occupy the same row as defined above). The surfaces left and right of the stack were termed sides. Planes parallel to these surfaces defined side pools (SP). Each side pool consists of BACs in the same column which share the same x coordinate. The three remaining pool types were sections taken at an angle through the stack. Row pools (RP) were established as follows: A BAC in row R (y), plate P (z) is a member of row pool R + P (y + z) (i.e., all of the wells from plate 1, row 2 are combined with those from plate 2, row 1 to form RP3 and so on for other combinations). To keep the number of wells in each row pool constant (768 wells per row pool), wrapping occurred. That is, when the R + P value was greater than 32 (the longest of the y and z dimensions), then 32 was subtracted from R + P to give the correct row pool (i.e., wells in plate 32, row 1 are a member of row pool 1 [32 + 1 = 33, 33>32, therefore 33 − 32 = 1]). Column pools (CP) were established in a manner similar to that of row pools. A BAC in column C (x), plate P (z) was added to column pool C + P (x + z). Thus, all the wells from plate 1, column 2 and plate 2, column 1 are in CP3. As with RPs, when the C + P value was greater than 32 (the longest of the z dimension), wrapping occurred by subtracting 32 from the C + P value to determine the correct column pool. The final pool type are called diagonal pools (DP). All wells in row R (y), column C (x) belong to diagonal pool R + C (x + y) (i.e., the well at row 1, column 1 in every plate [z] is in DP2). When the R + C value was greater than 32, wrapping occurred as described above such that the well at row 32, column 1 on every plate belongs to DP1.
The six pool types resulted in 184 BAC pools. Five of the six pool types (PP, FP, RP, CP, and DP) were composed of 32 pools each containing 768 BACs. The sixth pool type (SP) was composed of 24 pools each containing 1024 BACs. Thus every well in the stack was present in each of the six pool types and no well was present more than once in any pool type.
BAC DNA Isolation from Pools
Each time one of the six pool types was prepared, the 256 96-well microtiter plates comprising the pooling stack were inoculated with BAC stocks and the BAC DNA isolated. BACs were inoculated from a frozen stock plate using a 96-well pin tool into microtiter plates containing 200 μl TB media plus 12.5 μg/ml chloramphenicol per well and the plates incubated for 20–24 h at 37° C (without shaking). The next day the plates were arranged into the stack design and sampled in one of the six distinct ways to generate a particular pool type (see above). For BAC pooling, approx. 130 μl of culture was removed from each well using a multichannel pipette. The BAC cultures were placed in sterile containers with each container defining a given BAC pool. Each pool contained approx. 100 ml of culture. Individually, pooled cultures were transferred to 250-ml centrifuge bottles and incubated on ice for 15 min. The cells were collected by centrifugation at 4400 g for 15 min at 4° C. The supernatant was removed and the pellets drained. Cell pellets were respun for 5 min at 4400 g and the remaining TB media removed by aspiration. The cell pellet was resuspended in 2.4 ml of solution I (50 mM Tris-HCl pH 7.5, 50 mM EDTA pH 7.5) by vortexing and pipetting the solution up and down. The resuspended cell pellet was transferred to a 30-ml centrifuge tube and 4.8 ml of freshly prepared solution II (0.2 M NaOH, 1% SDS) was added. The sample was mixed by inversion 10–12 times and incubated 15 min at room temperature. Next, 3.6 ml of ice-cold solution III (3 M potassium acetate pH 5.2) was added and the sample mixed by inversion 12 times. Following incubation in an ice-water bath for 20 min, the sample was centrifuged at 31000 g for 20 min at 30° C. The supernatant was transferred to a new tube and respun at 31,000 g for 20 min at 30° C to completely clarify the supernatant. The supernatant was filtered through miracloth into a tube containing 1 vol (approx. 10 ml) of isopropanol, mixed, and incubated overnight at -20° C. The precipitated nucleic acid was centrifuged at 13,800 g for 30 min at 4° C, washed twice (70% ethanol first wash, 80% ethanol second wash), air-dried, and resuspended in TE buffer containing 20 μg/ml RNase A. The sample was incubated at 37° C for 1.5 h to degrade RNA, transferred to a 1.5-ml microcentrifuge tube and centrifuged at 15,000 g for 5 min to remove any residual insoluble material. The supernatant was transferred to a new tube and the BAC DNA quantified by fluorimetry. Typical yields ranged between 1.5–2.0 μg BAC DNA/ml of starting culture.
A portion of each pooled DNA sample was further purified by phenol:chloroform extraction. Three hundred μl of each pooled sample (approx. 30–60 μg BAC DNA) was transferred to a phase-lock gel tube (5 prime→3 prime Inc. Boulder, CO) and phenol extracted. Following purification, BAC DNA was resuspended in TE buffer containing 20 μg/ml RNase A and the DNA quantified by fluorimetry.
PCR-based Screening of BAC Pools
STS and SSR Screening
Primers that amplify sorghum SSRs and STSs were initially used to screen the BAC DNA pools. SSR primers were obtained from Dr. G. Hart (TAMU, College Station, Texas). Sequencing sorghum RFLP clones (Xu et al. 1994) generated STSs. Plasmid DNA for sequencing was prepared using a Wizard SV + miniprep DNA isolation kit (Promega). Plasmid DNA was sequenced using a Thermo Sequenase dye terminator cycle sequencing kit (Amersham Life Science Inc., Arlington Heights, Illinois) according to the manufacturer's protocol. Sequencing was performed on an ABI Prism 377XL sequencer (PE Applied Biosystems, Foster City, California). STS primers were designed from the sequence generated from each RFLP clone using the Oligo 5.0 software program (Molecular Biology Insights Inc., Cascade, Colorado) and were obtained from Sigma-Genosys (The Woodlands, Texas).
To screen the BAC DNA pools for PCR-based STS and SSR markers, a hot start amplification strategy utilizing AmpliTaq Gold DNA polymerase (Perkin Elmer, Foster City, California) was employed as described by Klein et al. (1998). Following amplification, loading dye was added and the samples denatured for 5 min at 95° C. Samples were electrophoresed on 4% polyacrylamide gels containing 8 M urea at 85 W for 2.5–4 h. Gels were dried and exposed to X-ray films for 1–3 days.
A subset of the SSRs was amplified using fluorescent IRD-labeled primers obtained from LI-COR. The IRD-labeled primers were diluted to 1 pmol/μl according to the manufacturer's recommendation. Amplification reactions were performed in a total volume of 10 μl containing 1× Perkin Elmer buffer II, 2.5 mM MgCl2, 200 μM dNTPs, 1 pmol IRD-labeled primer, 1 pmol unlabeled primer, 0.4 U AmpliTaq Gold polymerase and 20 ng pooled BAC DNA or BTx623 DNA. Amplification conditions were as previously described (Klein et al. 1998). The fluorescent-labeled amplification products were analyzed on a LI-COR dual-dye sequencer as described above for AFLP amplification products. BAC DNA pools containing STS or SSR markers were identified manually from the autoradiograms or TIFF images and the individual BACs containing the marker were identified (see below).
AFLP Screening
Template preparation for AFLPs using DNA from the 184 BAC pools was as described above for the RIL mapping population. Phenol/chloroform–extracted pooled BAC DNA (500 ng) was restricted with EcoRI and MseI and the resulting fragments ligated to EcoRI and MseI adapters. Preamplification (+0/+1 primers) and selective amplification (+3/+3 primers) reactions were performed as described above for genetic linkage analysis. When analyzing candidate BAC clones for the presence of a marker the selective amplification reaction was modified as follows: the 10-μl reaction contained 1× Taq buffer, 2.5 mM MgCl2, 200 μM dNTPs, 1.87 ng MseI selective primer, 0.03 μl IRD-labeled EcoRI selective primer, 0.2 U Taq polymerase, and 2 μl dilute preamplified BAC DNA (8 pg). AFLP amplification products from BAC DNA pools or individual BACs were analyzed on LI-COR DNA sequencers along with amplification products from the parental lines of the mapping population to serve as controls. The TIFF images were imported into Bionumerics software and band analysis performed.
Data Analysis
A Unix-based application was written in the Perl programming language to assist in resolving candidate BAC clones from the marker data obtained using the BAC DNA pools. Although there are six pool types, there are only three degrees of freedom (x, y, z) in the system. The redundancy makes it possible to use the presence of a signal (i.e., a PCR amplification product) in two pool types to predict the presence of the same signal in a third pool type. Specifically, (1) plate pools and face pools predict row pools, (2) plate pools and side pools predict column pools, and (3) face pools and side pools predict diagonal pools. When the three predictions are applied to real data, it is possible to relate signals appearing in the six pool types to unique wells within the stacked plates, a task that is nearly impossible with only three pool types. The confirmation process eliminates many alternative addresses and requires that a signal must be detected multiple times in the appropriate pools before it is considered meaningful. This feature greatly reduces the frequency of false-positives but also means that some BACs will be missed using this approach (false-negatives).The perl script and documentation are available at http://ars-genome.cornell.edu/software.html.
Acknowledgments
The authors are indebted to Dr. Gary Hart for providing SSR primer sequences, RFLP clones, and the framework sorghum genetic linkage map used in this study. This work was supported in part by an Advanced Technology Program grant 999902–059 (J.E.M.), a USDA-NRI grant 96–35300–3651 (J.E.M.), the Texas Agricultural Experiment Station (J.E.M. and P.E.K.), the Perry Adkisson Chair (J.E.M.) and the USDA-ARS (R.R.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jmullet@tamu.edu; FAX (409) 862–4718.
REFERENCES
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molec Biol Reporter. 1991;9:208–218. [Google Scholar]
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N. Human BAC library: construction and rapid screening. Gene. 1997;191:69–79. doi: 10.1016/s0378-1119(97)00044-9. [DOI] [PubMed] [Google Scholar]
- Avramova Z, Tikhonov A, SanMiguel P, Jin Y-K, Liu C, Woo S-S, Wing RA, Bennetzen JL. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- Barillot E, Lacroix B, Cohen D. Theoretical analysis of library screening using a N-dimensional pooling strategy. Nucl Acids Res. 1991;19:6241–6247. doi: 10.1093/nar/19.22.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin K, Deu M, Rami J-F, Trouche G, Hamon P. Towards a saturated sorghum map using RFLP and AFLP markers. Theor Appl Genet. 1999;98:320–328. [Google Scholar]
- Brenner S, Livak KJ. DNA fingerprinting by sampled sequencing. Proc Natl Acad Sci. 1989;86:8902–8906. doi: 10.1073/pnas.86.22.8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno WJ, Knill E, Balding DJ, Bruce DC, Doggett NA, Sawhill WW, Stallings RL, Whittaker CC, Torney DC. Efficient pooling designs for library screening. Genomics. 1995;26:21–30. doi: 10.1016/0888-7543(95)80078-z. [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA, Matz EC, Romero-Severson J. Pinning down loose ends: Mapping telomeres and factors affecting their length. Plant Cell. 1992;4:953–960. doi: 10.1105/tpc.4.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kang HL, Xu X, Wang M, Dho SH, Huh JR, Lee B-J, Kalush F, Bocskai D, Ding Y, et al. A 12-Mb complete coverage BAC contig map in human chromosome 16p13.1-p11.2. Genome Res. 1999;9:763–774. [PMC free article] [PubMed] [Google Scholar]
- Chittenden LM, Schertz KF, Lin Y-R, Wing RA, Paterson AH. A detailed RFLP map of Sorghum bicolor x S. propinquum, suitable for high-density mapping, suggests ancestral duplication of Sorghum chromosomes or chromosomal segments. Theor Appl Genet. 1994;87:925–933. doi: 10.1007/BF00225786. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tomé P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Doebley J, Durbin M, Golenberg EM, Clegg MT, Ma DP. Evolutionary analysis of the large subunit of carboxylase (rbcL) nucleotide sequence among the grasses (Gramineae) Evolution. 1990;44:1097–1108. doi: 10.1111/j.1558-5646.1990.tb03828.x. [DOI] [PubMed] [Google Scholar]
- Doggett H. Sorghum. 2nd ed. New York: John Wiley; 1988. [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Green ED, Olson MV. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci. 1990;87:1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Howell GR, Bentley DR. Genome mapping by fluorescent fingerprinting. Genome Res. 1997;7:1162–1168. doi: 10.1101/gr.7.12.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu S-H, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S-S, Wing RA, Gill BS, Ward DC. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Klein RR, Morishige DT, Klein PE, Dong J, Mullet JE. High throughput BAC DNA isolation for physical map construction of sorghum (Sorghum bicolor) Plant Molec Biol Reporter. 1998;16:351–364. [Google Scholar]
- Kong, L., J. Dong, and G.E. Hart. 2000. Isolation, characterization, and linkage mapping of Sorghum bicolor (L.) Moench DNA simple-sequence-repeats (SSRs). Theor. Appl. Genet. (In press).
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JT, Dong F, Jackson SA, Song J, Jiang J. Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics. 1998a;150:1615–1623. doi: 10.1093/genetics/150.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JT, Jackson SA, Nasuda S, Gill BS, Wing RA, Jiang J. Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor Appl Genet. 1998b;96:832–839. [Google Scholar]
- Peng Y, Schertz KF, Cartinhour S, Hart GE. Comparative genome mapping of Sorghum bicolor (L.) Moench using an RFLP map constructed in a population of recombinant inbred lines. Plant Breeding. 1999;118:225–235. [Google Scholar]
- Presting GG, Frary A, Pillen K, Tanksley SD. Telomere-homologous sequences occur near the centromeres of many tomato chromosomes. Mol Gen Genet. 1996;251:526–531. doi: 10.1007/BF02173641. [DOI] [PubMed] [Google Scholar]
- Qi X, Stam P, Lindhout P. Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor Appl Genet. 1998;96:376–384. doi: 10.1007/s001220050752. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Goodman HM, Ausubel FM. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucl Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucl Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R. FPC: a system for building contigs from restriction fingerprinted clones. Comput Applic Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Gregory S, Dunham I. Sequence ready clones. In: Bishop MJ, editor. Guide to Human Genome Computing. San Diego, CA: Academic Press; 1998. pp. 151–179. [Google Scholar]
- Soderlund C. FPC V4: User's manual fingerprinted contigs. Technical Report SC-01–99. Cambridge, UK: The Sanger Centre; 1999. [Google Scholar]
- Springer PS, Zimmer EA, Bennetzen JL. Genomic organization of the ribosomal DNA of sorghum and its close relatives. Theor Appl Genet. 1989;77:844–850. doi: 10.1007/BF00268337. [DOI] [PubMed] [Google Scholar]
- Sulston J, Mallett F, Staden R, Durbin R, Horsnell T, Coulson A. Software for genome mapping by fingerprinting techniques. Comput Applic Biosci. 1988;4:125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- Sulston J, Mallett F, Durbin R, Horsnell Terry. Image analysis of restsriction enzyme fingerprint autoradiograms. Comput Applic Biosci. 1989;5:101–106. doi: 10.1093/bioinformatics/5.2.101. [DOI] [PubMed] [Google Scholar]
- Tao Q, Qian Y, Zhao H, Yu S, Qiu L, Wu B, Zhu J, Yu D, Liu X, Hong G. Construction of Oryza sativa genome contigs by fingerprint strategy. Cell Res. 1995;5:263–271. [Google Scholar]
- Tao Q, Zhang H-B. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucl Acids Res. 1998;26:4901–4909. doi: 10.1093/nar/26.21.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Jaramillo-Babb VL. A sequence-ready BAC clone contig of a 2.2-Mb segment of human chromosome 1q24. Genome Res. 1999;9:150–157. [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: A new technique for DNA fingerprinting. Nucl Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuylsteke M, Mank R, Antonise R, Bastiaans E, Senior ML, Stuber CW, Melchinger AE, Lübberstedt T, Xia XC, Stam P, et al. Two high-density AFLP linkage maps of Zea mays L.: Analysis of distribution of AFLP markers. Theor Appl Genet. 1999;99:921–935. [Google Scholar]
- Williams CE, Ronald PC. PCR template-DNA isolated quickly from monocot and dicot leaves without tissue homogenization. Nucl Acids Res. 1994;22:1917–1918. doi: 10.1093/nar/22.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucl Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-W, Magill CW, Schertz KF, Hart GE. A RFLP linkage map of Sorghum bicolor (L.) Moench. Theor Appl Genet. 1994;89:139–145. doi: 10.1007/BF00225133. [DOI] [PubMed] [Google Scholar]
- Young WP, Schupp JM, Keim P. DNA methylation and AFLP marker distribution in the soybean genome. Theor Appl Genet. 1999;99:785–790. [Google Scholar]
- Zhu H, Blackmon BP, Sasinowski M, Dean RA. Physical map and organization of chromosome 7 in the rice blast fungus, Magnaporthe grisea. Genome Res. 1999;9:739–750. [PMC free article] [PubMed] [Google Scholar]