Abstract
The Myc-deregulating chromosomal T(12;15)(Igh–Myc) translocation, the hallmark mutation of inflammation- and interleukin 6-dependent mouse plasmacytoma (PCT), is the premier model of cancer-associated chromosomal translocations because it is the only translocation in mice that occurs spontaneously (B lymphocyte lineage) and with predictably high incidence (~85% of PCT), and has a direct counterpart in humans: Burkitt lymphoma t(8;14)(q24;q32) translocation. Here, we report on the development of a genetic system for the detection of T(12;15)(Igh–Myc) translocations in plasma cells of a mouse strain in which an enhanced green fluorescent protein (GFP)-encoding reporter gene has been targeted to Myc. Four of the PCTs that developed in the newly generated translocation reporter mice, designated iGFP5′Myc, expressed GFP consequent to naturally occurring T(12;15) translocation. GFP expression did not interfere with tumor development or the deregulation of Myc on derivative 12 of translocation, der (12), because the reporter gene was allocated to the reciprocal product of translocation, der (15). Although the described reporter gene approach requires refinement before T(12;15) translocations can be quantitatively detected in vivo, including in B lymphocyte lineage cells that have not yet completed malignant transformation, our findings provide proof of principle that reporter gene tagging of oncogenes in gene-targeted mice can be used to elucidate unresolved questions on the occurrence, distribution and trafficking of cells that have acquired cancer-causing chromosomal translocations of great relevance for humans.
Keywords: mouse plasmacytoma T(12;15) translocation, human Burkitt lymphoma t(8;14)(q24;q32) translocation, green fluorescence protein
Reciprocal chromosomal translocations that illegitimately recombine cellular oncogenes with immunoglobulin genes have long been recognized as hallmarks of both B-cell and plasma-cell neoplasms in humans (Willis and Dyer, 2000; Kuppers and Dalla-Favera, 2001). The t(8;14)(q24;q32) translocation, most commonly observed in the post-germinal center B-cell tumor, Burkitt lymphoma, is of special interest because it results in the deregulated expression of a gene that is among the most important cancer genes—MYC (Eisenman, 2001; Levens, 2003); it is the first cancer-associated translocation to have been characterized at the molecular level—balanced (reciprocal) genetic exchange of MYC with the Ig heavy-chain locus, IGH (Dalla-Favera et al., 1982); it is widely believed to be a very early, if not initiating, transforming event in neoplastic B-cell development; and, unlike the great majority of oncogene-activating translocations found in human lymphoma and leukemia, it has a direct counterpart in mice—T(12;15)(Igh–Myc)—that is itself the hallmark of peritoneal BALB/c plasmacytoma (PCT). Like human t(8;14)(q24;q32) in Burkitt lymphoma, mouse T(12;15) in PCT is thought to be a tumor-initiating mutation (Janz et al., 1993). Despite exciting recent progress in our understanding of the molecular mechanism that underlies Igh–Myc translocation (Ramiro et al., 2004; Robbiani et al., 2008; Wang et al., 2009; Gostissa et al., 2009b), fundamental questions on the biology of this translocation remain unresolved, primarily because of the difficulty in detecting individual translocation-bearing cells in vivo.
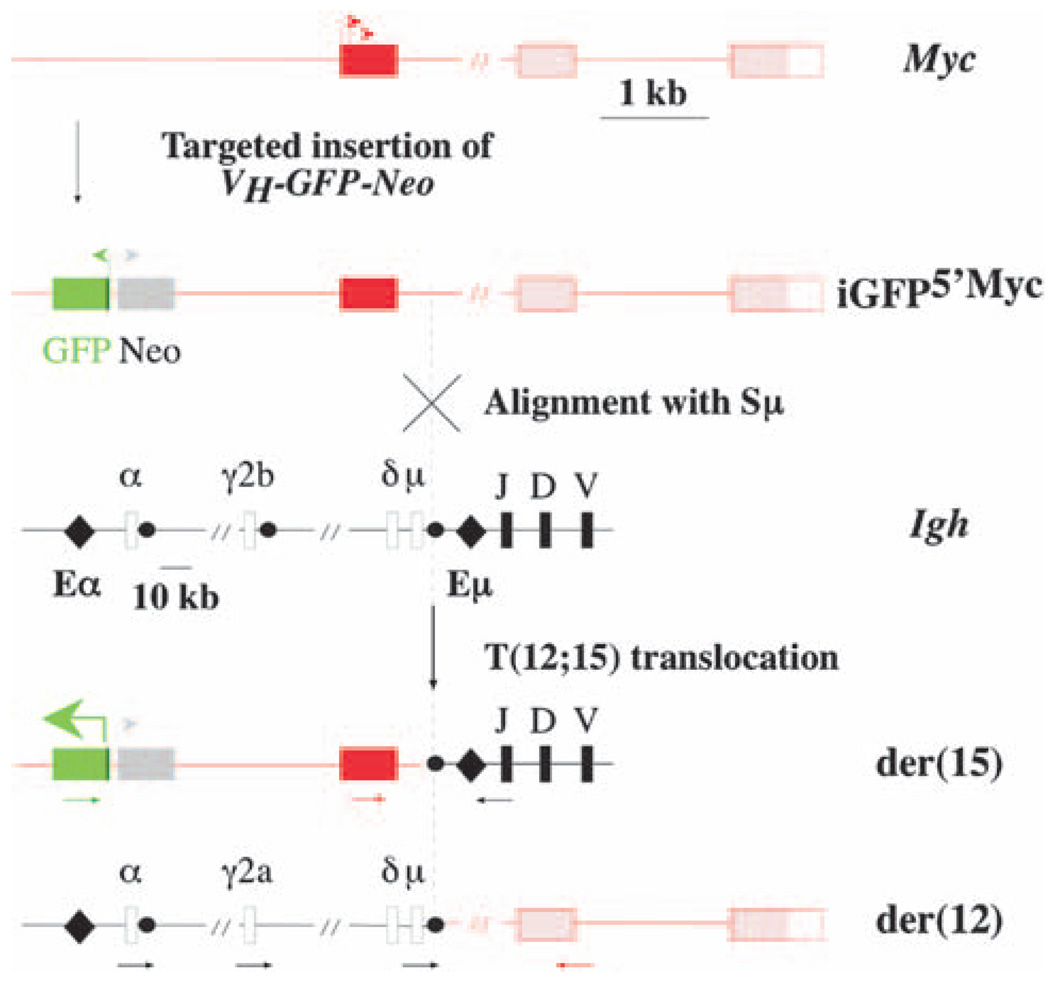
We have taken an important first step to overcome this difficulty by developing a genetic method for the detection of individual T(12;15)-carrying cells in mice. Using established gene-targeting methods in embryonic stem cells, we generated iGFP5′Myc gene-insertion mice that harbor in the near upstream flank of the Myc locus a cDNA gene that encodes the enhanced GFP protein under control of the mouse immunoglobulin heavy-chain (Igh) variable gene promoter, VH (Figure 1, top). The GFP reporter has been inserted ~1.5 kb 5′ of Myc exon 1, to make it possible to specifically detect derivative 15, der(15), of the T(12;15) translocation. Unlike der(12), which contains the deregulated Myc gene that drives tumor development (Eisenman, 2001; Levens, 2003) and is subject to secondary changes in the course of plasmacytomagenesis, including those caused by class switch recombination on the rearranged Igh–Myc domain (Kovalchuk et al., 1997), der(15) is thought to be inconsequential for malignant cell transformation and not affected by secondary structural modification. Moreover, cytogenetic analysis of hundreds of mouse PCTs has shown that der(15) is rarely, if ever, lost in T(12;15)+ tumors. These features render der(15) a particularly attractive host for a translocation reporter gene, such as the VH-GFP that we use here. Because the GFP gene insertion site in iGFP5′Myc mice is just upstream of the known 5′ boundary of the translocation breakpoint cluster in Myc, the reporter gene will invariably be allocated to der(15), irrespective of the actual break site in Myc, which occurs in most cases in intron 1 or exon 1 of the gene and, less frequently, in its near 5′ flank.
Figure 1.
Genetic system for detection of the T(12;15) translocation in B-lineage cells of iGFP5′Myc gene-insertion mice. Shown above are schematic representations of the normal mouse Myc locus on chromosome 15, and of the targeted Myc locus carrying the inserted VH-GFP-Neo gene in iGFP5′Myc knock-in mice. The non-coding first exon of Myc is depicted by a red box, with the P1 and P2 Myc promoters indicated by two red arrows pointing right. The coding region of Myc (exons 2 and 3) and the 3′ untranslated region of Myc exon 3 are depicted by two pink boxes and one white box, respectively. The 1.6-kb first intron of Myc is not drawn to scale (as indicated by the short, oblique double line). The transcriptional orientations of the inserted GFP and Neo genes are indicated by colored arrows at transcriptional start sites. Shown in center is a schematic representation of the Igh locus on chromosome 12. The Igh variable (VH), diversity (D) and joining (JH) regions are represented by thick vertical lines labeled as such. The Igh constant region (CH), which is flanked by the intronic heavy-chain enhancer (Eµ) and the 3′ Cα heavy-chain enhancer (Eα; indicated by two black diamonds), is only partially represented by four CH genes: Cµ, Cα, Cγ2b and Cα (white, labeled boxes). The corresponding switch regions are indicated by black dots (except in the case of Cδ, which does not have a canonical switch region). Four additional genes in the mouse CH cluster (Cγ1, Cγ2a, Cγ3, Cε) are not shown. The Igh locus and the targeted Myc locus are aligned at a crossover site typically used to generate the T(12;15) translocation: the switch µ region on chromosome 12 and the first intron of Myc on chromosome 15. This is denoted by a cross-labeled alignment with Sµ. The actual site of DNA double-strand breakage and reciprocal transchromosomal recombination is indicated by a vertical, dashed line and an arrow-labeled T(12;15) translocation. Shown at bottom are schematic representations of the reciprocal products of the translocation: der(15) and der(12). Juxtaposition of Eµ to the VH promoter of the GFP gene leads to the expression of GFP on der(15), as indicated by a thick green arrow pointing left. Annealing sites for PCR primers in Igh, Myc and VH-GFP used to detect reciprocal Igh–Myc junctions on der(12) and Igh–Myc–GFP junctions on der(15) are indicated by horizontal arrows that are colored black, red and green, respectively.
According to the scheme presented in Figure 1, GFP remains silent in B cells of strain iGFP5′Myc mice that have not undergone T(12;15) exchange because the VH promoter of the GFP gene on chromosome 15 cannot be activated in trans by Igh enhancers on chromosome 12 (Figure 1, top). However, in B cells that do undergo T(12;15) exchange (Figure 1, bottom), GFP is expressed upon the juxtaposition of VH–GFP to an Igh enhancer in cis. Assembly of the targeting vector, generation of iGFP5′Myc ‘knock-in’ mice, and the development of genomic PCR methods for genotyping transgenic off-spring are described online in Supplementary Methods and illustrated in Supplementary Figure 1.
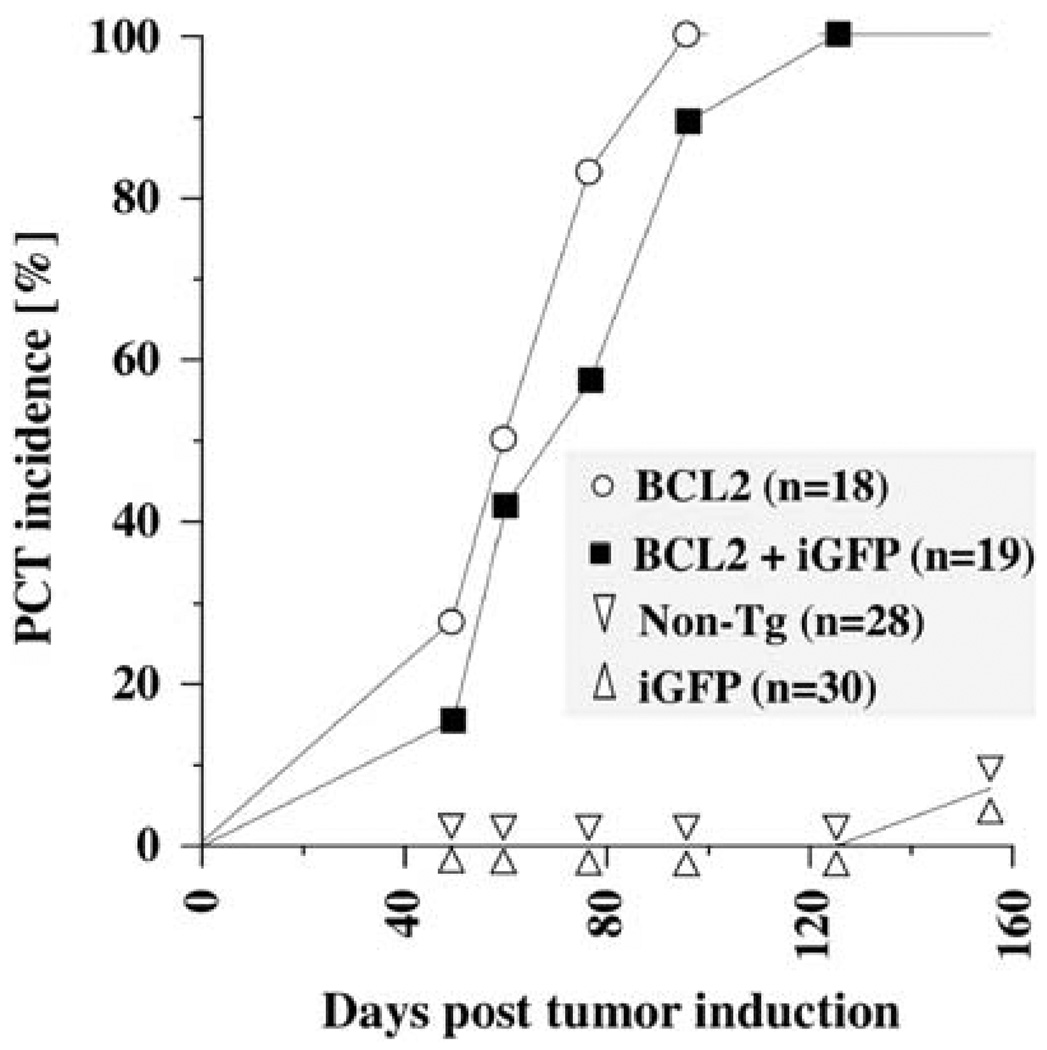
Reporter genes targeted to oncogenes may either promote or inhibit tumorigenesis—by changing the structure, accessibility or biological function of the modified genetic loci. To evaluate whether either is true in the case of strain iGFP5′Myc, we performed a PCT induction study in these mice. To accelerate tumor development, we combined the iGFP5′Myc reporter gene with a human BCL2 transgene that was shown in previous work to lead to T(12;15)-harboring plasma cell tumors with short onset and full penetrance (Silva et al., 2003). Figure 2 shows that upon induction of chronic peritoneal inflammation using pristane (Anderson and Potter, 1969), PCTs arose as rapidly (onset 50–120 days) and with the same incidence (100%) in double-transgenic iGFP5′Myc/BCL2 mice as in single-transgenic BCL2 mice. Characteristic features of mouse PCTs that are detectable by histomorphological criteria, by the expression of certain cell-surface markers including CD138 and by monoclonal Ig production, were also indistinguishable in iGFP5′Myc/BCL2 and BCL2 mice (results not shown).
Figure 2.
The iGFP5′Myc translocation reporter does not affect the genetic susceptibility of mice to plasmacytoma development. Depicted are onset and incidence of inflammation (pristane)-induced peritoneal plasmacytoma (PCT) in double-transgenic C.iGFP5′Myc/BCL2 mice (squares) and single-transgenic C.BCL2 mice (circles). In both strains, the BCL2 transgene accelerated PCT development with the same efficiency as observed in a previous study (Silva et al., 2003). Single transgenic C.iGFP5′Myc mice (triangles pointing up) and non-transgenic littermates (triangles pointing down), in which tumor development began just before termination of the study, on day 160 after tumor induction with pristane were included as controls. In all mice, PCTs were detected by finding neoplastic plasma cells in ascites-cell specimens that had been stained according to Wright–Giemsa.
To evaluate whether the proclivity of PCT to undergo T(12;15) translocation was comparable in iGFP5′Myc-transgenic mice and mice lacking the reporter transgene, we employed genomic PCR to analyze 13 tumors from iGFP5′Myc/BCL2 mice for the presence of chimeric Igh–Myc junction fragments (Kovalchuk et al., 2000b). The remaining six tumors included in Figure 2 were not available for molecular studies. Using the Myc and Igh primers depicted in Figure 1, we readily detected T(12;15)-typical junctions in 11 of 13 PCTs: der(12)-typical junctions were found in 10 tumors, and der(15)-typical junctions were seen in nine (Table 1, columns 3–4). The translocation status of two tumors remained undetermined, presumably because they contained an unusual T(12;15) exchange that was not detectable with the PCR methods used here, harbored a variant Myc translocation that relied on an immunoglobulin light-chain instead of the Igh locus, or did not harbor an Myc translocation at all. The detection of T(12;15) in 85% of iGFP5′Myc-carrying PCT was consistent with the incidence of this translocation in PCTs from normal mice of strain C (Janz, 2006).
Table 1.
Analysis of PCT
| PCTa | GFPb | der(12)c | der(15)d | GFP-Myce |
|---|---|---|---|---|
| 1 | + | + | + | + |
| 2 | + | + | + | + |
| 3 | + | ND | + | + |
| 4 | + | + | + | + |
| 5 | − | + | ND | ND |
| 6 | − | + | + | − |
| 7 | − | + | + | − |
| 8 | − | + | + | − |
| 9 | − | + | ND | ND |
| 10 | − | + | + | + |
| 11 | − | + | + | + |
Abbreviations: PCT, plasmacytomas; GFP, green fluorescent protein; Igh–Myc, Myc-deregulating chromosomal T(12;15); ND, not detected.
Periotoneal PCTs produced by injecting double-transgenic C.iGFP5′Myc/BCL2 mice intraperitoneally with 0.2 ml pristane (2,6,10,14-tetramethylpentadecane). Tumor incidence and onset are depicted in Figure 2.
GFP expression in tumor cells as assessed by FACS analysis of ascites cell specimens and immunofluorescence microscopy of peritoneal granulomas, as illustrated in Figure 3 and Supplementary Figure 2.
Igh–Myc junctions indicative of the Myc-deregulating product of the T(12;15) translocation, der(12), as detected by PCR analysis using the Myc exon 2 and Igh CH PCR primers indicated in Figure 1, bottom.
Igh–Myc junctions indicative of the reciprocal product of the T(12;15) translocation, der(15), as detected by PCR analysis using the Myc exon 1 (red) and JH (black) PCR primers indicated in Figure 1, bottom.
Igh–Myc junctions indicative of the reciprocal product of the T(12;15) translocation, der(15), as detected by PCR analysis using the GFP (green) and JH (black) primers indicated in Figure 1, bottom.
The results presented above suggested that the GFP reporter gene in strain iGFP5′Myc is a passive genomic passenger that neither affects PCT development nor diminishes the likelihood that the T(12;15) translocation will be generated. However, because the present sample of translocation-bearing tumors is small (n = 11), a follow-up study producing a larger data set is necessary before it can be unambiguously decided whether the GFP reporter does or does not cause bias with regard to translocation occurrence and tumor development. Including a side-by-side comparison of hemizygous transgenic iGFP5’Myc mice (containing one targeted and one normal Myc locus) and their homozygous transgenic counterparts (lacking the normal Myc locus) in that study may be particularly informative.
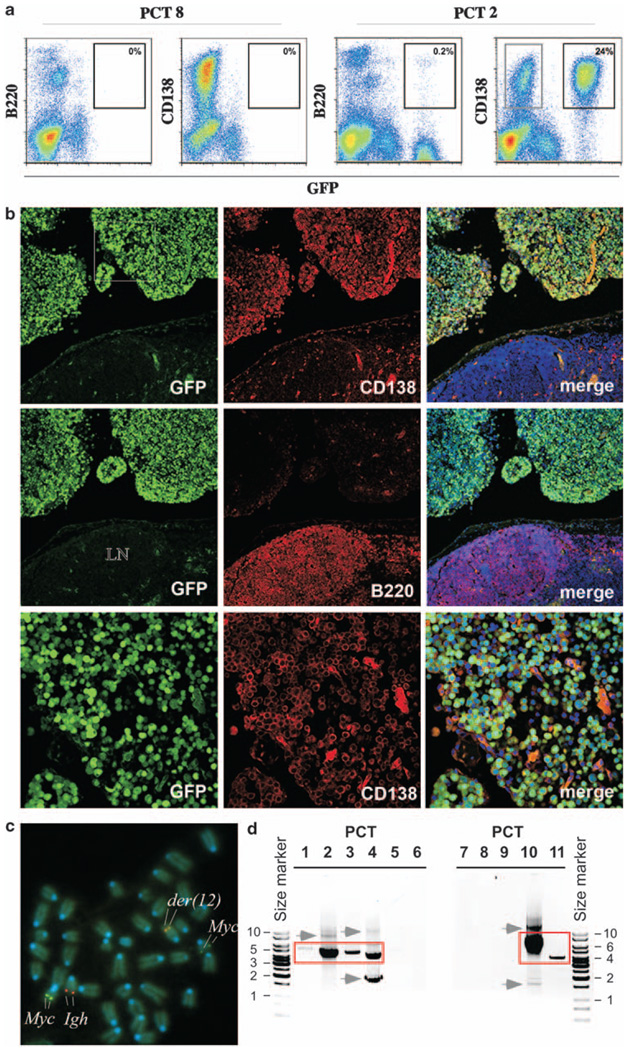
To identify PCTs in which T(12;15) is associated with GFP expression, we performed FACS analysis of the 11 tumors that showed PCR-based evidence of translocation. We stained PCT-containing ascites specimens with antibody to CD138 (syndecan-1, marker of plasma cells) and B220 (CD45R, marker of B cells) to distinguish the malignant plasma cells from populations of normal cells including peritoneal B cells. We found that 4 of 11 (36%) tumors expressed high levels of GFP (Table 1, column 2). This is illustrated in Figure 3a, in which PCT2 serves as a GFP+ example, and PCT8 as a GFP− example. In the case of PCT2, 24% of ascites cells were neoplastic plasma cells, as indicated by the co-expression of GFP and CD138 (GFP+CD138+) and a lack of B220 expression (B220−). The fraction of GFP+ tumor cells in the three other cases characterized by high levels of GFP (PCT1, 3, 4) ranged from 6 to 31% (Supplementary Figure 2). To confirm GFP expression in these cells by an independent method that additionally provides insight into the location and distribution of translocation-bearing cells in vivo, we performed immunofluorescence microscopy with antibodies to CD138 and B220, on serial tissue sections of peritoneal granulomas, the sites of PCT development. In all four cases (PCT1–4), GFP+CD138+ tumor cells that infiltrated the granulomatous tissue (Figure 3b, top and bottom rows) were readily distinguished from GFP−B220+ B lymphocytes (Figure 3b, center row). Normal CD138+ plasmablasts and plasma cells residing in spleen, gut villi and bone marrow were GFP-negative (not shown).
Figure 3.
Plasma-cell tumors in iGFP5’Myc mice express GFP consequent to T(12;15) translocation. (a) GFP expression in tumor cells, as detected by flow cytometry. Shown are scatter plots of ascites cells from mice bearing PCT8 (left, negative control) and PCT2 (right). Cells were treated with an Fc blocker (antibody to CD16/CD32) and stained with an antibody to B220 (CD45R; RA3–6B2) or CD138 (281–2). Isotype-matched antibodies of unrelated specificity confirmed the specificity of the B220 and CD138 signals. Cells were analyzed on a Beckman Coulter FC 500 instrument (Beckman Coulter Inc., Fullerton, CA, USA) using FlowJo software. Note the substantially expanded CD138+GFP− population of tumor cells (indicated by grey rectangle; 9% of all cells) that co-existed with the predominant CD138+GFP+ tumor cell clone (24%) in the PCT2-harboring mouse. (b) GFP expression in T(12;15)+CD138+ tumor cells, as detected by immunofluorescence microscopy. Shown in the top and center rows are confocal microscopy images of serial tissue sections of a peritoneal granuloma that was labeled with PE-conjugated antibody to CD138 or B220, followed by counterstaining with DAPI (blue). These images are shown at low magnification. Adjacent to the granuloma (upper half of each image) is a piece of the mesenteric lymph node (MLN, lower half of each image). The three images in the top row show that plasma cells expressing GFP (left panel) and CD138 (center panel) had infiltrated the granuloma in large numbers, but were rare (scattered) in the MLN. The images in the center row reveal B220+GFP− B cells that populate the MLN but not the granuloma. The bottom-row images present the CD138+GFP− tumor cells included in the white square in the upper left-hand panel taken at higher magnification. (c) Presence of the T(12;15), as detected by FISH. Metaphase chromosome spread of PCT 1 hybridized to FISH probes for Myc (FITC, green, Chr 15) and Igh (Cy5, red, Chr 12). der(12) is indicated by co-localization of FISH signals, whereas der(15) is visualized by the weak Myc signal to the upper right of the image. Chromosomes were counter-stained with DAPI (blue). (d) Rearrangement of GFP-targeted Myc allele with Igh as detected by PCR analysis. Shown are der(15)-typical GFP–Myc–Igh junction fragments (red rectangles) detected in PCT1–4 and PCT10–11 by PCR analysis of genomic DNA. Junction fragments were size fractionated in an agarose gel and confirmed to be clonotypic by DNA sequence analysis of the underlying DNA recombination (not shown). PCR fragments outside of the red boxes, the most prominent of which are indicated by grey arrows pointing right, are either artifacts or bona fide translocation junction fragments indicating the clonal diversity of incipient PCTs in strain C mice. Size markers (kb) are indicated.
To further show that GFP expression in PCT1–4 was caused by the T(12;15) translocation, we performed fluorescence in situ hybridyzation (FISH) of Myc and Igh as described in Supplementary Methods. FISH detects distinct Myc and Igh signals in normal cells, but a co-localized or merged Igh–Myc signal in T(12;15)+ cells. In the case of PCT1, 11 of 15 (73%) tumor cells analyzed at the metaphase stage of the cell cycle showed merged FISH signals on der(12) (Figure 3c). In the case of PCT2, this was true for 10 of 10 (100%) tumor cells. As PCT3 and PCT4 contained less than the 10 metaphase cells commonly required for reliable cytogenetic diagnosis, we analyzed these tumors by interphase FISH. In the case of PCT3, co-localization of Myc and Igh signals was seen in 34 of 154 (22.1%) cells from an ascites specimen that contained 35% tumor cells and 65% normal (mostly inflammatory) cells (Supplementary Figure 3). The observed co-localization frequency was significantly elevated compared with that in normal splenic B lymphocytes: 1.7% (3/180 cells). Similar findings were obtained for PCT4 (not shown).
All of the PCTs included in Table 1 arose in mice that were hemizygous for the iGFP5′Myc transgene, thus carrying one targeted Myc locus and one normal Myc locus. To show directly that the GFP+ tumors used the targeted Myc allele for genetic recombination with Igh, we refined our PCR analysis of der(15) by replacing the Myc primer used for detecting the underlying Igh–Myc junction with a GFP primer that annealed further upstream (~2 kb) of Myc (Figure 1, green arrow below the scheme of der[15]). We combined the GFP primer with a PCR primer that annealed to the JH portion of Igh (black arrow) and were readily able, in all four tumors (PCTs 1–4), to elongate the clonotypic Igh–Myc junction previously generated using the JH and Myc exon 1 primer pair (red arrow). The der(12)-specific GFP–Myc–JH fragments are shown in Figure 3d, lanes 1–4.
These results showed that in four independent tumors (PCTs 1–4), GFP activation was caused by juxtaposition of the VH-GFP gene to Igh enhancers following chromosomal T(12;15) translocation.
We predicted that, in contrast to the four GFP+ tumors described above, the GFP− neoplasms had used the normal rather than the targeted Myc allele for translocation with Igh. Consistent with this possibility, we failed to detect GFP–Myc–JH junctions in PCTs 6–8 (Figure 3d). In the case of PCTs 10 and 11, however, clonotytpic GFP–Myc–JH junctions were obtained (Figure 3d). This suggested that the targeted Myc allele had recombined with Igh, yet GFP had not been activated. Lack of GFP expression could potentially be due to (1) epigenetic changes that diminish or extinguish GFP expression by silencing the Igh–Myc chromatin domain or (2) genetic changes such as translocation breakpoints disrupting the Eµ enhancer or—in keeping with the role of activation-induced cytidine deaminase in the origin of the T(12;15) translocation (Ramiro et al., 2004; Franco et al., 2006)—crippling activation-induced cytidine deaminase-mediated point mutations in the VH promoter or the coding sequence of the GFP gene (Muramatsu et al., 2000).
As molecular analyses, including DNA sequencing of translocation breakpoint regions, have not supported any of these possibilities thus far, alternative explanations must be considered to account for the apparent absence of GFP expression in PCTs 10 and 11. The most likely of those postulates the co-existence of a frank GFP− neoplasm and incipient GFP+ tumor cell clones in the same mouse. According to this explanation, the mice contained an overt CD138+ plasma cell tumor, designated PCT 10 or 11, that was readily detected by flow cytometry, yet lacked GFP expression either because it contained an unusual T(12;15) exchange that was not detectable with the PCR methods used here, harbored a variant Myc translocation that used an immunoglobulin light-chain instead of the Igh locus, or did not harbor an Myc translocation at all. Be this as it may, both mice additionally contained at least one T(12;15)+ cell clone that had used the GFP-targeted Myc allele for recombination with Igh (based on the PCR results shown in Figure 3d) and, hence, expressed GFP. However, due to the small size of the underlying cell clone, the GFP+ cells remained undetected by FACS. Consistent with this theory, clonal diversity of T(12;15)+ PCTs has been repeatedly documented in strain C mice undergoing peritoneal tumor induction (Janz et al., 1993; Müller et al., 1996, 1997a; Kovalchuk et al., 2000a, b, 2003). Furthermore, it is well established that PCR analysis of clonotypic Myc-Igh junctions detects T(12;15) translocation-bearing cells with much greater sensitivity than possible with FACS analysis of GFP expression in iGFP5′Myc mice. PCR routinely detects one T(12;15)+ cell among 105–106 normal cells (Janz, 2006), whereas conventional flow cytometry detects one GFP+ cell among 102–103 normal cells. The large difference in the detection limit (two to three orders of magnitude) of scoring T(12;15)+ cells by PCR versus FACS analysis would thus explain the findings in PCT10/11-carrying mice without postulating that the GFP reporter had failed.
As stated above, a follow-up study producing a larger sample of T(12;15)+ tumors in iGFP5′Myc mice is necessary before it can be determined with which frequency, if any, the GFP-targeted Myc locus undergoes T(12;15) exchange without activating the GFP reporter. Using to that end homozygous transgenic iGFP5′Myc mice may be particularly productive, because in these mice only GFP-targeted Myc alleles are available for T(12;15) exchange.
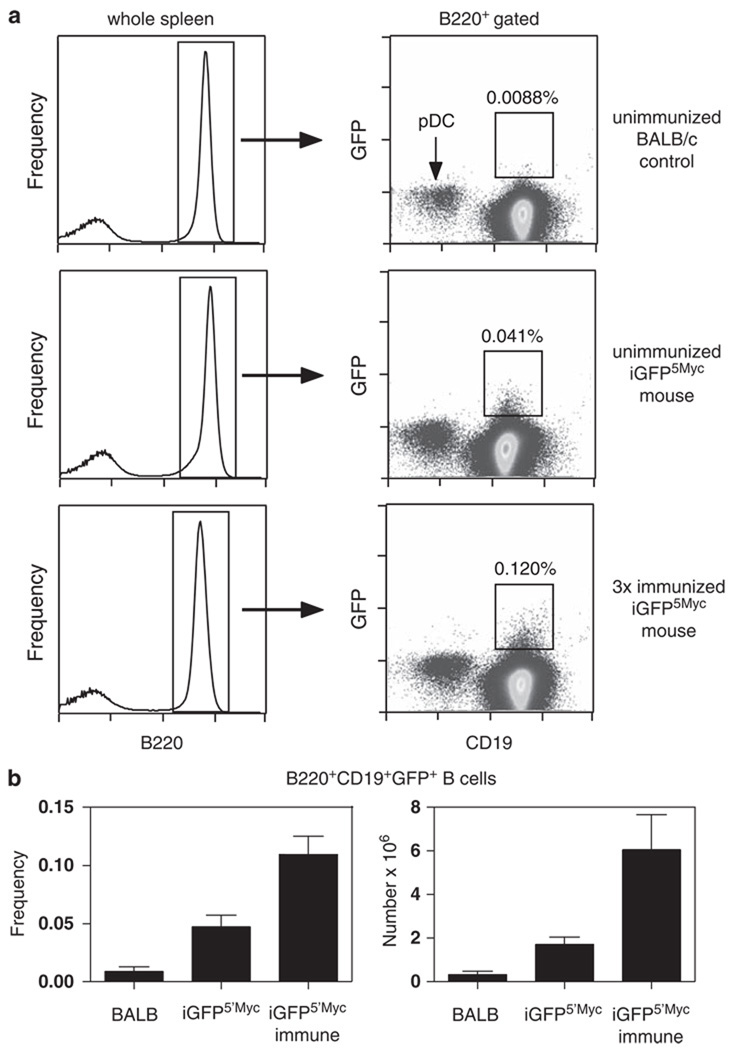
The newly developed iGFP5’Myc mice provide proof of principle that the targeted insertion of a transgenic reporter in the vicinity of an oncogene is a viable approach for detecting, enumerating and studying translocation-bearing cells in their normal tissue context. Combined with sensitive in vivo imaging techniques, such as two-photon microscopy (Germain et al., 2005; Wessels et al., 2007), the iGFP5′Myc mice may afford the possibility to visualize the migration and trafficking of Igh–Myc translocation-carrying cells in vivo. Additional questions that can be readily addressed with the assistance of the GFP reporter include the abundance and tissue distribution of T(12;15)+ tumor cells, using fluorescence microscopy, immunohistochemistry, flow cytometry and whole-body fluorescence imaging. Still more interesting are questions that might be attacked in future studies on translocation-carrying B-lineage cells that have not yet completed malignant transformation. Figure 4, which depicts our most recent findings in tumor-free iGFP5′Myc mice, suggests that the GFP reporter may have great potential in that regard. Thus, 8 to 9-month-old tumor-free iGFP5′Myc mice hyper-immunized with the T-cell-dependent antigen, sheep red blood cells, showed a nearly threefold increase in the number of GFP-expressing B220+CD19+ splenocytes (0.12%) compared with age-matched non-immunized iGFP5′Myc mice (0.041%) and normal C mice (0.0088%) used as background controls. The result implicates the germinal center reaction in the promotion of GFP+ cells and, hence, T(12;15)+ B-lymphocytes that can be operationally defined as pre-malignant PCT precursors.
Figure 4.
Flow cytometric detection of GFP+ splenic B cells from C.iGFP5′Myc mice. Viable mononuclear spleen cell suspensions from unimmunized BALB/c (C) control, unimmunized C.iGFP5′Myc, or SRBC hyperimmunized C.iGFP5′Myc mice were prepared and stained with anti-B220 and anti-CD19 mAbs, as described in Supplementary Methods. Cells were analyzed on a BD FACSCanto with 3 × 106 events collected per sample to ensure detection of rare events. Using FlowJo analysis software, viable single cells were carefully selected using forward versus side scatter, followed by forward scatter versus forward scatter pulse width gating. (a) Representative plots from unimmunized C, unimmunized C.iGFP5′Myc and hyperimmunized C.iGFP5′Myc mice are shown. Representative CD19 versus GFP bivariate plots (right panels) derived from the B220+ gate (left panels) are illustrated. The square gates used to calculate the frequency of B220+CD19+GFP+ events are shown on the CD19 versus GFP plots. Note that CD19− plasmacytoid dendritic cells (pDC) are present within the B220+ gate. Their identity was confirmed with CD11c staining (data not shown). (b) Graphs represent the frequency of CD19+GFP+ B cells within the B220+ gated population (left panel) and the total number of B220+CD19+GFP+ cells per spleen (right panel). BALB, unimmunized C control; iGFP5′Myc, unimmunized C.iGFP5′Myc; iGFP5′Myc immune, hyperimmunized C.iGFP5′Myc. Values obtained from C control mice represent background levels. Bar graphs = mean ± s.d.; n = 3–4 per group.
Relatively simple questions to be asked initially in follow-up studies include whether ‘pre-malignant’ T(12;15)+ cells are scattered in lymphoid tissues or clustered in certain anatomical sites, such as germinal centers. A similarly trivial question is whether T(12;15)+ cells can also reside in non-lymphoid tissues; for example, the lamina propria of the gut, as suggested by PCR results obtained in our laboratory (Müller et al., 1997a, b). Somewhat more involved will be attempts to pinpoint the stage(s) of B-cell differentiation at which translocations take place: T(12;15) caused by V(D)J recombination may occur in immature B cells in the bone marrow, whereas translocations caused by aborted isotype switching and VDJ hypermutation may take place in mature B cells in secondary lymphoid tissues (Wang et al., 2009). A third set of studies will focus on the mechanism of translocation (Gostissa et al., 2009a); for example, crossing the iGFP5′Myc reporter on genetic backgrounds that contain certain activation-induced cytidine deaminase (Cheng et al., 2009) or DNA double-strand break repair mutations (Nussenzweig et al., 1997; Ouyang et al., 1997; Difilippantonio et al., 2000) may provide important insights into the role of these pathways in the origin of T(12;15). Similarly, breeding the iGFP reporter onto certain cytokine including IL-6 transgenic (Suematsu et al., 1992) backgrounds may enhance our understanding of growth, differentiation and survival requirements of aberrant T(12;15)+ cell clones.
In conclusion, extension of the present reporter gene insertion approach to the pre-malignant state (Igh–Myc-bearing tumor precursors) and to other types of oncogene-activating or fusion-gene translocations (Mitelman et al., 2007) may lead to fundamental new insights into the nature, developmental stage and site of origin of tumor progenitors that are of great relevance for human cancer.
Supplementary Material
Acknowledgements
We thank our colleagues from NCI and NIAID, NIH for their contributions to this project: Tina Willington, Vaishali Jarral and Wendy DuBois for genotyping and assistance with the mouse experiments; Eileen Southon for gene targeting; Dr Alexander L Kovalchuk for advice on PCR analysis and providing primers; Drs Sung Sup Park and Santiago Silva for contributions to early stages of this project; and Drs Michael Potter and Beverly Mock for stimulating discussion and laboratory support. For assistance with transfer, cryopreservation, maintenance and rederivation of mice under SPF conditions, we thank: Ling Hu, CCOM, Iowa City, Iowa; the staff of the Jackson Laboratory, Bar Harbor, Maine; and the Office of Animal Resources, CCOM, particularly Dr Kem Singletary and James Hynes. We also thank Lorraine Tygrett, CCOM for assistance with flow cytometry. This work was supported, in part, by the Intramural Research Program of the NIH (LT, RC, TR) and Award Number P50CA097274 from the National Cancer Institute (SJ).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Anderson PN, Potter M. Induction of plasma cell tumours in BALB-c mice with 2,6,10,14-tetramethylpentadecane (pristane) Nature. 1969;222:994–995. doi: 10.1038/222994a0. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, et al. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci USA. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Germain RN, Castellino F, Chieppa M, Egen JG, Huang AY, Koo LY, et al. An extended vision for dynamic high-resolution intravital immune imaging. Semin Immunol. 2005;17:431–441. doi: 10.1016/j.smim.2005.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Ranganath S, Bianco JM, Alt FW. Chromosomal location targets different MYC family gene members for oncogenic translocations. Proc Natl Acad Sci USA. 2009a;106:2265–2270. doi: 10.1073/pnas.0812763106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature. 2009b;462:803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz S. Myc translocations in B cell and plasma cell neoplasms. DNA Repair (Amst) 2006;5:1213–1224. doi: 10.1016/j.dnarep.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Janz S, Müller J, Shaughnessy J, Potter M. Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci USA. 1993;90:7361–7365. doi: 10.1073/pnas.90.15.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk AL, Kim JS, Janz S. E mu/S mu transposition into Myc is sometimes a precursor for T(12;15) translocation in mouse B cells. Oncogene. 2003;22:2842–2850. doi: 10.1038/sj.onc.1206345. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Kishimoto T, Janz S. Lymph nodes and Peyer’s patches of IL-6 transgenic BALB/c mice harbor T(12;15) translocated plasma cells that contain illegitimate exchanges between the immunoglobulin heavy-chain mu locus and c-myc. Leukemia. 2000a;14:1127–1135. doi: 10.1038/sj.leu.2401767. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Müller JR, Janz S. Deletional remodeling of c-myc-deregulating translocations. Oncogene. 1997;15:2369–2377. doi: 10.1038/sj.onc.1201409. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Mushinski EB, Janz S. Clonal diversification of primary BALB/c plasmacytomas harboring T(12;15) chromosomal translocations. Leukemia. 2000b;14:909–921. doi: 10.1038/sj.leu.2401676. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Levens DL. Reconstructing MYC. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Müller JR, Jones GM, Janz S, Potter M. Migration of cells with immunoglobulin/c-myc recombinations in lymphoid tissues of mice. Blood. 1997a;89:291–296. [PubMed] [Google Scholar]
- Müller JR, Jones GM, Potter M, Janz S. Detection of immunoglobulin/c-myc recombinations in mice that are resistant to plasmacytoma induction. Cancer Res. 1996;56:419–423. [PubMed] [Google Scholar]
- Müller JR, Mushinski EB, Jones GM, Williams JA, Janz S, Hausner PF, et al. Generation of immunoglobulin/c-myc recombinations in murine Peyer’s patch follicles. Curr Top Microbiol Immunol. 1997b;224:251–255. doi: 10.1007/978-3-642-60801-8_25. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci USA. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Kovalchuk AL, Kim JS, Klein G, Janz S. BCL2 accelerates inflammation-induced BALB/c plasmacytomas and promotes novel tumors with coexisting T(12;15) and T(6;15) translocations. Cancer Res. 2003;63:8656–8663. [PubMed] [Google Scholar]
- Suematsu S, Matsusaka T, Matsuda T, Ohno S, Miyazaki J, Yamamura K, et al. Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Proc Natl Acad Sci USA. 1992;89:232–235. doi: 10.1073/pnas.89.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels JT, Busse AC, Mahrt J, Dullin C, Grabbe E, Mueller GA. In vivo imaging in experimental preclinical tumor research—a review. Cytometry A. 2007;71:542–549. doi: 10.1002/cyto.a.20419. [DOI] [PubMed] [Google Scholar]
- Willis TG, Dyer MJ. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808–822. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.